Poxvirus Interactions with the Host Ubiquitin System
Abstract
1. Introduction
2. The Ubiquitin System
3. The Role of the UPS in Poxvirus Uncoating and Replication
4. Viral Interaction with the Cul-RING E3 Ub Ligases
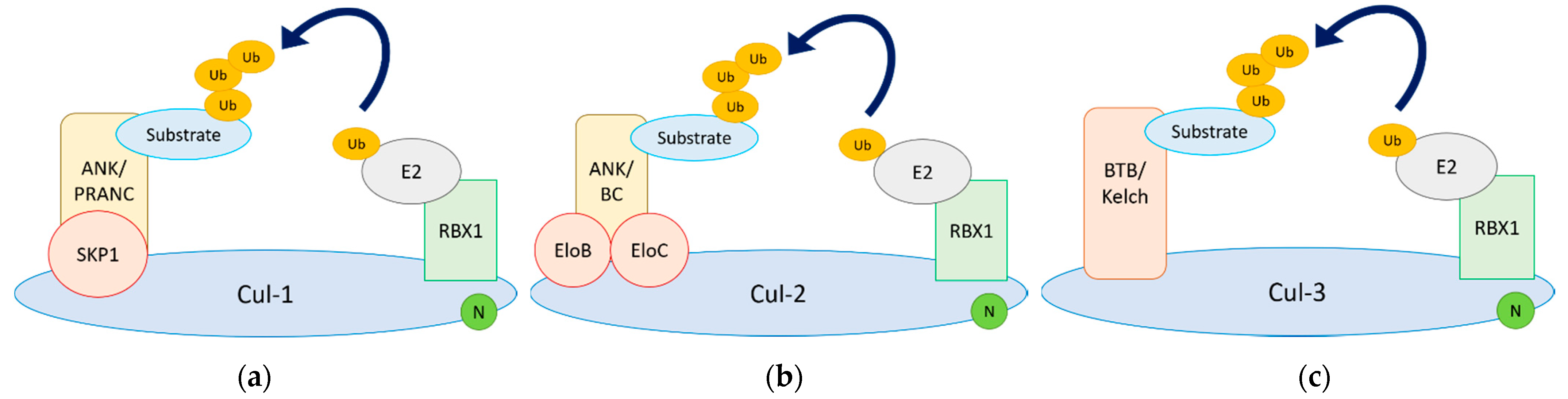
4.1. Functional Architecture of Cul-RING Ub Ligases
4.2. Viral Cul-1 Adaptors
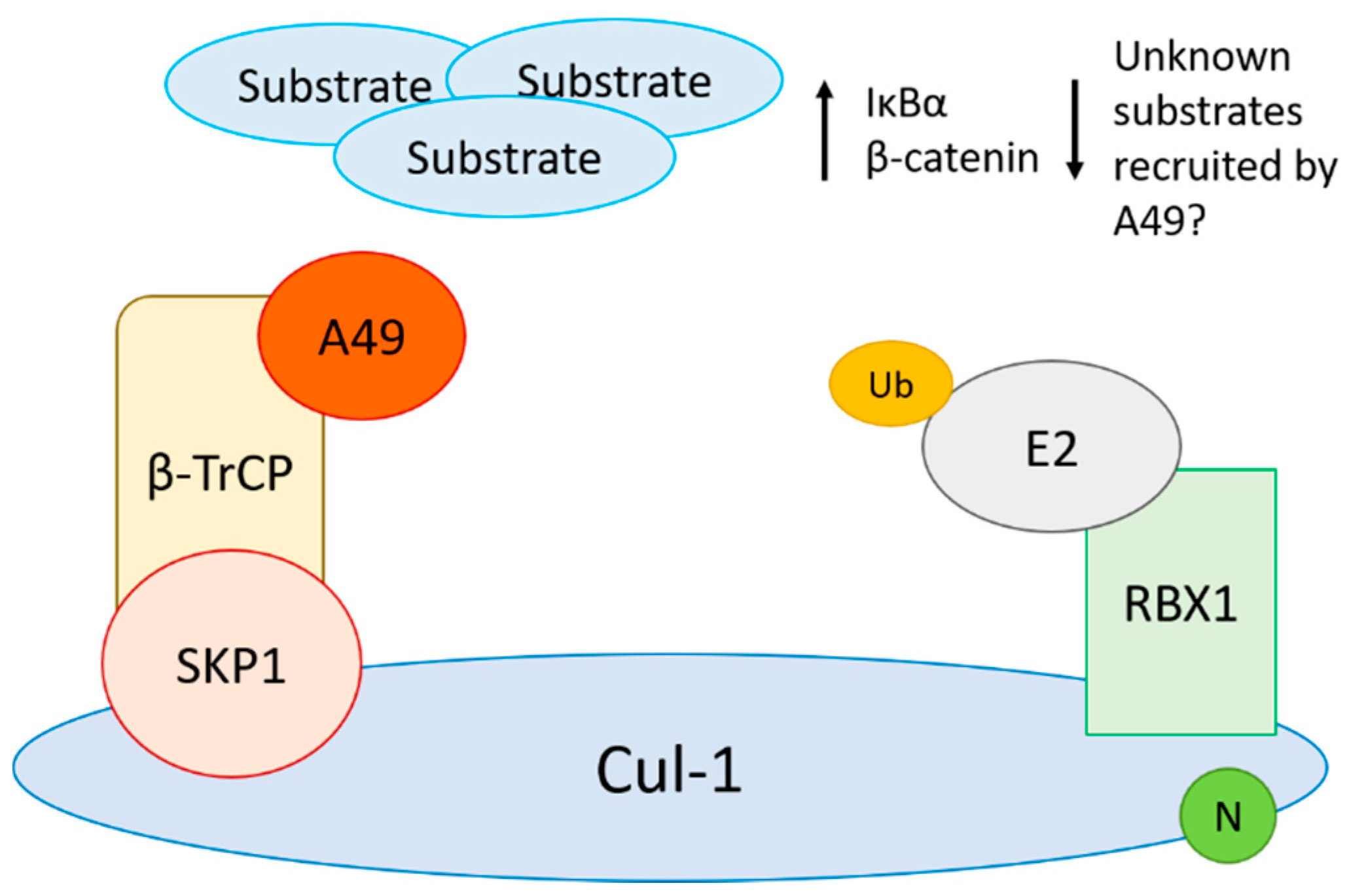
4.3. Viral Inhibitors of CRL1 Function
4.4. Viral Cul-2 Adaptors
4.5. Viral Cul-3 Adaptors
4.6. Targeting of Other CRLs
5. Poxvirus-Encoded E3 Ub Ligases and Ub Genes
6. Ubiquitylation of Viral Proteins
7. Other Interactions with the Ub System
8. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Moss, B. Poxviridae. In Fields Virology; Knipe, D.M., Howley, P., Eds.; Lippincott-Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 2, pp. 2129–2159. [Google Scholar]
- Smith, G.L.; Benfield, C.T.; Maluquer de Motes, C.; Mazzon, M.; Ember, S.W.; Ferguson, B.J.; Sumner, R.P. Vaccinia virus immune evasion: Mechanisms, virulence and immunogenicity. J. Gen. Virol. 2013, 94, 2367–2392. [Google Scholar] [CrossRef]
- El-Jesr, M.; Teir, M.; Maluquer de Motes, C. Vaccinia Virus Activation and Antagonism of Cytosolic DNA Sensing. Front. Immunol. 2020, 11, 568412. [Google Scholar] [CrossRef]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef]
- Buetow, L.; Huang, D.T. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2016, 17, 626–642. [Google Scholar] [CrossRef]
- Zhang, L.; Villa, N.Y.; McFadden, G. Interplay between poxviruses and the cellular ubiquitin/ubiquitin-like pathways. FEBS Lett. 2009, 583, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Barry, M.; van Buuren, N.; Burles, K.; Mottet, K.; Wang, Q.; Teale, A. Poxvirus exploitation of the ubiquitin-proteasome system. Viruses 2010, 2, 2356–2380. [Google Scholar] [CrossRef] [PubMed]
- Shchelkunov, S.N. Interaction of orthopoxviruses with the cellular ubiquitin-ligase system. Virus Genes 2010, 41, 309–318. [Google Scholar] [CrossRef]
- McClellan, A.J.; Laugesen, S.H.; Ellgaard, L. Cellular functions and molecular mechanisms of non-lysine ubiquitination. Open Biol. 2019, 9, 190147. [Google Scholar] [CrossRef] [PubMed]
- Dikic, I. Proteasomal and Autophagic Degradation Systems. Annu. Rev. Biochem. 2017, 86, 193–224. [Google Scholar] [CrossRef]
- Oh, E.; Akopian, D.; Rape, M. Principles of Ubiquitin-Dependent Signaling. Annu. Rev. Cell Dev. Biol. 2018, 34, 137–162. [Google Scholar] [CrossRef] [PubMed]
- Dittmar, G.; Winklhofer, K.F. Linear Ubiquitin Chains: Cellular Functions and Strategies for Detection and Quantification. Front. Chem. 2019, 7, 915. [Google Scholar] [CrossRef] [PubMed]
- Tracz, M.; Bialek, W. Beyond K48 and K63: Non-canonical protein ubiquitination. Cell. Mol. Biol. Lett. 2021, 26, 1. [Google Scholar] [CrossRef] [PubMed]
- Isaacson, M.K.; Ploegh, H.L. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe 2009, 5, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Randow, F.; Lehner, P.J. Viral avoidance and exploitation of the ubiquitin system. Nat. Cell Biol. 2009, 11, 527–534. [Google Scholar] [CrossRef]
- Gustin, J.K.; Moses, A.V.; Fruh, K.; Douglas, J.L. Viral takeover of the host ubiquitin system. Front. Microbiol. 2011, 2, 161. [Google Scholar] [CrossRef]
- Munyon, W.; Paoletti, E.; Grace, J.T., Jr. RNA polymerase activity in purified infectious vaccinia virus. Proc. Natl. Acad. Sci. USA 1967, 58, 2280–2287. [Google Scholar] [CrossRef] [PubMed]
- Kates, J.R.; McAuslan, B.R. Messenger RNA synthesis by a “coated” viral genome. Proc. Natl. Acad. Sci. USA 1967, 57, 314–320. [Google Scholar] [CrossRef]
- Joklik, W.K. The Intracellular Uncoating of Poxvirus DNA. Ii. The Molecular Basis of the Uncoating Process. J. Mol. Biol. 1964, 8, 277–288. [Google Scholar] [CrossRef]
- Satheshkumar, P.S.; Anton, L.C.; Sanz, P.; Moss, B. Inhibition of the ubiquitin-proteasome system prevents vaccinia virus DNA replication and expression of intermediate and late genes. J. Virol. 2009, 83, 2469–2479. [Google Scholar] [CrossRef]
- Teale, A.; Campbell, S.; Van Buuren, N.; Magee, W.C.; Watmough, K.; Couturier, B.; Shipclark, R.; Barry, M. Orthopoxviruses require a functional ubiquitin-proteasome system for productive replication. J. Virol. 2009, 83, 2099–2108. [Google Scholar] [CrossRef]
- Mercer, J.; Snijder, B.; Sacher, R.; Burkard, C.; Bleck, C.K.; Stahlberg, H.; Pelkmans, L.; Helenius, A. RNAi screening reveals proteasome- and Cullin3-dependent stages in vaccinia virus infection. Cell Rep. 2012, 2, 1036–1047. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.S.; Chen, C.H.; Ho, M.Y.; Huang, C.Y.; Liao, C.L.; Chang, W. Vaccinia virus proteome: Identification of proteins in vaccinia virus intracellular mature virion particles. J. Virol. 2006, 80, 2127–2140. [Google Scholar] [CrossRef]
- Doellinger, J.; Schaade, L.; Nitsche, A. Comparison of the Cowpox Virus and Vaccinia Virus Mature Virion Proteome: Analysis of the Species-and Strain-Specific Proteome. PLoS ONE 2015, 10, e0141527. [Google Scholar] [CrossRef]
- Resch, W.; Hixson, K.K.; Moore, R.J.; Lipton, M.S.; Moss, B. Protein composition of the vaccinia virus mature virion. Virology 2007, 358, 233–247. [Google Scholar] [CrossRef]
- Kilcher, S.; Schmidt, F.I.; Schneider, C.; Kopf, M.; Helenius, A.; Mercer, J. siRNA screen of early poxvirus genes identifies the AAA+ ATPase D5 as the virus genome-uncoating factor. Cell Host Microbe 2014, 15, 103–112. [Google Scholar] [CrossRef]
- Evans, E.; Klemperer, N.; Ghosh, R.; Traktman, P. The vaccinia virus D5 protein, which is required for DNA replication, is a nucleic acid-independent nucleoside triphosphatase. J. Virol. 1995, 69, 5353–5361. [Google Scholar] [CrossRef]
- De Silva, F.S.; Lewis, W.; Berglund, P.; Koonin, E.V.; Moss, B. Poxvirus DNA primase. Proc. Natl. Acad. Sci. USA 2007, 104, 18724–18729. [Google Scholar] [CrossRef]
- Liu, B.; Panda, D.; Mendez-Rios, J.D.; Ganesan, S.; Wyatt, L.S.; Moss, B. Identification of Poxvirus Genome Uncoating and DNA Replication Factors with Mutually Redundant Roles. J. Virol. 2018, 92, e02152-17. [Google Scholar] [CrossRef] [PubMed]
- Sperling, K.M.; Schwantes, A.; Staib, C.; Schnierle, B.S.; Sutter, G. The orthopoxvirus 68-kilodalton ankyrin-like protein is essential for DNA replication and complete gene expression of modified vaccinia virus Ankara in nonpermissive human and murine cells. J. Virol. 2009, 83, 6029–6038. [Google Scholar] [CrossRef] [PubMed]
- Sperling, K.M.; Schwantes, A.; Schnierle, B.S.; Sutter, G. The highly conserved orthopoxvirus 68k ankyrin-like protein is part of a cellular SCF ubiquitin ligase complex. Virology 2008, 374, 234–239. [Google Scholar] [CrossRef]
- Soucy, T.A.; Smith, P.G.; Milhollen, M.A.; Berger, A.J.; Gavin, J.M.; Adhikari, S.; Brownell, J.E.; Burke, K.E.; Cardin, D.P.; Critchley, S.; et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 2009, 458, 732–736. [Google Scholar] [CrossRef]
- Petroski, M.D.; Deshaies, R.J. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005, 6, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Skaar, J.R.; Pagan, J.K.; Pagano, M. Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 2013, 14, 369–381. [Google Scholar] [CrossRef]
- Duda, D.M.; Borg, L.A.; Scott, D.C.; Hunt, H.W.; Hammel, M.; Schulman, B.A. Structural insights into NEDD8 activation of cullin-RING ligases: Conformational control of conjugation. Cell 2008, 134, 995–1006. [Google Scholar] [CrossRef]
- Saha, A.; Deshaies, R.J. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell 2008, 32, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Feldman, R.M.; Correll, C.C.; Kaplan, K.B.; Deshaies, R.J. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 1997, 91, 221–230. [Google Scholar] [CrossRef]
- Bai, C.; Sen, P.; Hofmann, K.; Ma, L.; Goebl, M.; Harper, J.W.; Elledge, S.J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 1996, 86, 263–274. [Google Scholar] [CrossRef]
- Pierce, N.W.; Lee, J.E.; Liu, X.; Sweredoski, M.J.; Graham, R.L.; Larimore, E.A.; Rome, M.; Zheng, N.; Clurman, B.E.; Hess, S.; et al. Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins. Cell 2013, 153, 206–215. [Google Scholar] [CrossRef]
- Herbert, M.H.; Squire, C.J.; Mercer, A.A. Poxviral ankyrin proteins. Viruses 2015, 7, 709–738. [Google Scholar] [CrossRef]
- Mercer, A.A.; Fleming, S.B.; Ueda, N. F-box-like domains are present in most poxvirus ankyrin repeat proteins. Virus Genes 2005, 31, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Sonnberg, S.; Fleming, S.B.; Mercer, A.A. Phylogenetic analysis of the large family of poxvirus ankyrin-repeat proteins reveals orthologue groups within and across chordopoxvirus genera. J. Gen. Virol. 2011, 92, 2596–2607. [Google Scholar] [CrossRef] [PubMed]
- Sonnberg, S.; Fleming, S.B.; Mercer, A.A. A truncated two-alpha-helix F-box present in poxvirus ankyrin-repeat proteins is sufficient for binding the SCF1 ubiquitin ligase complex. J. Gen. Virol. 2009, 90, 1224–1228. [Google Scholar] [CrossRef]
- Sonnberg, S.; Seet, B.T.; Pawson, T.; Fleming, S.B.; Mercer, A.A. Poxvirus ankyrin repeat proteins are a unique class of F-box proteins that associate with cellular SCF1 ubiquitin ligase complexes. Proc. Natl. Acad. Sci. USA 2008, 105, 10955–10960. [Google Scholar] [CrossRef]
- Van Buuren, N.; Couturier, B.; Xiong, Y.; Barry, M. Ectromelia virus encodes a novel family of F-box proteins that interact with the SCF complex. J. Virol. 2008, 82, 9917–9927. [Google Scholar] [CrossRef]
- Johnston, J.B.; Wang, G.; Barrett, J.W.; Nazarian, S.H.; Colwill, K.; Moran, M.; McFadden, G. Myxoma virus M-T5 protects infected cells from the stress of cell cycle arrest through its interaction with host cell cullin-1. J. Virol. 2005, 79, 10750–10763. [Google Scholar] [CrossRef]
- Bratke, K.A.; McLysaght, A.; Rothenburg, S. A survey of host range genes in poxvirus genomes. Infect. Genet. Evol. 2013, 14, 406–425. [Google Scholar] [CrossRef]
- Buttigieg, K.; Laidlaw, S.M.; Ross, C.; Davies, M.; Goodbourn, S.; Skinner, M.A. Genetic screen of a library of chimeric poxviruses identifies an ankyrin repeat protein involved in resistance to the avian type I interferon response. J. Virol. 2013, 87, 5028–5040. [Google Scholar] [CrossRef]
- Laidlaw, S.M.; Robey, R.; Davies, M.; Giotis, E.S.; Ross, C.; Buttigieg, K.; Goodbourn, S.; Skinner, M.A. Genetic screen of a mutant poxvirus library identifies an ankyrin repeat protein involved in blocking induction of avian type I interferon. J. Virol. 2013, 87, 5041–5052. [Google Scholar] [CrossRef]
- Chen, D.Y.; Fabrizio, J.A.; Wilkins, S.E.; Dave, K.A.; Gorman, J.J.; Gleadle, J.M.; Fleming, S.B.; Peet, D.J.; Mercer, A.A. Ankyrin Repeat Proteins of Orf Virus Influence the Cellular Hypoxia Response Pathway. J. Virol. 2017, 91, e01430-16. [Google Scholar] [CrossRef] [PubMed]
- Mazzon, M.; Peters, N.E.; Loenarz, C.; Krysztofinska, E.M.; Ember, S.W.; Ferguson, B.J.; Smith, G.L. A mechanism for induction of a hypoxic response by vaccinia virus. Proc. Natl. Acad. Sci. USA 2013, 110, 12444–12449. [Google Scholar] [CrossRef] [PubMed]
- Lamb, S.A.; Rahman, M.M.; McFadden, G. Recombinant myxoma virus lacking all poxvirus ankyrin-repeat proteins stimulates multiple cellular anti-viral pathways and exhibits a severe decrease in virulence. Virology 2014, 464–465, 134–145. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Barrett, J.W.; Stanford, M.; Werden, S.J.; Johnston, J.B.; Gao, X.; Sun, M.; Cheng, J.Q.; McFadden, G. Infection of human cancer cells with myxoma virus requires Akt activation via interaction with a viral ankyrin-repeat host range factor. Proc. Natl. Acad. Sci. USA 2006, 103, 4640–4645. [Google Scholar] [CrossRef] [PubMed]
- Werden, S.J.; Barrett, J.W.; Wang, G.; Stanford, M.M.; McFadden, G. M-T5, the ankyrin repeat, host range protein of myxoma virus, activates Akt and can be functionally replaced by cellular PIKE-A. J. Virol. 2007, 81, 2340–2348. [Google Scholar] [CrossRef] [PubMed]
- Camus-Bouclainville, C.; Fiette, L.; Bouchiha, S.; Pignolet, B.; Counor, D.; Filipe, C.; Gelfi, J.; Messud-Petit, F. A virulence factor of myxoma virus colocalizes with NF-kappaB in the nucleus and interferes with inflammation. J. Virol. 2004, 78, 2510–2516. [Google Scholar] [CrossRef]
- Blanie, S.; Gelfi, J.; Bertagnoli, S.; Camus-Bouclainville, C. MNF, an ankyrin repeat protein of myxoma virus, is part of a native cellular SCF complex during viral infection. Virol. J. 2010, 7, 56. [Google Scholar] [CrossRef] [PubMed]
- Odon, V.; Georgana, I.; Holley, J.; Morata, J.; Maluquer de Motes, C. Novel class of viral Ankyrin proteins targeting the host E3 ubiquitin ligase Cullin-2. J. Virol. 2018, 92, e01374-18. [Google Scholar] [CrossRef]
- Ramsey-Ewing, A.L.; Moss, B. Complementation of a vaccinia virus host-range K1L gene deletion by the nonhomologous CP77 gene. Virology 1996, 222, 75–86. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Perkus, M.E.; Goebel, S.J.; Davis, S.W.; Johnson, G.P.; Limbach, K.; Norton, E.K.; Paoletti, E. Vaccinia virus host range genes. Virology 1990, 179, 276–286. [Google Scholar] [CrossRef]
- Liu, J.; Wennier, S.; Zhang, L.; McFadden, G. M062 is a host range factor essential for myxoma virus pathogenesis and functions as an antagonist of host SAMD9 in human cells. J. Virol. 2011, 85, 3270–3282. [Google Scholar] [CrossRef] [PubMed]
- Sivan, G.; Ormanoglu, P.; Buehler, E.C.; Martin, S.E.; Moss, B. Identification of Restriction Factors by Human Genome-Wide RNA Interference Screening of Viral Host Range Mutants Exemplified by Discovery of SAMD9 and WDR6 as Inhibitors of the Vaccinia Virus K1L-C7L- Mutant. mBio 2015, 6, e01122. [Google Scholar] [CrossRef]
- Meng, X.; Zhang, F.; Yan, B.; Si, C.; Honda, H.; Nagamachi, A.; Sun, L.Z.; Xiang, Y. A paralogous pair of mammalian host restriction factors form a critical host barrier against poxvirus infection. PLoS Pathog. 2018, 14, e1006884. [Google Scholar] [CrossRef]
- Zhang, F.; Meng, X.; Townsend, M.B.; Satheshkumar, P.S.; Xiang, Y. Identification of CP77 as the Third Orthopoxvirus SAMD9 and SAMD9L Inhibitor with Unique Specificity for a Rodent SAMD9L. J. Virol. 2019, 93, e00225-19. [Google Scholar] [CrossRef]
- Chang, S.J.; Hsiao, J.C.; Sonnberg, S.; Chiang, C.T.; Yang, M.H.; Tzou, D.L.; Mercer, A.A.; Chang, W. Poxvirus host range protein CP77 contains an F-box-like domain that is necessary to suppress NF-kappaB activation by tumor necrosis factor alpha but is independent of its host range function. J. Virol. 2009, 83, 4140–4152. [Google Scholar] [CrossRef] [PubMed]
- Van Buuren, N.; Burles, K.; Schriewer, J.; Mehta, N.; Parker, S.; Buller, R.M.; Barry, M. EVM005: An ectromelia-encoded protein with dual roles in NF-kappaB inhibition and virulence. PLoS Pathog. 2014, 10, e1004326. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, M.R.; Rahman, M.M.; Lanchbury, J.S.; Shattuck, D.; Neff, C.; Dufford, M.; van Buuren, N.; Fagan, K.; Barry, M.; Smith, S.; et al. Proteomic screening of variola virus reveals a unique NF-kappaB inhibitor that is highly conserved among pathogenic orthopoxviruses. Proc. Natl. Acad. Sci. USA 2009, 106, 9045–9050. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, M.R.; Rahman, M.M.; Rice, A.; Moyer, R.W.; Werden, S.J.; McFadden, G. Cowpox virus expresses a novel ankyrin repeat NF-kappaB inhibitor that controls inflammatory cell influx into virus-infected tissues and is critical for virus pathogenesis. J. Virol. 2009, 83, 9223–9236. [Google Scholar] [CrossRef]
- Liu, Z.; Nailwal, H.; Rector, J.; Rahman, M.M.; Sam, R.; McFadden, G.; Chan, F.K. A class of viral inducer of degradation of the necroptosis adaptor RIPK3 regulates virus-induced inflammation. Immunity 2021, 54, 247–258.e7. [Google Scholar] [CrossRef]
- Veyer, D.L.; Carrara, G.; Maluquer de Motes, C.; Smith, G.L. Vaccinia virus evasion of regulated cell death. Immunol. Lett. 2017, 186, 68–80. [Google Scholar] [CrossRef]
- Suraweera, C.D.; Hinds, M.G.; Kvansakul, M. Poxviral Strategies to Overcome Host Cell Apoptosis. Pathogens 2020, 10, 6. [Google Scholar] [CrossRef]
- Liu, R.; Moss, B. Vaccinia Virus C9 Ankyrin Repeat/F-Box Protein Is a Newly Identified Antagonist of the Type I Interferon-Induced Antiviral State. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Liu, R.; Olano, L.R.; Mirzakhanyan, Y.; Gershon, P.D.; Moss, B. Vaccinia Virus Ankyrin-Repeat/F-Box Protein Targets Interferon-Induced IFITs for Proteasomal Degradation. Cell Rep. 2019, 29, 816–828.e6. [Google Scholar] [CrossRef]
- Diamond, M.S.; Farzan, M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 2013, 13, 46–57. [Google Scholar] [CrossRef]
- Brady, G.; Bowie, A.G. Innate immune activation of NFkappaB and its antagonism by poxviruses. Cytokine Growth Factor Rev. 2014, 25, 611–620. [Google Scholar] [CrossRef]
- Kanarek, N.; Ben-Neriah, Y. Regulation of NF-kappaB by ubiquitination and degradation of the IkappaBs. Immunol. Rev. 2012, 246, 77–94. [Google Scholar] [CrossRef]
- Fagan-Garcia, K.; Barry, M. A vaccinia virus deletion mutant reveals the presence of additional inhibitors of NF-kappaB. J. Virol. 2011, 85, 883–894. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Oie, K.L.; Pickup, D.J. Cowpox virus and other members of the orthopoxvirus genus interfere with the regulation of NF-kappaB activation. Virology 2001, 288, 175–187. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mansur, D.S.; Maluquer de Motes, C.; Unterholzner, L.; Sumner, R.P.; Ferguson, B.J.; Ren, H.; Strnadova, P.; Bowie, A.G.; Smith, G.L. Poxvirus targeting of E3 ligase beta-TrCP by molecular mimicry: A mechanism to inhibit NF-kappaB activation and promote immune evasion and virulence. PLoS Pathog. 2013, 9, e1003183. [Google Scholar] [CrossRef]
- Sumner, R.P.; Maluquer de Motes, C.; Veyer, D.L.; Smith, G.L. Vaccinia virus inhibits NF-kappaB-dependent gene expression downstream of p65 translocation. J. Virol. 2014, 88, 3092–3102. [Google Scholar] [CrossRef]
- Neidel, S.; Maluquer de Motes, C.; Mansur, D.S.; Strnadova, P.; Smith, G.L.; Graham, S.C. Vaccinia virus protein A49 is an unexpected member of the B-cell Lymphoma (Bcl)-2 protein family. J. Biol. Chem. 2015, 290, 5991–6002. [Google Scholar] [CrossRef]
- Maluquer de Motes, C.; Smith, G.L. Vaccinia virus protein A49 activates Wnt signalling by targetting the E3 ligase beta-TrCP. J. Gen. Virol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Neidel, S.; Ren, H.; Torres, A.A.; Smith, G.L. NF-kappaB activation is a turn on for vaccinia virus phosphoprotein A49 to turn off NF-kappaB activation. Proc. Natl. Acad. Sci. USA 2019, 116, 5699–5704. [Google Scholar] [CrossRef]
- Kamura, T.; Maenaka, K.; Kotoshiba, S.; Matsumoto, M.; Kohda, D.; Conaway, R.C.; Conaway, J.W.; Nakayama, K.I. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004, 18, 3055–3065. [Google Scholar] [CrossRef]
- Mahrour, N.; Redwine, W.B.; Florens, L.; Swanson, S.K.; Martin-Brown, S.; Bradford, W.D.; Staehling-Hampton, K.; Washburn, M.P.; Conaway, R.C.; Conaway, J.W. Characterization of Cullin-box sequences that direct recruitment of Cul2-Rbx1 and Cul5-Rbx2 modules to Elongin BC-based ubiquitin ligases. J. Biol. Chem. 2008, 283, 8005–8013. [Google Scholar] [CrossRef] [PubMed]
- Kamura, T.; Sato, S.; Haque, D.; Liu, L.; Kaelin, W.G., Jr.; Conaway, R.C.; Conaway, J.W. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes Dev. 1998, 12, 3872–3881. [Google Scholar] [CrossRef]
- Zollman, S.; Godt, D.; Prive, G.G.; Couderc, J.L.; Laski, F.A. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc. Natl. Acad. Sci. USA 1994, 91, 10717–10721. [Google Scholar] [CrossRef] [PubMed]
- Bardwell, V.J.; Treisman, R. The POZ domain: A conserved protein-protein interaction motif. Genes Dev. 1994, 8, 1664–1677. [Google Scholar] [CrossRef] [PubMed]
- Stogios, P.J.; Downs, G.S.; Jauhal, J.J.; Nandra, S.K.; Prive, G.G. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005, 6, R82. [Google Scholar] [CrossRef] [PubMed]
- Wilton, B.A.; Campbell, S.; Van Buuren, N.; Garneau, R.; Furukawa, M.; Xiong, Y.; Barry, M. Ectromelia virus BTB/kelch proteins, EVM150 and EVM167, interact with cullin-3-based ubiquitin ligases. Virology 2008, 374, 82–99. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Pallett, M.A.; Croll, T.I.; Smith, G.L.; Graham, S.C. Molecular basis of cullin-3 (Cul3) ubiquitin ligase subversion by vaccinia virus protein A55. J. Biol. Chem. 2019, 294, 6416–6429. [Google Scholar] [CrossRef] [PubMed]
- Beard, P.M.; Froggatt, G.C.; Smith, G.L. Vaccinia virus kelch protein A55 is a 64 kDa intracellular factor that affects virus-induced cytopathic effect and the outcome of infection in a murine intradermal model. J. Gen. Virol. 2006, 87, 1521–1529. [Google Scholar] [CrossRef] [PubMed]
- Froggatt, G.C.; Smith, G.L.; Beard, P.M. Vaccinia virus gene F3L encodes an intracellular protein that affects the innate immune response. J. Gen. Virol. 2007, 88, 1917–1921. [Google Scholar] [CrossRef] [PubMed]
- Pires de Miranda, M.; Reading, P.C.; Tscharke, D.C.; Murphy, B.J.; Smith, G.L. The vaccinia virus kelch-like protein C2L affects calcium-independent adhesion to the extracellular matrix and inflammation in a murine intradermal model. J. Gen. Virol. 2003, 84, 2459–2471. [Google Scholar] [CrossRef] [PubMed]
- Balinsky, C.A.; Delhon, G.; Afonso, C.L.; Risatti, G.R.; Borca, M.V.; French, R.A.; Tulman, E.R.; Geary, S.J.; Rock, D.L. Sheeppox virus kelch-like gene SPPV-019 affects virus virulence. J. Virol. 2007, 81, 11392–11401. [Google Scholar] [CrossRef]
- Kochneva, G.; Kolosova, I.; Maksyutova, T.; Ryabchikova, E.; Shchelkunov, S. Effects of deletions of kelch-like genes on cowpox virus biological properties. Arch. Virol. 2005, 150, 1857–1870. [Google Scholar] [CrossRef] [PubMed]
- Pallett, M.A.; Ren, H.; Zhang, R.Y.; Scutts, S.R.; Gonzalez, L.; Zhu, Z.; Maluquer de Motes, C.; Smith, G.L. Vaccinia Virus BBK E3 Ligase Adaptor A55 Targets Importin-Dependent NF-kappaB Activation and Inhibits CD8(+) T-Cell Memory. J. Virol. 2019, 93, e00051-19. [Google Scholar] [CrossRef]
- Wang, Q.; Burles, K.; Couturier, B.; Randall, C.M.; Shisler, J.; Barry, M. Ectromelia virus encodes a BTB/kelch protein, EVM150, that inhibits NF-kappaB signaling. J. Virol. 2014, 88, 4853–4865. [Google Scholar] [CrossRef]
- Ren, H.; Ferguson, B.J.; Maluquer de Motes, C.; Sumner, R.P.; Harman, L.E.; Smith, G.L. Enhancement of CD8(+) T-cell memory by removal of a vaccinia virus nuclear factor-kappaB inhibitor. Immunology 2015, 145, 34–49. [Google Scholar] [CrossRef]
- Mathew, A.; O’Bryan, J.; Marshall, W.; Kotwal, G.J.; Terajima, M.; Green, S.; Rothman, A.L.; Ennis, F.A. Robust intrapulmonary CD8 T cell responses and protection with an attenuated N1L deleted vaccinia virus. PLoS ONE 2008, 3, e3323. [Google Scholar] [CrossRef]
- Di Pilato, M.; Mejias-Perez, E.; Sorzano, C.O.S.; Esteban, M. Distinct Roles of Vaccinia Virus NF-kappaB Inhibitor Proteins A52, B15, and K7 in the Immune Response. J. Virol. 2017, 91, e00575-17. [Google Scholar] [CrossRef] [PubMed]
- Sumner, R.P.; Ren, H.; Ferguson, B.J.; Smith, G.L. Increased attenuation but decreased immunogenicity by deletion of multiple vaccinia virus immunomodulators. Vaccine 2016, 34, 4827–4834. [Google Scholar] [CrossRef]
- Garcia-Arriaza, J.; Arnaez, P.; Gomez, C.E.; Sorzano, C.O.; Esteban, M. Improving Adaptive and Memory Immune Responses of an HIV/AIDS Vaccine Candidate MVA-B by Deletion of Vaccinia Virus Genes (C6L and K7R) Blocking Interferon Signaling Pathways. PLoS ONE 2013, 8, e66894. [Google Scholar] [CrossRef]
- Angers, S.; Li, T.; Yi, X.; MacCoss, M.J.; Moon, R.T.; Zheng, N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 2006, 443, 590–593. [Google Scholar] [CrossRef]
- Fischer, E.S.; Scrima, A.; Bohm, K.; Matsumoto, S.; Lingaraju, G.M.; Faty, M.; Yasuda, T.; Cavadini, S.; Wakasugi, M.; Hanaoka, F.; et al. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell 2011, 147, 1024–1039. [Google Scholar] [CrossRef]
- He, Y.J.; McCall, C.M.; Hu, J.; Zeng, Y.; Xiong, Y. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. 2006, 20, 2949–2954. [Google Scholar] [CrossRef]
- Brady, G.; Haas, D.A.; Farrell, P.J.; Pichlmair, A.; Bowie, A.G. Poxvirus Protein MC132 from Molluscum Contagiosum Virus Inhibits NF-B Activation by Targeting p65 for Degradation. J. Virol. 2015, 89, 8406–8415. [Google Scholar] [CrossRef]
- Yu, H.; Peters, J.M.; King, R.W.; Page, A.M.; Hieter, P.; Kirschner, M.W. Identification of a cullin homology region in a subunit of the anaphase-promoting complex. Science 1998, 279, 1219–1222. [Google Scholar] [CrossRef]
- Zachariae, W.; Shevchenko, A.; Andrews, P.D.; Ciosk, R.; Galova, M.; Stark, M.J.; Mann, M.; Nasmyth, K. Mass spectrometric analysis of the anaphase-promoting complex from yeast: Identification of a subunit related to cullins. Science 1998, 279, 1216–1219. [Google Scholar] [CrossRef]
- Nikolaev, A.Y.; Li, M.; Puskas, N.; Qin, J.; Gu, W. Parc: A cytoplasmic anchor for p53. Cell 2003, 112, 29–40. [Google Scholar] [CrossRef]
- Dias, D.C.; Dolios, G.; Wang, R.; Pan, Z.Q. CUL7: A DOC domain-containing cullin selectively binds Skp1.Fbx29 to form an SCF-like complex. Proc. Natl. Acad. Sci. USA 2002, 99, 16601–16606. [Google Scholar] [CrossRef]
- Castro, A.; Bernis, C.; Vigneron, S.; Labbe, J.C.; Lorca, T. The anaphase-promoting complex: A key factor in the regulation of cell cycle. Oncogene 2005, 24, 314–325. [Google Scholar] [CrossRef]
- Mo, M.; Fleming, S.B.; Mercer, A.A. Cell cycle deregulation by a poxvirus partial mimic of anaphase-promoting complex subunit 11. Proc. Natl. Acad. Sci. USA 2009, 106, 19527–19532. [Google Scholar] [CrossRef]
- Mo, M.; Fleming, S.B.; Mercer, A.A. Orf virus cell cycle regulator, PACR, competes with subunit 11 of the anaphase promoting complex for incorporation into the complex. J. Gen. Virol. 2010, 91, 3010–3015. [Google Scholar] [CrossRef]
- Mo, M.; Shahar, S.; Fleming, S.B.; Mercer, A.A. How viruses affect the cell cycle through manipulation of the APC/C. Trends Microbiol. 2012, 20, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Fruh, K.; Bartee, E.; Gouveia, K.; Mansouri, M. Immune evasion by a novel family of viral PHD/LAP-finger proteins of gamma-2 herpesviruses and poxviruses. Virus Res. 2002, 88, 55–69. [Google Scholar] [CrossRef]
- Guerin, J.L.; Gelfi, J.; Boullier, S.; Delverdier, M.; Bellanger, F.A.; Bertagnoli, S.; Drexler, I.; Sutter, G.; Messud-Petit, F. Myxoma virus leukemia-associated protein is responsible for major histocompatibility complex class I and Fas-CD95 down-regulation and defines scrapins, a new group of surface cellular receptor abductor proteins. J. Virol. 2002, 76, 2912–2923. [Google Scholar] [CrossRef]
- Barry, M.; Lee, S.F.; Boshkov, L.; McFadden, G. Myxoma virus induces extensive CD4 downregulation and dissociation of p56lck in infected rabbit CD4+ T lymphocytes. J. Virol. 1995, 69, 5243–5251. [Google Scholar] [CrossRef]
- Mansouri, M.; Bartee, E.; Gouveia, K.; Hovey Nerenberg, B.T.; Barrett, J.; Thomas, L.; Thomas, G.; McFadden, G.; Fruh, K. The PHD/LAP-domain protein M153R of myxomavirus is a ubiquitin ligase that induces the rapid internalization and lysosomal destruction of CD4. J. Virol. 2003, 77, 1427–1440. [Google Scholar] [CrossRef]
- Bartee, E.; McCormack, A.; Fruh, K. Quantitative membrane proteomics reveals new cellular targets of viral immune modulators. PLoS Pathog. 2006, 2, e107. [Google Scholar] [CrossRef]
- Zuniga, M.C.; Wang, H.; Barry, M.; McFadden, G. Endosomal/lysosomal retention and degradation of major histocompatibility complex class I molecules is induced by myxoma virus. Virology 1999, 261, 180–192. [Google Scholar] [CrossRef]
- Coscoy, L.; Ganem, D. Kaposi’s sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc. Natl. Acad. Sci. USA 2000, 97, 8051–8056. [Google Scholar] [CrossRef]
- Ishido, S.; Wang, C.; Lee, B.S.; Cohen, G.B.; Jung, J.U. Downregulation of major histocompatibility complex class I molecules by Kaposi’s sarcoma-associated herpesvirus K3 and K5 proteins. J. Virol. 2000, 74, 5300–5309. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, P.G.; Efstathiou, S.; Doherty, P.C.; Lehner, P.J. Inhibition of MHC class I-restricted antigen presentation by gamma 2-herpesviruses. Proc. Natl. Acad. Sci. USA 2000, 97, 8455–8460. [Google Scholar] [CrossRef] [PubMed]
- Ogbomo, H.; Zemp, F.J.; Lun, X.; Zhang, J.; Stack, D.; Rahman, M.M.; McFadden, G.; Mody, C.H.; Forsyth, P.A. Myxoma virus infection promotes NK lysis of malignant gliomas in vitro and in vivo. PLoS ONE 2013, 8, e66825. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.M.; Rahman, M.M.; McFadden, G. Oncolytic myxoma virus: The path to clinic. Vaccine 2013, 31, 4252–4258. [Google Scholar] [CrossRef] [PubMed]
- Upton, C.; Schiff, L.; Rice, S.A.; Dowdeswell, T.; Yang, X.; McFadden, G. A poxvirus protein with a RING finger motif binds zinc and localizes in virus factories. J. Virol. 1994, 68, 4186–4195. [Google Scholar] [CrossRef]
- Senkevich, T.G.; Koonin, E.V.; Buller, R.M. A poxvirus protein with a RING zinc finger motif is of crucial importance for virulence. Virology 1994, 198, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, R.D.; Gray, T.A. Cellular source of the poxviral N1R/p28 gene family. Virus Genes 2004, 29, 359–364. [Google Scholar] [CrossRef]
- Senkevich, T.G.; Wolffe, E.J.; Buller, R.M. Ectromelia virus RING finger protein is localized in virus factories and is required for virus replication in macrophages. J. Virol. 1995, 69, 4103–4111. [Google Scholar] [CrossRef]
- Huang, J.; Huang, Q.; Zhou, X.; Shen, M.M.; Yen, A.; Yu, S.X.; Dong, G.; Qu, K.; Huang, P.; Anderson, E.M.; et al. The poxvirus p28 virulence factor is an E3 ubiquitin ligase. J. Biol. Chem. 2004, 279, 54110–54116. [Google Scholar] [CrossRef]
- Nerenberg, B.T.; Taylor, J.; Bartee, E.; Gouveia, K.; Barry, M.; Fruh, K. The poxviral RING protein p28 is a ubiquitin ligase that targets ubiquitin to viral replication factories. J. Virol. 2005, 79, 597–601. [Google Scholar] [CrossRef]
- Brick, D.J.; Burke, R.D.; Schiff, L.; Upton, C. Shope fibroma virus RING finger protein N1R binds DNA and inhibits apoptosis. Virology 1998, 249, 42–51. [Google Scholar] [CrossRef]
- Mottet, K.; Bareiss, B.; Milne, C.D.; Barry, M. The poxvirus encoded ubiquitin ligase, p28, is regulated by proteasomal degradation and autoubiquitination. Virology 2014, 468–470, 363–378. [Google Scholar] [CrossRef]
- Brick, D.J.; Burke, R.D.; Minkley, A.A.; Upton, C. Ectromelia virus virulence factor p28 acts upstream of caspase-3 in response to UV light-induced apoptosis. J. Gen. Virol. 2000, 81, 1087–1097. [Google Scholar] [CrossRef]
- Afonso, C.L.; Tulman, E.R.; Lu, Z.; Oma, E.; Kutish, G.F.; Rock, D.L. The genome of Melanoplus sanguinipes entomopoxvirus. J. Virol. 1999, 73, 533–552. [Google Scholar] [CrossRef]
- Bawden, A.L.; Glassberg, K.J.; Diggans, J.; Shaw, R.; Farmerie, W.; Moyer, R.W. Complete genomic sequence of the Amsacta moorei entomopoxvirus: Analysis and comparison with other poxviruses. Virology 2000, 274, 120–139. [Google Scholar] [CrossRef] [PubMed]
- Tulman, E.R.; Afonso, C.L.; Lu, Z.; Zsak, L.; Kutish, G.F.; Rock, D.L. The genome of canarypox virus. J. Virol. 2004, 78, 353–366. [Google Scholar] [CrossRef]
- Offerman, K.; Carulei, O.; van der Walt, A.P.; Douglass, N.; Williamson, A.L. The complete genome sequences of poxviruses isolated from a penguin and a pigeon in South Africa and comparison to other sequenced avipoxviruses. BMC Genom. 2014, 15, 463. [Google Scholar] [CrossRef]
- Carulei, O.; Douglass, N.; Williamson, A.L. Comparative analysis of avian poxvirus genomes, including a novel poxvirus from lesser flamingos (Phoenicopterus minor), highlights the lack of conservation of the central region. BMC Genom. 2017, 18, 947. [Google Scholar] [CrossRef]
- Grossegesse, M.; Doellinger, J.; Fritsch, A.; Laue, M.; Piesker, J.; Schaade, L.; Nitsche, A. Global ubiquitination analysis reveals extensive modification and proteasomal degradation of cowpox virus proteins, but preservation of viral cores. Sci. Rep. 2018, 8, 1807. [Google Scholar] [CrossRef] [PubMed]
- Wickramasekera, N.T.; Traktman, P. Structure/Function analysis of the vaccinia virus F18 phosphoprotein, an abundant core component required for virion maturation and infectivity. J. Virol. 2010, 84, 6846–6860. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, F.I.; Bleck, C.K.; Reh, L.; Novy, K.; Wollscheid, B.; Helenius, A.; Stahlberg, H.; Mercer, J. Vaccinia virus entry is followed by core activation and proteasome-mediated release of the immunomodulatory effector VH1 from lateral bodies. Cell Rep. 2013, 4, 464–476. [Google Scholar] [CrossRef] [PubMed]
- Najarro, P.; Traktman, P.; Lewis, J.A. Vaccinia virus blocks gamma interferon signal transduction: Viral VH1 phosphatase reverses Stat1 activation. J. Virol. 2001, 75, 3185–3196. [Google Scholar] [CrossRef]
- Bidgood, S.R.; Mercer, J. Cloak and Dagger: Alternative Immune Evasion and Modulation Strategies of Poxviruses. Viruses 2015, 7, 4800–4825. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.W.; Watson, J.C.; Jacobs, B.L. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc. Natl. Acad. Sci. USA 1992, 89, 4825–4829. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.C.; Chang, H.W.; Jacobs, B.L. Characterization of a vaccinia virus-encoded double-stranded RNA-binding protein that may be involved in inhibition of the double-stranded RNA-dependent protein kinase. Virology 1991, 185, 206–216. [Google Scholar] [CrossRef]
- Xiang, Y.; Condit, R.C.; Vijaysri, S.; Jacobs, B.; Williams, B.R.; Silverman, R.H. Blockade of interferon induction and action by the E3L double-stranded RNA binding proteins of vaccinia virus. J. Virol. 2002, 76, 5251–5259. [Google Scholar] [CrossRef]
- Koehler, H.; Cotsmire, S.; Langland, J.; Kibler, K.V.; Kalman, D.; Upton, J.W.; Mocarski, E.S.; Jacobs, B.L. Inhibition of DAI-dependent necroptosis by the Z-DNA binding domain of the vaccinia virus innate immune evasion protein, E3. Proc. Natl. Acad. Sci. USA 2017, 114, 11506–11511. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Santamaria, J.; Campagna, M.; Garcia, M.A.; Marcos-Villar, L.; Gonzalez, D.; Gallego, P.; Lopitz-Otsoa, F.; Guerra, S.; Rodriguez, M.S.; Esteban, M.; et al. Regulation of vaccinia virus E3 protein by small ubiquitin-like modifier proteins. J. Virol. 2011, 85, 12890–12900. [Google Scholar] [CrossRef]
- Maluquer de Motes, C.; Schiffner, T.; Sumner, R.P.; Smith, G.L. Vaccinia virus virulence factor N1 can be ubiquitylated on multiple lysine residues. J. Gen. Virol. 2014, 95, 2038–2049. [Google Scholar] [CrossRef]
- Cooray, S.; Bahar, M.W.; Abrescia, N.G.A.; McVey, C.E.; Bartlett, N.W.; Chen, R.A.; Stuart, D.I.; Grimes, J.M.; Smith, G.L. Functional and structural studies of the vaccinia virus virulence factor N1 reveal a Bcl-2-like anti-apoptotic protein. J. Gen. Virol. 2007, 88, 1656–1666. [Google Scholar] [CrossRef]
- Maluquer de Motes, C.; Cooray, S.; Ren, H.; Almeida, G.M.; McGourty, K.; Bahar, M.W.; Stuart, D.I.; Grimes, J.M.; Graham, S.C.; Smith, G.L. Inhibition of apoptosis and NF-kappaB activation by vaccinia protein N1 occur via distinct binding surfaces and make different contributions to virulence. PLoS Pathog. 2011, 7, e1002430. [Google Scholar] [CrossRef] [PubMed]
- Veyer, D.L.; Maluquer de Motes, C.; Sumner, R.P.; Ludwig, L.; Johnson, B.F.; Smith, G.L. Analysis of the anti-apoptotic activity of four vaccinia virus proteins demonstrates that B13 is the most potent inhibitor in isolation and during viral infection. J. Gen. Virol. 2014, 95, 2757–2768. [Google Scholar] [CrossRef] [PubMed]
- DiPerna, G.; Stack, J.; Bowie, A.G.; Boyd, A.; Kotwal, G.; Zhang, Z.; Arvikar, S.; Latz, E.; Fitzgerald, K.A.; Marshall, W.L. Poxvirus protein N1L targets the I-kappaB kinase complex, inhibits signaling to NF-kappaB by the tumor necrosis factor superfamily of receptors, and inhibits NF-kappaB and IRF3 signaling by toll-like receptors. J. Biol. Chem. 2004, 279, 36570–36578. [Google Scholar] [CrossRef]
- Soday, L.; Lu, Y.; Albarnaz, J.D.; Davies, C.T.R.; Antrobus, R.; Smith, G.L.; Weekes, M.P. Quantitative Temporal Proteomic Analysis of Vaccinia Virus Infection Reveals Regulation of Histone Deacetylases by an Interferon Antagonist. Cell Rep. 2019, 27, 1920–1933.e7. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Stuart, J.H.; Talbot-Cooper, C.; Agrawal-Singh, S.; Huntly, B.; Smid, A.I.; Snowden, J.S.; Dupont, L.; Smith, G.L. Histone deacetylase 4 promotes type I interferon signaling, restricts DNA viruses, and is degraded via vaccinia virus protein C6. Proc. Natl. Acad. Sci. USA 2019, 116, 11997–12006. [Google Scholar] [CrossRef]
- Unterholzner, L.; Sumner, R.P.; Baran, M.; Ren, H.; Mansur, D.S.; Bourke, N.M.; Randow, F.; Smith, G.L.; Bowie, A.G. Vaccinia virus protein C6 is a virulence factor that binds TBK-1 adaptor proteins and inhibits activation of IRF3 and IRF7. PLoS Pathog. 2011, 7, e1002247. [Google Scholar] [CrossRef]
- Stuart, J.H.; Sumner, R.P.; Lu, Y.; Snowden, J.S.; Smith, G.L. Vaccinia Virus Protein C6 Inhibits Type I IFN Signalling in the Nucleus and Binds to the Transactivation Domain of STAT2. PLoS Pathog. 2016, 12, e1005955. [Google Scholar] [CrossRef]
- Sumner, R.P.; Ren, H.; Smith, G.L. Deletion of immunomodulator C6 from vaccinia virus strain Western Reserve enhances virus immunogenicity and vaccine efficacy. J. Gen. Virol. 2013, 94, 1121–1126. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Arriaza, J.; Najera, J.L.; Gomez, C.E.; Tewabe, N.; Sorzano, C.O.; Calandra, T.; Roger, T.; Esteban, M. A candidate HIV/AIDS vaccine (MVA-B) lacking vaccinia virus gene C6L enhances memory HIV-1-specific T-cell responses. PLoS ONE 2011, 6, e24244. [Google Scholar] [CrossRef]
- Marin, M.Q.; Perez, P.; Gomez, C.E.; Sorzano, C.O.S.; Esteban, M.; Garcia-Arriaza, J. Removal of the C6 Vaccinia Virus Interferon-beta Inhibitor in the Hepatitis C Vaccine Candidate MVA-HCV Elicited in Mice High Immunogenicity in Spite of Reduced Host Gene Expression. Viruses 2018, 10, 414. [Google Scholar] [CrossRef] [PubMed]
| Orthologue Group 1 | CPXV (BR) | VACV (Cop) | VARV (B75) | ECTV (Mos) | MPXV (Z) | CMPV (CMS) |
|---|---|---|---|---|---|---|
| I | 006/225 | C19L | G1R | 002 | J1R | 003L |
| II | 008/223 | C17L | D1L | 004L | ||
| III | 011 | 005 | ||||
| IV | 016 | 010 | ||||
| V | 017 | D1L | ||||
| VI | 019 | |||||
| VII | 025 | D8L | D7L | |||
| VIII | 027 | C9L | D9L | |||
| IX | 039 | M1L | O1L | 021 | O1L | |
| X | 041 | K1L | C1L | 022 | C1L | |
| XI | 198 | B4R | B5R | 154 | B5R | 177R |
| XII | 200 | B6R | ||||
| XIII | 211 | B18R | B16R | 165 | B17R | 197R |
| XIV | 213 | B18R | ||||
| XV | 220 |
| CPXV (GRI). | VACV (Cop) | ECTV (Mos) | MPXV (Z) | CMPV (CMS) |
|---|---|---|---|---|
| A57R | A55 | 150 | 172R | |
| C18L | C2 | 018 | 24L | |
| G3L | F3 | 027 | C9L | 38L |
| D11L | ||||
| B19R | ||||
| B9R | 167 (C13R) | 186R | ||
| C5 | D12L | 21L |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lant, S.; Maluquer de Motes, C. Poxvirus Interactions with the Host Ubiquitin System. Pathogens 2021, 10, 1034. https://doi.org/10.3390/pathogens10081034
Lant S, Maluquer de Motes C. Poxvirus Interactions with the Host Ubiquitin System. Pathogens. 2021; 10(8):1034. https://doi.org/10.3390/pathogens10081034
Chicago/Turabian StyleLant, Sian, and Carlos Maluquer de Motes. 2021. "Poxvirus Interactions with the Host Ubiquitin System" Pathogens 10, no. 8: 1034. https://doi.org/10.3390/pathogens10081034
APA StyleLant, S., & Maluquer de Motes, C. (2021). Poxvirus Interactions with the Host Ubiquitin System. Pathogens, 10(8), 1034. https://doi.org/10.3390/pathogens10081034







