Abstract
Bartonella bacilliformis is the causal agent of Carrion’s disease, an overlooked illness endemic in the Andean Mountains with Peru being the most affected country. The diagnostic of this illness is a challenge due to the limited resources and the common symptomatology with other infectious diseases. The goal of this study was to identify immunogenic peptides from Pap31 and succinyl-CoA synthetase α (SCS-α) of B. bacilliformis that might be suitable for developing a serologic tool. The immunodominant character of Pap31 and SCS-α was determined by Western blotting and in-silico analysis. Subsequently, 35 peptides were selected for epitope mapping and their immunoreactivity was tested by enzyme-linked immunosorbent assay (ELISA). A total of 30 sera were tested including pre-exposed people with high IgM levels for Pap31/SCS-α (23 sera), patients (2 sera) as well as 5 sera with no reactivity to Pap31/SCS-α. The results indicate that Pap31-8 (187QAIGSAILKGTKDTGT202) and SCS-α-12 (59IFASVAEGKEKTGANA74) are the most immunogenic peptides, with Pap31-8 showing potential to discriminate between B. bacilliformis and the remaining Bartonella spp., and SCS-α-12 differentiating Bartonella spp. from other microorganisms.
1. Introduction
Bartonella bacilliformis is the microorganism responsible for Carrion’s disease, an overlooked illness restricted to the Andean region of Peru, Ecuador, and Colombia [1]. This illness is transmitted by the bite of a sandfly, predominantly Lutzomyia verrucarum, and up to now, humans are the only established reservoir [1]. There are three well-defined clinical manifestations of this disease: the acute phase or Oroya fever in which B. bacilliformis invades the erythrocytes and include the following main symptoms: hemolytic anemia, malaise, fever, headache, and mild chills, as well as pallor, hepatomegaly, abdominal pain, and other unspecific symptoms [1,2], with lethality of up to 88% in the absence of treatment [3]. Meanwhile, in the chronic phase, B. bacilliformis produces an abnormal proliferation of endothelial cells causing characteristic dermal eruptions named Peruvian warts [1]. In this phase, mortality is considered irrelevant [1]. Finally, in the asymptomatic cases, the individual does not present any symptoms but is an asymptomatic carrier, thereby perpetuating the illness [4,5]. In the studies by Chamberlin et al., 0.5% of the population living in endemic areas presented asymptomatic bacteremia, and an indirect fluorescence antibody (IFA) assay showed that 45% of the volunteers were seropositive for B. bacilliformis antibodies [4,6]. Studies using more sensitive techniques, such as real-time PCR, detected approximately 40% of asymptomatic carriers in post-outbreak and endemic areas [5].
The diagnosis of Oroya fever is mainly based on clinical parameters and/or Giemsa-stained peripheral blood smears [2,7]. The common symptomatology with other infectious diseases present in endemic areas and the low sensitivity of around 30% with microscopy makes it extremely difficult to correctly diagnose acute patients [7,8,9,10,11].
The identification and evaluation of B. bacilliformis antigenic proteins would aid in the development of better diagnostic tests and could be a prelude to future testing of candidates for vaccine antigens. Succinyl-CoA synthetase α (SCS-α) and Pap31 are 2 of the main antigenic proteins of B. bacilliformis [5,12]. Pap31 is a 31.15 kDa-sized protein homolog of a bacteriophage-associated protein in Bartonella henselae (also called HbpA) and was first identified as an antigen of B. bacilliformis by immunoblotting with a patient serum [12]. An ELISA using the recombinant protein has already been described in the literature [13], and Pap31 was also considered a potential antigen candidate for a synthetic vaccine [14]. SCS-α is an essential protein of 30.99 kDa involved in the tricarboxylic acid cycle, which has been identified as an immunogen in studies developed in Brucella spp. [15,16]. Both Pap31 and SCS-α antigens were selected for this study because these two antigens were identified using an anti-human IgM as a secondary antibody [5], demonstrating their potential use for the identification of acute cases of Carrion’s disease, the phase of the illness with the highest mortality.
In order to avoid cross-reactivity among B. bacilliformis and other Bartonella species or other organisms, more specific and conserved antigens are essential as candidates for use in the serological diagnosis of this infection. Therefore, the objective of this work was to identify which peptides from the SCS-α and Pap31 antigens are the most immunogenic.
2. Results
After cloning the complete proteins of Pap31 and SCS-α and their respective truncated constructs (Pap31–A, Pap31–B, Pap31–C, SCS α–A, SCS α–B and SCS α–C), we performed Western blotting showing that the immunogenic region of Pap31 antigen is contained in clones B and C, and for SCS-α, the immunogenic region is in clone A (Figure 1).
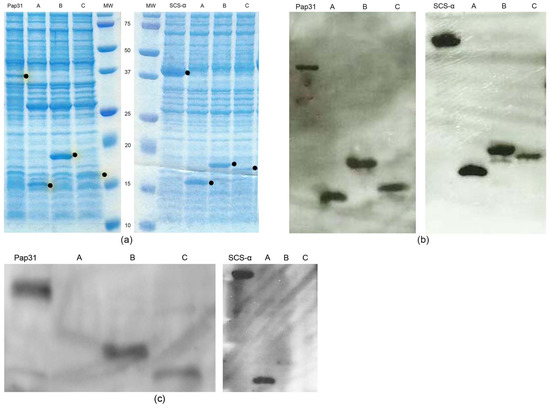
Figure 1.
Expression and detection of Pap31 and SCS- α and their respective truncated constructs; SDS-PAGE showing the expression of the proteins of interest, full-length Pap31 and SCS- α and respective clones A, B, and C, corresponding to the three truncated constructs. The proteins of interest are marked with a dot. (a); Western blotting performed with the anti-Xpress antibody signalizing the proteins of interest. (b); Example of Western blotting performed with serum from exposed people for Pap31 and SCS-α and respective clones A, B, and C (c).
The in-silico analysis using the IEDB Analysis Resource software confirmed the immunogenic region for SCS-α was on clone A, while that of Pap31 was between clones B and C as shown in Figure 2. This in-silico analysis allowed the immunogenic zone to be analyzed to be reduced, and a series of overlapping peptides from amino acids 166 to 238 (Pap31) and 26 to 89 (SCS-α) was designed for further determinations.
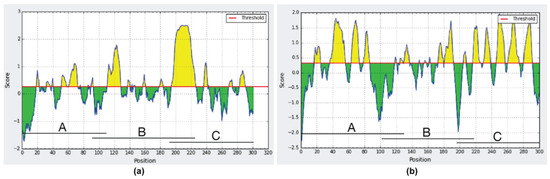
Figure 2.
In-silico analysis of the immunogenic regions of (a) Pap31 protein; (b) SCS-α protein.
The immunogenicity of the overlapping Pap31-B and Pap31-C regions, as well as SCSα–A, was tested by ELISA and the results are presented in Figure 3.
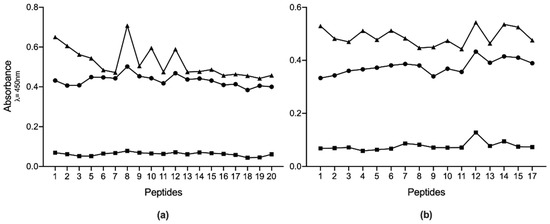
Figure 3.
Absorbance levels of IgM ELISAs for all the antigenic peptides studied. (a) results for Pap31, and (b) results for SCS-α. Squares are the mean results for non-reactive samples; circles are the mean results of recently exposed sera, and triangles are the mean results for the positive samples.
Two different analyses were performed to establish which peptides could be the best candidates for use in a future diagnostic test. First, for sera from recently exposed people, we determined in which peptides the mean absorbance was higher than the remaining sera. The results showed that Pap31-8 (187QAIGSAILKGTKDTGT202) and SCS-α-12 (59IFASVAEGKEKTGANA74) presented higher reactivity values (Abs450 0.502 and 0.433 respectively), followed by Pap31-12 and Pap31-9, and SCS-α-14 and SCS-α-15, respectively. The statistical analysis showed that Pap31-8 was significantly more immunogenic than all the Pap31 peptides, except for Pap31-12, Pap31-9 and Pap31-6. Meanwhile, SCS-α-12 was significantly more immunogenic than nine other SCS-α peptides (Figure 4).
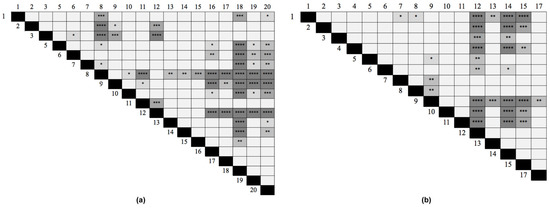
Figure 4.
Differences in the immunogenicity among peptides calculated with the Friedmann test. (a) Pap31, (b) SCS-α. p < 0.0001 (****); p < 0.0005 (***); p < 0.005 (**); p < 0.05 (*).
On the other hand, when we determined the peptides resulting in a higher difference between the mean values of pre-exposed and non-reactive sera, Pap31-8 was the most immunogenic Pap31 peptide, followed by Pap31-5 and Pap31-12 (both showing the same value) (Figure 5). These results confirm that the middle region of Pap31 is the most immunogenic, with Pap-31-B and Pap31-C overlapping. In this regard, in SCS-α, the peptide SCS-α-15 (68EKTGANASVIYVPPAG83) is the most immunogenic, followed by SCS-α-14 and SCS-α-17.
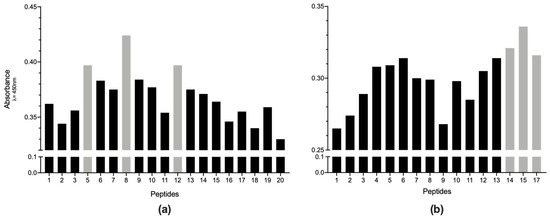
Figure 5.
Immunogenicity levels for each peptide of (a) Pap31 and (b) SCS-α. The immunogenicity level of each peptide corresponds to the difference between the mean values of pre-exposed and non-reactive sera.
The in-silico specificity analysis was done for the peptides that showed higher immunogenicity in both analyses (Pap31-8 and SCS-α-12 in the Friedman test and SCS-α-15 when we compared the mean values of pre-exposed and non-reactive sera). The results showed that Pap31-8 was conserved in almost all the B. bacilliformis genomes sequenced, with the exception of strain Ver-097 (Reference Sequence: WP_041848867) which differed in 1 amino acid. No identity higher than 65% was found with the remaining Bartonella spp. genomes.
A 100% identity was found between SCS-α-12 and the corresponding region of the SCS-α protein of 6 B. bacilliformis genomes, with slight differences (1 amino acid) with the remaining 10 B. bacilliformis genomes present in GenBank. Similarly, SCS-α-15 was fully conserved in 15 out of 16 B. bacilliformis genomes present in GenBank, while in the remaining genome (strain Ver097, Reference Sequence: WP_041849026), 1 amino acid difference was observed. In contrast to Pap-31-8, both SCS-α-12 and SCS-α-15 peptides showed high identity with other members of the Bartonella genus, including all the human pathogenic members (either withstanding nomenclature or Candidatus species) present in GenBank.
In any case, we detected an identity higher than 90% with microorganisms belonging to other bacterial genera. Nevertheless, SCS-α-12 showed 87.50% identity with the corresponding region of the SCS-α protein of members of the genus Brevundimonas and 81.50% with Brucella spp. (Table 1). It is important to highlight that the peptide SCS-α-15 presented an identity higher than 80% with the SCS-α of several pathogenic microorganisms, including Brucella spp., such as Brucella abortus or Brucella melitensis (87.50%), or Anaplasma spp., such as Anaplasma phagocytophilum, Brevundimonas spp. such as Brevundimonas diminuta, Enterobacteriales, such as Escherichia coli or Salmonella enterica, different Rickettsia spp., such as Rickettsia australis, Rickettsia prowazekii, Rickettsia rickettsii, Rickettsia sibirica or Rickettsia slovaca, among others, with an identity of 81.25% (data not shown).

Table 1.
Amino acid differences observed in the specificity analysis of the selected peptides.
When the peptides were compared with the corresponding human protein, the highest identity observed was for SCS-α-12 and SCS-α-15 with 75% of identity due to four amino acid substitutions (Table 1 and data not shown).
3. Discussion
The development of robust diagnostic tools is essential for the control of infectious diseases. In the last years, the design of diagnostic tools has involved a series of high throughput molecular approaches, including matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) and next-generation sequencing, among others [20,21]. Nonetheless, these techniques remain economically and technically inaccessible in most low- and middle-income countries, especially in remote areas where the majority of Carrion’s disease cases occur, and the lack of communication in these areas, the limited resources, and non-adequately equipped laboratories can be added to this challenging scenario [1]. While Carrion’s disease may seriously compromise a patient’s life [1,3], the initial symptoms of the disease are similar to other illnesses present in the same areas [7], often leading to misdiagnosis [7,8,9]. Furthermore, a high percentage of asymptomatic carriers acting as a B. bacilliformis reservoir have been described [4,5]. In these conditions, the development of a low-cost, easy-to-use diagnostic tool, able to detect acute cases and also sensitive enough to detect asymptomatic carriers is essential for the control and elimination of Carrion’s disease.
Previously, we identified Pap31 and SCS-α as B. bacilliformis antigens that are strongly reactive to sera from pre-exposed people when using an anti-human IgM as a secondary antibody [5]. These results indicated that antibodies to these proteins are strongly induced by B. bacilliformis infection and are therefore potential candidates for developing a diagnostic tool for recent infections. Nonetheless, the use of short antigenic peptides versus whole proteins may result in enhanced specificity eliminating false positives caused by cross-reaction with other antibodies [22]. Previous studies highlighted the potentiality of peptides as candidates for the development of diagnostic tools in sera [22,23]. The most common antigens are from the outer membrane proteins and in particular for Bartonella spp. infections, the VompA, VompB, PpI, and hemin-biding proteins from B. quintana [24,25] and Pap31 and BH11510 from B. henselae [26] have been proposed as possible candidates.
Access to proteins for the development of immunogenic assays designed to detect specific immunogenic regions is limited and cumbersome, but in the current “omic” era, accessibility to directly sequenced proteins or inferred amino acid sequences from genomic DNA analysis is immense. The use of in-silico computer tools might allow predicting the most promising immunogenic regions based on standardized parameters in a fast and reliable manner, facilitating both the study of proteins independently of their physical availability and the selection of the protein regions to be analyzed. Thus, after selecting the putative immunogenic regions of Pap31 and SCS-α, we included short overlapping peptides covering the desired area in epitope mapping studies and identified Pap31-8 and SCS-α-12 as the most immunogenic peptides of these proteins.
In other Bartonella species, such as B. quintana, Pap31 binds to environmental heme [27], highlighting the presence of this protein on the bacterial surface. In addition to B. bacilliformis, an immunogenic role of Pap31 has been proposed in B. henselae [26]. In-silico analysis has revealed that the immunogenic region of B. henselae Pap31 is placed around amino acid positions 60–80 (data not shown), different from the B. bacilliformis immunogenic peptide Pap31-8. This should support the lack of cross-reactivity to B. bacilliformis Pap31-8 from patients infected with B. henselae. Indeed, in a study by Angkasekwinai et al., an ELISA performed with recombinant Pap31 from B. bacilliformis showed no cross-reactivity when testing samples containing antibodies against Coxiella burnetti, Brucella spp., and B. henselae [13]. Further analysis showed several differences in the surroundings of peptide Pap31-8 between B. bacilliformis and the remaining Bartonella spp., highlighting the presence of a repetitive region just after Pap31-8 [28].
Together with SCS-α-12, the neighboring SCS-α-14 and SCS-α-15 were the second and third most immunogenic peptides for SCS-α, highlighting the immunogenicity of this region (SCS-α-15 being the peptide showing the highest difference in reactivity compared to non-reactive sera). While no data has been found about the cytoplasmatic, membrane, periplasmatic, or secretory nature of SCS-α in B. bacilliformis, the immunogenicity of either SCS-α or SCS-β, the two subunits of Succinyl-CoA Synthetase, have been highlighted in different microorganisms, such as Brucella spp. or Leptospira interrogans [15,16,29]. Furthermore, it has been suggested that SCS-α is a relevant factor involved in B. henselae interactions with host cells [30].
The in-silico analysis showed that while the peptide SCS-α-12 and SCS-α-15 have identity levels between 81.25 and 93.75% with the equivalent region of SCS-α from other Bartonella spp. (as well as with specific isolates of B. bacilliformis), Pap31-8 is highly specific to B. bacilliformis, with no identity with any other Bartonella spp. higher than 65%. It was of note that the B. bacilliformis strain Ver097, which showed lower levels of identity for both Pap31 and SCS-α, has been considered as a possible B. bacilliformis subspecies or a phylogenetically related species [18,19]. Although further studies are needed to establish the cross-reactivity with other Bartonella spp. as well as other pathogens, this finding suggests that Pap31-8 might be a reliable candidate for detecting B. bacilliformis infections. Identity levels higher than 80% with SCS-α-15 of B. bacilliformis were found among numerous human pathogenic microorganisms, several of which were phylogenetically close, such as Brucella spp. or Rickettsia spp., and also more distant, such as E. coli or S. enterica, likely hindering the usefulness of this peptide for diagnostic purposes.
Two main limitations of this study need to be considered; (1) the limited number of sera used from patients with acute infections (nonetheless, the sera used from recently exposed people showed only a slight decrease in reactivity as compared to the patient samples), and; (2) the lack of sera from people with other infections (including other Bartonella spp.), which would definitively confirm the specificity of the selected peptides.
In summary, we describe three short peptides able to detect the presence of B. bacilliformis antibodies. While further studies are needed, at least one of these immunogenic peptides, Pap31-8, seems to be a strong candidate for the future development of a rapid diagnostic tool, which is essential to improve the control, elimination, and eradication of Carrion’s disease, a neglected infectious disease.
4. Materials and Methods
4.1. Serum Samples
In total, 46 serum samples were used in this study; the majority (44 samples) were obtained during a 2014 study conducted in 5 different villages in the Piura department (northern Peru). The villages consisted of 1 endemic and 4 post-outbreak areas of Carrion’s disease [5]. Thirty-six of the sera samples were from people with a positive ELISA result performed with the full-length proteins of Pap31 or SCS-α as antigens and anti-human IgM as a secondary antibody and were, therefore, considered to be from people who had recently been exposed. Eight sera from people living in the endemic area with a negative result for the same assay [5] were also included and considered as non-recently exposed (thereafter, referred to as non-reactive). Additionally, 2 samples from patients with the acute phase of the disease collected a few months previously in the same department were also included (positive samples).
4.2. Bacterial Strains
The microorganisms and plasmids used in this study are listed in Table 2. B. bacilliformis (CIP 57.20) was cultured at 28 °C on Columbia agar with 5% sheep blood for 7 days. The E. coli strains for cloning were grown on Luria-Bertani medium, supplemented with 50 µg/mL of ampicillin at convenience.

Table 2.
Bacterial strains and plasmids used in the study.
4.3. Amplification, Cloning, and Purification of Antigenic Proteins and Respective Truncated Constructs
In order to identify the immunogenic portion of Pap31 and SCS-α, 3 truncated constructs of 110–130 amino acids of Pap31 (Pap31-A: amino acid 1 to 108; Pap31-B: amino acid 92 to 225 and Pap31-C: amino acid 193 to 301) and SCS-α (SCSα-A: amino acid 1 to 127; SCSα-B: amino acid 101 to 217 and SCSα-C: amino acid 196 to 301) were created. Genes coding for the antigenic proteins as well as the respective truncated constructs were amplified by PCR using the primers listed in Table 3. Of note, the pap31 and sucD genes of B. bacilliformis 57.20 were sequenced in a previous study, being identical to those belonging to reference strain KC583 [5].

Table 3.
Primers used in the study.
Amplified products were purified using the MinElute PCR purification Kit (QIAGEN, Hilden, Germany) and cloned in Champion pET Directional 100/D-TOPO vector (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions in order to generate Xpress-tagged versions of the candidate antigenic proteins. The DNA sequences were verified by Sanger sequencing using the following primers: M13F 5′-GTAAAACGACGGCCAG-3′ and M13R 5′-CAGGAAACAGCTATGAC-3′.
For the detection, E. coli isolates with the vector of interest were grown for 3 h in the presence of isopropyl ß-D-1-thiogalactopyranoside (IPTG) 1mM and thereafter suspended in lysis buffer (50 mM potassium phosphate, pH 7.8, 400 mM NaCl, 100 mM KCl, 10% glycerol, 0.5% Triton X-100, 10 mM imidazole). All proteins were separated on a 10–20% SDS-PAGE (FUJIFILM Wako Pure Chemicals Corporation, Osaka, Japan) and immunoblotted with anti-Xpress antibody (Invitrogen, Carlsbad, CA, USA). The complete proteins of Pap31 and SCS-α were previously purified using a His-Binding kit and following the manufacturer’s instructions (GE Healthcare, Buckinghamshire, UK) [5].
4.4. Western Blotting
Western blotting was performed to determine the immunogenic region of Pap31 and SCS-α. Both the purified complete proteins and their respective 3 truncated constructs were electrophoresed on 15% SDS-PAGE gels and electrotransferred onto a PVDF membrane (GE Healthcare, Buckinghamshire, UK) using a Bio-Rad Trans-Blot SD Cell apparatus. A blocking step for 1 h at room temperature using 5% skim milk in PBS was performed. The membranes were immunoblotted during 90 min with sera (1:50 dilution in Tris-buffered saline [TBS]-Tween with 1% bovine serum albumin [BSA]). Western blottings with at least 10 sera from exposed people were performed for both Pap31 and SCS-α. A commercial negative control (DAKO X0939) consisting of a pool of at least 30 sera from healthy blood donors was used. After washing 3 times with TBS-0.05% Tween20) for10 min, the membranes were incubated for 1 h with a polyclonal rabbit anti-human IgM 1:1000 dilution in TBS-T with 1% BSA conjugated with peroxidase (Dako, Glostrup, Denmark) using o-phenylenediamine tablets as substrate (Sigma, St. Louis, MO, USA).
4.5. In-Silico Analysis
An in-silico analysis using IEDB Analysis Resource software (http://tools.iedb.org/main/, accessed on 13 July 2021) was also performed. In addition to confirming the in vitro results, this in-silico analysis shortened the immunogenic zone of Pap31 and SCS-α.
4.6. Design and Synthesis of Biotinylated Peptides
A total of 35 overlapping peptides (Table 4) were purchased from GL Biochem Ltd. (Shanghai, China) corresponding to the immunogenic regions identified, 19 peptides corresponding to Pap31, and 16 peptides to SCS-α. The peptides were designed by Epitope Mapping, with a size of 16 amino acids each, offset by 3 amino acids relative to the former, 13 of which are therefore coincident between consecutive peptides. A molecule of biotin was coupled at the N-terminus of each peptide in order to obtain more signals when performing the ELISA technique. Peptide purity was quantified by the manufacturer, being at least 95%. The peptides were dissolved according to the instructions provided by GL Biochem and stored at −80 °C until use.

Table 4.
Peptides used in the study.
4.7. ELISA
A total of 30 sera were selected to be tested with each peptide. Of these, 23 were from pre-exposed people with high IgM levels for Pap31 or SCS-α (accordingly to the peptide to be tested), 2 from patients with Carrión’s disease, and 5 that had no reactivity to Pap31 or SCS-α. To determine the immunogenicity of each peptide, ELISA tests were carried out using the synthetic biotinylated peptides as an antigen. Briefly, 96-well Nunc-Immuno Maxisorp microtiter plates (Nalgene Nunc International, Roskilde, Denmark) were coated with 100 μL (0.67 μg/well) of NeutrAvidin biotin-binding protein (Invitrogen, Carlsbad, CA, USA) overnight at 4 °C. After washing with phosphate-buffered saline (PBS) 0.1% Tween-20, 100 μL of a single biotinylated peptide (28.6 μg/mL in each well) was added and incubated for 1 h. The plates were washed 3 times, and sera diluted 1:100 with TBS-Tween with 1% BSA were incubated for 2 h. The same negative control and secondary antibody used in the Western blottings were used here. The optical densities were measured as absorbance at 450 nm after using o-phenylenediamine tablets as substrate (Sigma, St. Louis, MO, USA). Each sample was analyzed in triplicate intra-plate.
4.8. Selection of the Best Immunogenic Candidates
The best immunogenic candidates were selected following two criteria of analysis. Thus, the means of the absorbance values obtained in the ELISA of the sera of recently-exposed people as well as the difference between the mean values of all pre-exposed and all non-reactive sera were determined. Peptides showing the highest mean absorbance when pre-exposed sera were tested and those presenting higher reactivity differences when pre-exposed sera and non-pre-exposed sera were compared were considered.
The specificity of the selected antigenic peptides was determined by comparison with GenBank. The identity levels were determined by comparing the peptide sequence with protein sequences of Bartonellaceae, bacterial pathogens, and humans.
4.9. Statistical Analysis
The statistical analysis was performed using GraphPad Prism. Normality of the data was accessed by the Shapiro test and the mean reactivity of each peptide was compared with the mean reactivity of every other peptide using the Friedman test. Significance was considered with p < 0.05.
Author Contributions
Conceptualization, C.G., M.M. and J.R.; methodology, C.G. and M.J.P.; formal analysis, C.G., M.J.P., M.M. and J.R.; investigation, C.G. and M.J.P.; resources, J.d.V.-M. and M.M.; writing—original draft preparation, C.G. and J.R.; writing—review and editing, C.G., M.J.P., J.d.V.-M., M.M. and J.R.; visualization, C.G., M.J.P. and J.R. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by the Programa Nacional de Innovación para la Competitividad y Productividad (Innóvate Perú), under the contract 117-PNICP-PIAP-2015. J.R. had a fellowship from the program I3, of the ISCIII [grant number: CES11/012]. C.G. had a PhD fellowship of the ISCIII [FI12/00561] and was recipient of a Canon Foundation Fellowship and a Small Grant from the Royal Society for Tropical Medicine and Hygiene [grant reference 000584]. MJP had a postdoctoral fellowship from CONCYTEC/FONDECYT [grant number: CG05-2013-FONDECYT].
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of Universidad Peruana de Ciencias Aplicadas, Lima, Peru (protocol code CEI/559-05-15 approved on May 2015), Hospital Regional de Cajamarca, Cajamarca, Peru (protocol code 1958851 approved on October 2015) and the Hospital Clinic, Barcelona, Spain (protocol code CPMP/ICH/135/95 approved on February 2014).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available in [Immunogenic peptides from Pap31 and SCS-α of Bartonella bacilliformis: one step closer to a rapid diagnostic tool for Carrion’s disease].
Acknowledgments
ISGlobal is a member of the CERCA Programme, Generalitat de Catalunya. We thank Donna Pringle for the grammar correction.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Gomes, C.; Ruiz, J. Carrion’s disease. The sound of silence. Clin. Microb. Rev. 2018, 31, e00056-17. [Google Scholar] [CrossRef]
- Pons, M.J.; Gomes, C.; del Valle, J.; Ruiz, J. Carrion’s Disease, more than a sandfly-vectored illness. PLoS Path. 2016, 12, e1005863. [Google Scholar] [CrossRef]
- Gray, G.C.; Johnson, A.A.; Thornton, S.A.; Smith, W.A.; Knobloch, J.; Kelley, P.W.; Obregon Escudero, L.; Arones Huayda, M.; Wignall, F.S. An epidemic of Oroya fever in the Peruvian Andes. Am. J. Trop. Med. Hyg. 1990, 42, 215–221. [Google Scholar] [CrossRef]
- Chamberlin, J.; Laughlin, L.W.; Romero, S.; Solórzano, N.; Gordon, S.; Andre, R.G.; Pachas, P.; Friedman, H.; Ponce, C.; Watts, D. Epidemiology of endemic Bartonella bacilliformis: A prospective cohort study in a Peruvian mountain valley community. J. Infect. Dis. 2002, 186, 983–990. [Google Scholar] [CrossRef] [PubMed]
- Gomes, C.; Palma, N.; Pons, M.J.; Magallón-Tejada, A.; Sandoval, I.; Tinco-Valdez, C.; Gutarra, C.; del Valle-Mendoza, J.; Ruiz, J.; Matsuoka, M. Succinyl-CoA synthetase: New antigen candidate of Bartonella bacilliformis. PLoS Negl. Trop. Dis. 2016, 10, e0004989. [Google Scholar] [CrossRef] [PubMed]
- Chamberlin, J.; Laughlin, L.; Gordon, S.; Romero, S.; Solórzano, N.; Regnery, R.L. Serodiagnosis of Bartonella bacilliformis infection by indirect fluorescence antibody assay: Test development and application to a population in an area of bartonellosis endemicity. J. Clin. Microbiol. 2000, 38, 4269–4271. [Google Scholar] [CrossRef]
- Maguiña, C.; García, P.J.; Gotuzzo, E.; Cordero, L.; Spach, D.H. Bartonellosis (Carrión’s disease) in the modern era. Clin. Infect. Dis. 2001, 33, 772–779. [Google Scholar] [CrossRef] [PubMed]
- Cornejo, A.; Gomes, C.; Suarez, L.; Martinez-Puchol, S.; Bustamante, P.; Pons, M.J.; Ruiz, J.; del Valle, J. An unidentified cluster of infection in the Peruvian Amazon region. J. Infect. Dev. Ctries 2015, 9, 524–529. [Google Scholar] [CrossRef]
- del Valle, J.; Silva, W.; Tinco, C.; Pons, M.J.; del Valle, L.J.; Champin Michelena, D.; Bazán Mayra, J.; Zavaleta Gavidea, V.; Vargas, M.; Ruiz, J. Diagnosis of Carrion’s disease by direct blood PCR in thin blood smear negative samples. PLoS ONE 2014, 9, e92283. [Google Scholar] [CrossRef]
- Ellis, B.A.; Rotz, L.D.; Leake, J.A.; Samalvides, F.; Bernable, J.; Ventura, G.; Padilla, C.; Villaseca, P.; Beati, L.; Regnery, R.; et al. An outbreak of acute bartonellosis (Oroya fever) in the Urubamba region of Peru, 1998. Am. J. Trop. Med. Hyg. 1999, 61, 344–349. [Google Scholar] [CrossRef]
- Sanchez Clemente, N.; Ugarte-Gil, C.A.; Solórzano, N.; Maguiña, C.; Pachas, P.; Blazes, D.; Bailey, R.; Mabey, D.; Moore, D. Bartonella bacilliformis: A systematic review of the literature to guide the research agenda for elimination. PLoS Negl. Trop. Dis. 2012, 6, e1819. [Google Scholar] [CrossRef]
- Taye, A.; Chen, H.; Duncan, K.; Zhang, Z.; Hendrix, L.; Gonzalez, J.; Ching, W. Production of recombinant protein Pap31 and its application for the diagnosis of Bartonella bacilliformis infection. Ann. N. Y. Acad. Sci. 2005, 1063, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Angkasekwinai, N.; Atkins, E.H.; Romero, S.; Grieco, J.; Chao, C.C.; Ching, W.M. An evaluation study of enzyme-linked immunosorbent assay (ELISA) using recombinant protein Pap31 for detection of antibody against Bartonella bacilliformis infection among the Peruvian population. Am. J. Trop. Med. Hyg. 2014, 90, 690–696. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Henriquez-Camacho, C.A.; Ventosilla, P.; Minnick, M.; Ruiz, J.; Maguiña, C. Proteins of Bartonella bacilliformis: Candidates for vaccine development. Int. J. Pept. 2015, 2015, 702784. [Google Scholar] [CrossRef] [PubMed]
- Teixeira-Gomes, A.P.; Cloeckaert, A.; Bézard, G.; Bowden, R.A.; Dubray, G.; Zygmunt, M.S. Identification and characterization of Brucella ovis immunogenic proteins using two-dimensional electrophoresis and immunoblotting. Electrophoresis 1997, 18, 1491–1497. [Google Scholar] [CrossRef]
- Teixeira-Gomes, A.P.; Cloeckaert, A.; Bézard, G.; Dubray, G.; Zygmunt, M.S. Mapping and identification of Brucella melitensis proteins by two-dimensional electrophoresis and microsequencing. Electrophoresis 1997, 18, 156–162. [Google Scholar] [CrossRef]
- Ruiz, J.; Pons, M.J. Revisiting Bartonella bacilliformis MLST. Infect. Genet. Evol. 2018, 63, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Loparev, V.N.; Dasch, G.A.; Knipe, K.M.; Rowe, L.A.; Lydy, S.L. Optical mapping and genome sequences of isolates of Bartonella bacilliformis. In Proceedings of the Program and Abstracts Book of International Conference on Emerging Infectious Diseases, Atlanta, GA, USA, 24–26 August 2015; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2015; p. 6. [Google Scholar]
- Paul, S.; Minnick, M.F.; Chattopadhyay, S. Mutation-driven divergence and convergence indicate adaptive evolution of the intracellular human-restricted pathogen, Bartonella bacilliformis. PLoS Negl. Trop. Dis. 2016, 10, e0004712. [Google Scholar] [CrossRef]
- Chan, W.S.; Au, C.H.; Leung, H.C.; Ho, D.N.; Li, D.; Chan, T.L.; Lam, T.W.; Ma, E.S.; Tang, B.S. Potential utility of metagenomic sequencing for improving etiologic diagnosis of infective endocarditis. Futur. Cardiol. 2019, 15, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Fournier, P.E.; Couderc, C.; Buffet, S.; Flaudrops, C.; Raoult, D. Rapid and cost-effective identification of Bartonella species using mass spectrometry. J. Med. Microbiol. 2009, 58 Pt 9, 1154–1159. [Google Scholar] [CrossRef]
- Cretich, M.; Gori, A.; D’Annessa, I.; Chiari, M.; Colombo, G. Peptides for infectious diseases: From probe design to diagnostic microarrays. Antibodies 2019, 8, 23. [Google Scholar] [CrossRef]
- Feliciano, N.D.; Ribeiro, V.S.; Gonzaga, H.T.; Santos, F.A.; Fujimura, P.T.; Goulart, L.R.; Costa-Cruz, J.M. Short epitope-based synthetic peptides for serodiagnosis of human strongyloidiasis. Immunol. Lett. 2016, 172, 89–93. [Google Scholar] [CrossRef]
- Boonjakuakul, J.K.; Gerns, H.L.; Chen, Y.T.; Hicks, L.D.; Minnick, M.F.; Dixon, S.E.; Hall, S.C.; Koehler, J.E. Proteomic and immunoblot analyses of Bartonella quintana total membrane proteins identify antigens recognized by sera from infected patients. Infect. Immun. 2007, 75, 2548–2561. [Google Scholar] [CrossRef]
- Matsuoka, M.; Sasaki, T.; Seki, N.; Kobayashi, M.; Sawabe, K.; Sasaki, Y.; Shibayama, K.; Sasaki, T.; Arakawa, Y. Hemin-binding proteins as potent markers for serological diagnosis of infections with Bartonella quintana. Clin. Vaccine Immunol. 2013, 20, 620–626. [Google Scholar] [CrossRef] [PubMed]
- Saisongkorh, W.; Kowalczewska, M.; Azza, S.; Decloquement, P.; Rolain, J.M.; Raoult, D. Identification of candidate proteins for the diagnosis of Bartonella henselae infections using an immunoproteomic approach. FEMS Microbiol. Lett. 2010, 310, 158–167. [Google Scholar] [CrossRef]
- Carroll, J.A.; Coleman, S.A.; Smitherman, L.S.; Minnick, M.F. Hemin-binding surface protein from Bartonella quintana. Infect. Immun. 2000, 68, 6750–6757. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J.; Gomes, C. In silico analysis of Pap31 from Bartonella bacilliformis and other Bartonella spp. Infect. Genet. Evol. 2020, 84, 104482. [Google Scholar] [CrossRef] [PubMed]
- Sakolvaree, Y.; Maneewatch, S.; Jiemsup, S.; Klaysing, B.; Tongtawe, P.; Srimanote, P.; Saengjaruk, P.; Banyen, S.; Tapchaisri, P.; Chonsa-nguan, M.; et al. Proteome and immunome of pathogenic Leptospira spp. revealed by 2DE and 2DE-immunoblotting with immune serum. Asian Pac. J. Allergy Immunol. 2007, 25, 53–73. [Google Scholar]
- Chang, C.C.; Chen, Y.J.; Tseng, C.S.; Lai, W.L.; Hsu, K.Y.; Chang, C.L.; Lu, C.C.; Hsu, Y.M. A comparative study of the interaction of Bartonella henselae strains with human endothelial cells. Vet. Microbiol. 2011, 149, 147–156. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).