Abstract
Wastewater-based epidemiology (WBE) has a long history of identifying a variety of viruses from poliovirus to coronaviruses, including novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). The presence and detection of SARS-CoV-2 in human feces and its passage into the water bodies are significant public health challenges. Hence, the hot issue of WBE of SARS-CoV-2 in the coronavirus respiratory disease (COVID-19) pandemic is a matter of utmost importance (e.g., SARS-CoV-1). The present review discusses the background, state of the art, actual status, and prospects of WBE, as well as the detection and quantification protocols of SARS-CoV-2 in wastewater. The SARS-CoV-2 detection studies have been performed in different water matrixes such as influent and effluent of wastewater treatment plants, suburban pumping stations, hospital wastewater, and sewer networks around the globe except for Antarctica. The findings revealed that all WBE studies were in accordance with clinical and epidemiological data, which correlates the presence of SARS-CoV-2 ribonucleic acid (RNA) with the number of new daily positive cases officially reported. This last was confirmed via Reverse Transcriptase-quantitative Polymerase Chain Reaction (RT-qPCR) testing which unfortunately is not suitable for real-time surveillance. In addition, WBE concept may act as a faster protocol to alert the public health authorities to take administrative orders (possible re-emerging infections) due to the impracticality of testing all citizens in a short time with limited diagnostic facilities. A comprehensive and integrated review covering all steps starting from sampling to molecular detection of SARS-CoV-2 in wastewater has been made to guide for the development well-defined and reliable protocols.
1. Background of Application of Wastewater Based Epidemiology
All the community’s physical, chemical, and biological substances are excreted to the sewer systems and transported to wastewater treatment plants (WWTPs) providing a pooled sample from a group of people in a specific geographical location at a point in time. Risks of emerging infectious diseases and increasing rates of antimicrobial resistance emphasize that infectious disease surveillance is still a fundamental piece of public health. There are some techniques to monitor spatial and temporal trends of diseases such as sentinel surveillance, surveys, mortality and morbidity rates, hospital admission data, human biomonitoring, and wastewater-based methodology (WBE) [1]. This last technique uses “water fingerprinting” to provide an objective and comprehensive assessment of both public and environmental health status in near real-time from water sources (surface waters and domestic wastewaters). Christian Daughton is considered the pioneer of WBE concept [2,3]. He postulated that the evaluation of drug residues (concentration) could be associated with population usage. In 2017, Andrés-Costa et al. estimated that WBE would be a promising tool to collect data on entire communities’ health (or at least in an important percentage) [4]. In this sense, the population’s health condition may be assessed by monitoring biomarkers that included endogenous and exogenous human metabolites as well as different substances. These biomarkers are identified and quantified in untreated wastewaters, and the samples are usually collected in influents of WWTPs that serve communities [5].
In recent decades, WBE, as a novel biomonitoring tool, has been successfully used to investigate polio circulation within the community, monitor the success of international poliovirus vaccine campaigns, investigate the use of some illicit drugs, and provide early warnings of hepatitis A virus and norovirus outbreaks [5,6,7]. Viruses are the main causative agent of several mortal illnesses such as gastroenteritis, hepatitis, and respiratory diseases. After the appearance of Severe Acute Respiratory Syndrome Coronavirus 1 (SARS-CoV-1) in 2003 and Middle East Respiratory Syndrome (MERS) in 2012, the environmental circulation of viruses has received more attention to detect/track human pathogen spread into communities. The importance of surveillance systems became more highlighted with the emergence of coronavirus respiratory disease (COVID-19) in December 2019 in Wuhan, China [1,8]. Since many Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) patients might exhibit few or non-specific symptoms, rapid and accurate diagnosis of potential virus carriers is a critical step to suppress the risk of disease transmission at an early stage [9]. With the first report of SARS-CoV-2 detection in both symptomatic and asymptomatic patient’s feces, several studies have been performed to assess WBE as an early indication tool for COVID-19 transmission and pandemic monitoring [10,11]. Researchers have achieved the identification of SARS-CoV-2 ribonucleic acid (RNA) in different wastewater samples in Netherlands, Australia, France, Brazil, New Zealand, the USA, Japan, and Canada [7,12]. For instance, presence of SARS-CoV-2 RNA has been confirmed in hospital and municipal wastewaters [13,14,15]. Therefore, the World Health Organization (WHO) guideline has been stated for the good management of wastewater [16].
Various studies show a positive correlation between COVID-19 cases and SARS-CoV-2 RNA identification in wastewater. This last confirms that WBE approach is a capable method for early detection, monitoring trends, evaluating the efficiency of public health measures, and tracking immunity of both infected and vaccinated people in response to COVID-19 [7,17]. With the increasing frequency of zoonotic epidemics, WBE can supply future action plans against such pandemics as an environmental surveillance approach [7,18]. As of 2019, only seven studies had investigated the coronaviruses’ presence in environmental compartments such as water, wastewater, and sludge. However, after the emergence of COVID-19, a significant number of studies were conducted over a year, highlighting the importance of WBE studies [7,18]. Although promising, continuous monitoring studies based on different methods are underway, they may provide conflicting results, which indicate that a critical search is necessary to establish a standard virus concentration and identification method for enveloped viruses such as SARS-CoV-2 in water matrix samples. This latter will enhance the accuracy of such surveillance approaches [19,20,21,22].
The present review discusses the background, state-of-the-art, actual status, and prospects of WBE. In addition, detection and quantification methods of SARS-CoV-2 in wastewater covering sampling, storage, inactivation, concentration, extraction and molecular assays are assessed to make a comprehensive and comparable list of studies.
2. Cutting-Edge of Wastewater Based Epidemiology Concept
WBE is based on the extraction, detection, analysis, and interpretation of chemical/biological compounds (biomarkers). Subsequently, this methodology gives information about health community and environmental exposure, as mentioned above. Genetic biomarkers are crucial for determining the disease incidence. A suitable genetic biomarker must have the following characteristics: stable (in sampling and storage), specific for a particular disease, consistent between distinct genders and ethnic groups, human-specific and excreted in urine or feces constantly, and not absorbable to particulate matter, for instance, deoxyribonucleic acid (DNA), RNA, or antibiotic resistance genes [23].
The main advantages of WBE method are: (i) asymptomatic and pre-symptomatic patients may be detected (keeping the anonymity of individuals); (ii) evidence of SARS-CoV-2 circulation to support public health measures and limit the transmission; (iii) helps to identify hotspots for further classical surveillance interventions; and (iv) a non-invasive, viable, and almost real-time method. However, the main disadvantage is the matrix complexity (wastewater) together with loss of biomarkers within the drain system and during storage. Another important consideration to keep in mind is the dynamic population (tourism/business activities) [24] and the weather variations (geographic location). Although WBE has many advantages, it should not be viewed as a replacement for clinical testing. Still, it can provide independent information to public health decisions.
SARS-CoV-2 DNA/RNA residues present in raw wastewater indicate that COVID-19 disease is circulating inside the community. Unfortunately, the lengthy incubation time and virus shedding have allowed the spreading of SARS-CoV-2 causing a poor containment. The knowledge of the occurrence of SARS-CoV-2 in wastewater and secondary sludge from WWTPs could predict the decreasing or rising trends (second or more infectious waves). Kumar et al. concluded that high viral concentration goes hand in hand with the high number of COVID-19 infected individuals [25]. Similar studies have reported the occurrence of SARS-CoV-2 in fecal and urine samples [26,27,28,29,30,31,32]. It is important to note that wastewater generally contains organic matter, particulate solids, micro and macro pollutants, and other pathogens. Moreover, SARS-CoV-2 contained in feces usually undergoes several transformations along the sewer network, and consequently, the dilution factor must be considered. Wastewater properties and sewer conditions cause a reasonable uncertainty (20–40%) [33]. In the same vein, disinfection technologies (thermal, chlorine species, ozonation, and UV radiation, among others) usually inactivate the SARS-CoV-2 virus due to the similarities in phylogenetic with SARS-CoV-1. Without active human cells as hosts in wastewater, the infectivity of SARS-CoV-2 could be reduced up to 90% in minutes. However, SARS-CoV-2 RNA was significantly more persistent (3–33 days) [33]. The resulting non-infectious virus could still be detected by Reverse Transcriptase Polymerase Chain Reaction (RT-PCR) based methods [34]. Zhang et al. investigated the presence of SARS-CoV-2 RNA by Reverse Transcriptase-quantitative Polymerase Chain Reaction (RT-qPCR) in influent and effluent of septic tanks located at a hospital in China. Although the occurrence of SARS-CoV-2 viral RNA was not found in the influent sample, the effluent of septic tank tested positive for such viral RNA, even after the second stage of disinfection [35]. This last is attributed to the shield provided by suspended solids by embedding viruses during disinfection. In conclusion, disinfection step may lead to viral loss; however, health and well-being of the operating/laboratory personnel is a priority. Moreover, studies to refine WBE procedures and, consequently, to improve estimates are urged.
According to the Commission Recommendation (EU) 2021/472 on a common approach to establish a systematic surveillance of SARS-CoV-2 and its variants in wastewaters in the EU [36], WBE surveillance should be taken into account as a complementary and independent tool to COVID-19 surveillance and testing strategies. Therefore, WBE requires to be included more systematically in the national testing strategies for tracing of the SARS-CoV-2 virus. In addition, the Member States are strongly encouraged to put in place as soon as possible and no later than 1 October 2021 a national wastewater surveillance system targeted at data collection of SARS-CoV-2 and its variants in wastewaters.
Finally, WBE serves to observe trends on SARS-CoV-2 spreading and could be an effective early warning for possible COVID-19 re-emerging infections. Moreover, local health authorities can use this knowledge to take administrative orders related to notify the potential health threat and diminish the possibility of transmission. Updated data have confirmed that surveillance of SARS-CoV-2 and its variants in wastewaters can supply a cost effective, rapid, and reliable source of information.
3. Actual Status of Wastewater Contamination with SARS-CoV-2
The COVID-19 wastewater-based epidemiology, other than the droplet or aerosol-based transmission, needs to be emphasized [37,38,39,40]. The SARS-CoV-2 genome load has been predicted to be 600,000 per mL of feces [41], and it is known that one COVID-19 patient can produce up to 370 L water per day [42]. The SARS-CoV-2 virus can remain intact in various media [43,44,45]. Therefore, the virus can be transported to water bodies. The importance of epidemiology based on different sources of wastewater together with its environmental implication has had a greater emphasis nowadays [46,47,48,49]. The change of position of the virus caused by water flows can produce a quick evolution of the same, as observed in the case of the subsequent waves of the COVID-19 disease in many countries [50,51]. It is clearly known that an infected person who sneezes, coughs, talks or sings can infect a healthy person through droplets of various sizes. Similarly, if COVID-19 patients use the sink, bathroom, toilet or basin, the possibility of transmitting the virus to the environment is logically accepted and studied [48,49,52,53].
Environmental surveillance for both the self-quarantined or isolated patients and from COVID-19 hospitals to identify and control the spreading of COVID-19 has been emphasized very early [41,48,54]. Worldwide reports, as observed below, have confirmed that the viral RNA exists in several wastewater samples. This last could make the situation worse for the disease’s outbreak. The RNA of SARS-CoV-2 was argued to come from waste such as stools, urine, washing materials, clothes, etc. of COVID-19 patients [52,53,54]. As per the infection rate observed in different cities around the globe (Figure 1), the viral loading about 100–300 million genomes in drainage would happen before the wastewater treatment from COVID-19 care centers and isolation homes. Data were well correlated as per the cases noticed in those cities.
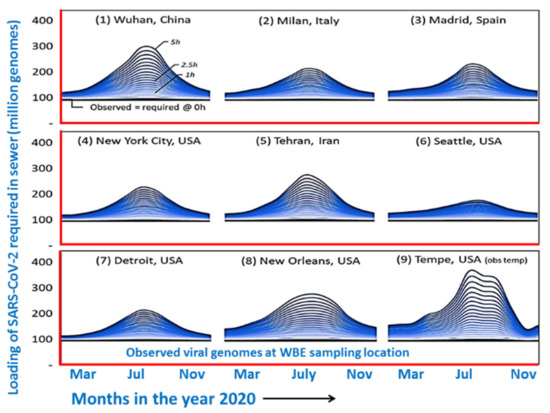
Figure 1.
Predicted water borne viral load in different cities around the world (Redrawn after Hart et al. [42] under CC BY-NC-ND 4.0 license).
Still now, the theory of zoonotic origin of the virus has been established. However, the urination, defecation, and sneezing activities of infected patients or asymptomatic persons cause the incidence of SARS-CoV-2 RNA in drainage systems. Then, healthy people may be infected via drinking water or other activities using contaminated water.
As vaccination campaigns increase, the viral load of SARS-CoV-2 in wastewater samples hardly relates to the reported cases due to the new asymptomatic cases [55]. However, medium or low SARS-CoV-2 RNA concentrations in wastewater may prevent possible new waves, and vaccines may be directed by governments to specific areas [56,57]. For instance, La Rosa et al. [58] confirmed the wastewater contamination by SARS-CoV-2 RNA two months before the disease outbreak in Milan and Turin. A similar report was also documented by Chavarria-Miro et al. [59] in Barcelona 41 days before the onset of the disease in 2020 [59]. In Bnei municipality area of Israel, a positive correlation was observed between the RT-PCR test and the wastewater samples of infected cases in March 2020 [60]. Wurtzer et al. [61] also observed a similar association between SARS-CoV-2 RNA detection in wastewater and the number of confirmed cases with an 8 day temporal shift in Paris. Interestingly, a similar exponential viral load was detected in municipal wastewater in Massachusetts from early January to May 2020 before the confirmed cases were documented [62].
Several analyses have been performed to detect the presence of SARS-CoV-2 in all continents (except Antarctica). As shown in Figure 2, the first detection of SARS-CoV-2 in different types of waters such as WWTPs, suburban pumping stations (PS), hospital wastewater (HW), and sewer networks (SN) around the globe are reported in Figure 2. All sampling dates were in 2020. Presence of SARS-CoV-2 RNA in wastewater was first reported in a WWTP at USA (Louisiana) from 13 January to 8 April. About 28.57% (2/7) of samples were reported positives [63]. In Canada (Ottawa and Gatineau), RT-qPCR analyses showed an increase in the rate of documentation of N1 and N2 genes primary in sludge (92.7, 90.6%) as compared to influent post grit solids samples (79.2, 82.3%). The authors mentioned that after statistical treatment of the data, a strong positive association was noticed between the copy number of the viral RNA and number of confirmed cases from April 2020 to June 2020 [64]. A similar report during inlet and outlet of preprocessing and disinfection in the sewage network from 19 to 24 February was also documented in China (Zhejiang). The samples showed 100% detection rate of SARS-CoV-2 RNA in the sewage water [15].
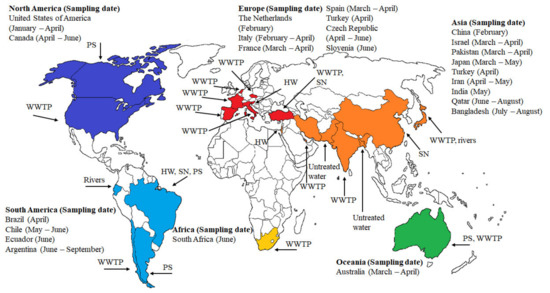
Figure 2.
Overview of the first detection of SARS-CoV-2 in wastewater around the world from January 2020 to February 2021. Sampling point: wastewater treatment plant (WWTP), suburban pumping station (PS), hospital wastewater (HW), and sewer network (SN).
The Netherlands (Haarlemmermeer) was the first European country where the SARS-CoV-2 viral RNA (RT-qPCR test) was detected in untreated wastewaters (from 17 to 24 February) [17]. Then, about 50% positive tests (6/12 samples) for the virus in WWTP were detected at Milan and Rome, Italy, from 3 February to 2 April [65]. Spain (Murcia) and France (Paris) analyzed samples in WWTPs from March to April. In Spain, the sampling points were the untreated wastewater, and the effluent after the secondary and tertiary processes. Viral RNA detections in influent, secondary effluent, and tertiary effluent were 83.33% (35/42 samples), 11.11% (2/18 samples), 0.0% (0/12 samples), respectively. The data revealed that community shedding of SARS-CoV-2 RNA via fecal discharge was occurring in many cities before the wastewaters were sampled for testing [14]. In a WWTP located in Paris, viral RNA detection was 100.0% (3/3 samples) [61]. Samples in WWTPs form Turkey (Istanbul, from 21 to 25 April) and Czech Republic (from April to June) indicated a 71.42% (5/7 samples) and 27.3% (9/33 samples) of the viral RNA detection, respectively [66,67].
Adding to the count, hospital wastewaters samples from Slovenia (Ljubljana, 1 to 15 June), Brazil (Rio de Janeiro, April), and Israel (Tel Aviv, 10 March to 21 April) were positive detected as 66.7% (10/15 samples), 41.6% (5/12 samples), and 38.4%, respectively [68,69,70]. The reports were good correlated with the high COVID-19 patients cases reported during the mentioned periods. Surprisingly, studies from Japan (Yamanashi Prefecture, from 17 March to 7 May) and Ecuador (Quito, June) revealed that the SARS-CoV-2 RNA was present in natural waters (rivers). Adding to the fact in Japan, the viral RNA detection was 20% (1/5 samples) when the COVID-19 cases peaked in the community [71], whereas in Ecuador, the viral RNA detection was documented in all the three samples [72]. Studies in South America (Chile and Argentina) reported the occurrence of the virus in untreated wastewater and suburban pumping stations. In Chile (Santiago, from March to June), the viral RNA was noticed in the influent and effluent sampling points. In Argentina (Buenos Aires, from 5 June to 7 September), all 11 samples collected from wastewater showed a viral load [73]. In this case, it is important to highlight that SARS-CoV-2 RNA was not detected during the first two months of analyses. However, in May and June, SARS-CoV-2 viral load was progressively increased in wastewater bodies [74].
The occurrence of viral SARS-CoV-2 RNA has been analyzed in the Gomoti river of India [75]. In Asia, India (Ahmedabad, from 8 to 27 May), Qatar (Doha, from 21 June to 30 August), and Iran (Tehran, Qom, and Anzali, from 4 April to 2 May), the presence of the virus in WWTPs was noticed. Viral RNA detection analyses showed 100% (2/2 samples), 100% (43/43 samples), and 66.66% (4/6 samples) in influent samples collected from the above countries, respectively [25,76,77]. The trend of the detection of SARS-CoV-2 RNA analyzed by RT-qPCR testing in wastewater in these countries was matched by the number of new daily COVID-19 positive cases. Bangladesh (Noakhali, from 10 July to 29 August) was entered into the club when viral RNA (75.0% (12/16 samples)) in sewage waste tanks, passage drains, and toilets was detected [13]. Finally, in Pakistan (Islamabad, from March to April), the viral RNA detection was 27% (21/78 samples) in the sewage network [78]. Therefore, almost all South Asian countries have well documented the SARS-CoV-2 viral load in wastewater.
Lastly, in Oceania (Queensland, Australia, from 27 March to 1 April), through Monte Carlo simulation, the number of COVID-19 cases estimated in the catchment agreed with clinical observations. Viral RNA detection was reported as 22.2% (2/9 samples) [79]; and in Africa (Western Cape, South Africa, June), the Viral RNA detection was found to be 100.0% (5/5 samples) in WWTPs. The results showed that the presence of SARS-CoV-2 data has corresponded with the COVID-19 cases [80].
By way of conclusion, the detection of SARS-CoV-2 RNA in wastewater indicates a strong association of the COVID-19 disease via WBE [81]. Proper hygienic knowledge and implications of COVID-19 guidelines are put forth by many world-class health organizations including the Centers for Disease Control and Prevention of US (CDC-US), WHO, etc. Just as the use of masks is mandatory to restrict the mode of infection transmitted by air, the treatment of wastewater before discharge into the tanks is also equally important. Strong policies and awareness should be adopted for the treatment of wastewater matrices of COVID-19 patients as well as to their corpses and materials for daily use to control such disease.
4. Molecular Detection of SARS-CoV-2 in Wastewater and Treatment Plant Sludge
Until today, many WBE surveillance studies have confirmed the detection of SARS-CoV-2 genetic material in wastewater and solids/sludge originated from WWTPs located in cities all over the world. In these studies, the followed protocols mainly encompassed the following steps: (i) the selection of an urbanized area drained by the sewer system, (ii) wastewater samples collection, (iii) biomarker selection (e.g., RNA, anti-inflammatories, IgM/IgG), (iv) sample pretreatment and instrumental analysis, (v) quantifying of SARS-CoV-2 RNA copies in wastewater (concentration methods) and data analysis, (vi) comparison between WBE and other data sources (validation), (vii) comparison of WBE results with other studies (other countries), and (viii) analysis of uncertainties and limitations. Some steps are introduced and discussed in detail in the following subsections.
4.1. Sampling
Wastewater samples were collected from different sites in WWTPs or manholes (MH). Some research groups performed their SARS-CoV-2 detection and quantification studies using composite samples, which were prepared by combining portions of multiple grab samples or using automatic sampling devices. Others preferred to use grab samples, as indicated in Table 1 and Table 2. The solid and sludge samples associated with wastewater treatment were taken as grab samples representing a snapshot in all studies.

Table 1.
The methods used for SARS-CoV-2 detection in wastewater and sewage sludge.

Table 2.
The protocols applied to wastewater and sewage sludge samples for detecting and quantifying SARS-CoV-2.
4.2. Storage
Storage and shipment of the sample are of great importance in terms of virus survival. Most samples were shipped on ice or with cold packs [101]. Some samples were stored in a refrigerator at 4 °C, while others were frozen in a freezer at a ranged temperature between −20 °C and −80 °C, after the concentration step, for further processing and analysis, as shown in Table 1 and Table 2. As of now, few studies have investigated the effect of the storage temperature on the SARS-CoV-2 RNA copies in wastewater [76,77]. The persistence of SARS-CoV-2 genetic material at 4, −20, and −75 °C was assessed using RT-qPCR assays targeting E-Sarbeco and N2 for a time interval of 29, 64, and 84 days [77]. Findings confirmed the stability of the SARS-CoV-2 RNA in cold storage, particularly at frozen conditions. Data of Baldovin et al. [76] also confirmed the presence of SARS-CoV-2 RNA in the refrigerated samples for a 24 h storage period.
4.3. Inactivation of SARS-CoV-2
A heating protocol, screening and elevating temperature from 56 to 92 °C, was performed using clinical samples collected from COVID-19 patients. This protocol was used to ensure laboratory personnel and the environment’s safety [102]. A lower inactivation performance (5 log10 reduction) was exhibited at 60 °C for 60 min. However, a drastic reduction in RNA copies detection was determined at 92 °C for 15 min, which yielded a total inactivation of SARS-CoV-2 virus. Heating protocol allowed the detection and quantification of RNA copies in an acceptable interval. Based on results, the author has recommended the inactivation protocols at 56 °C for 30 min and 60 °C for 60 min for SARS-CoV-2 virus [103]. A few studies initiated the inactivation of SARS-CoV-2 virus at 60 °C for 90 min [60,64,67].
4.4. Preconditioning
Most wastewater samples, before the concentration step to remove bacterial debris and large particles, use centrifugation and/or filtration methods. As shown in Table 1 and Table 2, the samples have been centrifuged at a varied speed from 1200× g to 5000× g for a time interval from 5 min to 45 min. Only one research performed the centrifugation method at an extreme speed of 24,000× g at 4 °C for 30 min [104]. Some molecular assays performed filtration process using a 0.22 μm pore size filter. Generally, the partition of SARS-CoV-2 virus to the solids in wastewater was ignored, except for some studies [49,85,99]. Kocamemi et al. [99] shook the sludge samples produced after the wastewater treatment. These samples were stirred at 100 rpm and 4 °C for 30 min to transfer SARS-CoV-2 viruses into the liquid phase before the pre-centrifugation at 7471× g for 30 min. Kitamura et al. [85] and Westhaus et al. [49] analyzed the solids obtained from the pre-centrifugation step separately. A loss of solids has already been mainly reported for enveloped viruses [105].
4.5. Concentration
Since the viral load is severely dilute in a huge volume of wastewater, the concentration method utilized for the recovery of SARS-CoV-2 genetic material plays an integral role to maximize the overall performance of molecular assays. Until now, several concentration methods such as (i) ultrafiltration (UF), (ii) precipitation with polyethylene glycol (PEG), (iii) electronegative or electropositive membrane filtration, (iv) the aluminum-driven flocculation, and (v) the skimmed-milk flocculation (SMF) have been applied. The Figure 3 shows the percentage distribution of mentioned techniques. It is worth emphasizing that all of them have been well established and documented for the non-enveloped viruses such as polioviruses, noroviruses, and adenoviruses [105,106,107]. Within the context of the WBE surveillance, UF and PEG are the most utilized techniques for the concentration of SARS-CoV-2 genetic material from wastewater [17,25,49,63,76,77,79,83,84,85,86,87,88,90,91,93,95,99,104,108]. These studies are compiled from the recent literature and presented in Table 1 and Table 2.
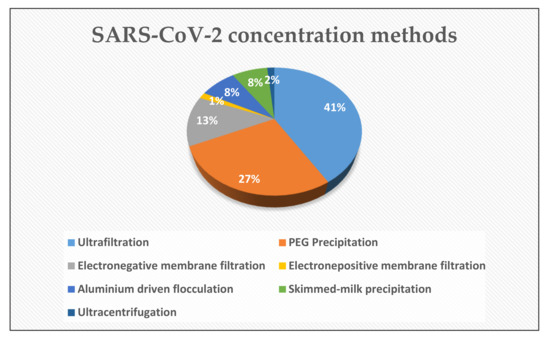
Figure 3.
Pie chart showing the SARS-CoV-2 concentration methods used within the context of the WBE surveillance.
SARS-CoV-2 genetic material has been commonly concentrated from wastewater via centrifugal filters with a nominal molecular weight limits (NMWL) ranging from 10 kDa [26,66,76,77,83,84,89,90], 30 kDa [88,93,97,98,101], 50 kDa [109], and 100 kDa [23,92] to 150 kDa [87,93] at centrifugation speeds varying in the range from 3000× g–4000× g. The protocols followed by these studies are summarized in Table 2. Additional to the centrifugal ultrafiltration (CeUF), hollow fiber ultrafilters (HFUF) composed of polysulfone PVP high flow pipettes were also tested [93,95]. Data obtained from CeUF and HFUF proved that UF is one of the most applicable and sensitive promising concentration techniques for the recovery of SARS-CoV-2 genetic material from wastewater and wastewater treatment plant sludge. However, the sensitivity of UF could be improved by its double application [84]. The application of either CeUF or PEG precipitation to HFUF concentrated is an unsuccessful and unnecessary secondary concentration method, according to Gerrity et al. [95].
The PEG precipitation method was firstly proposed by Albertsson and Frick [110]. This technique is based on the partition of some proteins in a liquid two-phase system, composed of dextran, methylcellulose, and water. This method has been tailored, well-documented, and successfully applied for the concentration of enteroviruses in groundwater, river water, tap water, and wastewater since 1960s [111,112,113]. WHO has also recommended a modified version incorporating dextran addition of PEG precipitation for poliovirus circulation’s environmental surveillance [114]. Due to its well-known success in the detection of poliovirus, many researchers have applied PEG precipitation with slightly different implementations or modifications to the composite, grab wastewater, or sewage sludge samples collected from different sources (manholes and WWTPs) located in the Republic of Turkey [66,99], India [25,75], Italy [58], USA [62,115], Argentina [73], Japan [97], Spain [14,59,90,93], Israel [60], Canada [64], the United Arab Emirates [86], and France [87] to recover SARS-CoV-2 genetic material as indicated in Table 2. Moreover, it has been successfully detected and quantified by its different implication like overnight standing [25,71,87,91,97,116]. Positive results were also achieved by the PEG-Dextran method [114] and its modification [100,117,118]. Some research groups employed another modified version of PEG precipitation proposed by Wu et al. [62,85,87,115,118]. However, this modified version yielded inconsistent data but lower inconsistency than other concentration techniques such electronegative membrane filtration (ENMF) [85] or UF [85,86].
Virus adsorption–elution (VIRADEL) utilizes electrostatically charged microporous materials as filtration media [105,111]. Based on their surface charges, VIRADEL can be classified as electropositive membrane filtration (EPMF) and ENMF. The RNA recovery efficiency of ENMF can be enhanced by adding MgCl2 or NaCl to support the attachment of virus particles onto a cellulose nitrate membrane filter with a pore size of 0.45 µm via salt-bridging. VIRADEL has been adopted as a standard method “US EPA Method 1623: Cryptosporidium and Giardia in Water by Filtration/IMS/FA” by US EPA [118]. In few studies, ENMF with/without support with MgCl2 was used and suggested as a suitable concentration technique for the WBE SARS-CoV-2 surveillance [13,71,83,119]. Only one study performed this technique to concentrate SARS-CoV-2 RNA from wastewater using an electropositive NanoCeram column filter [119].
Based on the viruses’ elution with glycine alkaline buffer prior to organic flocculation with skimmed-milk flocculent, SMF, is another concentration technique tested within the context of SARS-CoV-2 WBE surveillance. This technique exhibited superior performance to concentrate SARS-CoV-2 genetic material from wastewater [72,120]. Finally, aluminum-driven flocculation based on the capability of freshly formed Al(OH)3 in the adsorption of the viruses provided an efficient SARS-CoV-2 RNA recovery [14,96].
4.6. Extraction
Isolation of SARS-CoV-2 RNA from the concentrated sample without damage is another important step that may drastically affect the overall detection and quantification performance. Several techniques based on extraction with organic solvents, silica membrane-based spin column, and the use of paramagnetic particles [121,122] have been adopted and refined for this purpose. As seen in Table 1 and Table 2, all extraction approaches use acid guanidinium thiocyanate–phenol–chloroform (TRIzol-chloroform), commercial kits based on solvent extraction utilizing TRIzol-chloroform, lysis buffer/TRIzol LS, or silica membrane-based spin column except the paramagnetic particle’s method. The CDC-US has qualified and validated several commercial RNA extraction kits for SARS-CoV-2 which are mentioned on the webpage of CDC-US [123].
4.7. Detection and Quantification
Quantification of SARS-CoV-2 genetic material is the last and the utmost important step of a molecular assay. RT-PCR and RT-qPCR have been globally accepted as standard methods for quantifying RNA viruses [13,62,124,125,126]. These molecular tests offer high sensitivity and specificity. However, these analyses require quite complex sample handling in the laboratory, expertise personnel, and a long time of data processing and analysis (4–6 h). Likewise, Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and digital PCR (dPCR) have been evaluated [26]. Findings indicated that the reverse transcription droplet digital PCR (RT-ddPCR) is also an alternative technique for the detection and quantification the SARS-CoV-2 RNA in wastewater. The research groups have generally used RT-PCR and/or RT-qPCR (Table 1 and Table 2). A few research groups have carried out the molecular assays using RT-ddPCR [64,87,104,127]. Some of them also compared the performance of R-ddPCR with RT-qPCR [66,87,95]. Different sets of primers/probes targeting different parts of viral particles were used to amplify SARS-CoV-2 RNA extracted from wastewater/sludge samples. For instance, 2019-nCoV_N1(-F; -R; -P), 2019-nCoV_N2(-F; -R; -P), 2019-nCoV_N3(-F; -R; -P), E_Sarbeco(_F; _R; _P1), Cor-p-F2(+), and Cor-p-R1 (-) were used to target nucleocapsid (N) and envelope (E). In some studies, RdRp (ORF1a and ORF1b) and S genes were also targeted. A list of “Only 2019-Novel Coronavirus (2019-nCoV) Real-time RT-PCR Primers and Probes” has already been published by CDC-US (CDC, 2019) for respiratory virus surveillance and research purposes.
To validate the SARS-CoV-2 detection and quantification, molecular assays have been performed using domestic wastewater samples [14,49,91,100,128,129] as well as clinical samples collected from the COVID-19 patients [130,131,132]. Studies focused on designing these modern molecular techniques have been already continued [116,133,134] to develop a gold standard for the detection and qualification of SARS-CoV-2.
Additionally, the aforementioned molecular assays do not provide information on viability of SARS-CoV-2 being present in the water matrix [77]. To gather information on the virus viability, specific molecular techniques such as ethidium monoazide (EMA)-RT-qPCR, propidium monoazide (PMA)- RT-qPCR, or integrated cell culture-RT-qPCR, could be established.
In summary, modern molecular techniques produce accurate data when validation and standardization studies are completed. However, these techniques are not suitable for real-time surveillance due to their skilled operator requirement and expensive initial costs. Therefore, the development of easy-to-operate, cost-effectively, and fast response equipment for online monitoring is another urgent issue to be searched for the WBE SARS-CoV-2 surveillance.
5. Conclusions
The conclusions and future directions of the present review can be drawn as follows:
- Recent data proved that SARS-CoV-2 RNA can be detected in human feces from a few days to a week before the onset of symptoms. Therefore, the monitoring of SARS-CoV-2 genetic signals in wastewater samples seems to be a useful methodology for prediction of COVID-19 outbreaks. However, at the early stage of COVID-19 prevalence, its surveillance in sewage network may not be quite accurate due to the extremely low concentration of virus in wastewater.
- One liter of wastewater is like an ocean of information that can provide valuable data about the timing and prevalence of COVID-19 outbreaks in communities, especially in low-income countries with low clinical COVID-19 testing rates.
- Periodically collecting wastewater samples from sewer networks for the trace of the SARS-CoV-2 virus can be effective as a primary non-clinical warning tool for early detection.
- Considering that SARS-CoV-2 RNA has already been detected in the activated sludge, the dissociation of SARS-CoV-2 genetic material from solids/semisolids in wastewater arises urgent issue to be sought for accurate quantification. In this sense, a protocol tailored to SARS-CoV-2 is another crucial need.
- Although the sampling method is one of the most important steps of WBE SARS-CoV-2 surveillance, there is no real consensus on adequate and representative wastewater sampling technique. Then, further studies are required to find out a properly designed and well-defined sampling procedure.
- Among the concentration techniques, CeUF, double UF, and PEG precipitation seem to be promising concentration techniques for SARS-CoV-2 RNA enrichment from wastewater/sludge after re-partitioning.
- Findings prevailed that more standardization and quality control studies covering (i) selection of appropriate sample volume, (ii) improvement of preconditioning step including the significant contribution of the solid fraction of raw wastewater, (iii) spike controls, (iv) PCR inhibitor removal, and (v) selection of suitable extraction approach must be performed to maximize the SARS-CoV-2 recovery efficiency.
- An urgent issue to be faced is the development of molecular techniques providing information on the viability of the virus in the sewage system for worker safety.
- Recent data have revealed a correlation between genetic material in wastewater and the number of clinically reported positive cases confirming that WBE surveillance could be utilized as a sensitive tool to trace the circulation of SARS-CoV-2 virus in the population. Nevertheless, an accurate mathematical model considering several factors such as wastewater characteristics, climate, and sewage system structure, among others, should be developed.
- WBE for SARS-CoV-2 is a fast and effective surveillance system with the clearest potential to avoid and control the infectious disease outbreak.
Author Contributions
M.M.: conceptualization, visualization and project administration; M.M., R.A., B.P., I.K., M.H., M.A.S. and K.D.: data curation, methodology, software, validation, formal analysis, investigation, writing—original draft preparation; M.M. and M.M.E.: writing—review, and editing of the manuscript, as well as revision of the manuscript; M.M.E.: funding acquisition, resources, supervision; M.A.S., Z.F. and S.S.: writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.
Acknowledgments
The support of the Research Vice-Chancellor of Qazvin University of Medical Sciences (IR.QUMS.REC.1399.543) is wholeheartedly appreciated. Milad Mousazadeh thanks the Iran’s National Elites Foundation (INEF) for granting the Elite Award, No. 15/11529. Miguel A. Sandoval is grateful to Agencia Nacional de Investigación y Desarrollo (ANID-FONDECYT, Chile) for granting the postdoctoral scholarship, No. 3200274.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sims, N.; Kasprzyk-Hordern, B. Future perspectives of wastewater-based epidemiology: Monitoring infectious disease spread and resistance to the community level. Environ. Int. 2020, 139, 105689. [Google Scholar] [CrossRef] [PubMed]
- Daughton, C. The international imperative to rapidly and inexpensively monitor community-wide Covid-19 infection status and trends. Sci. Total. Environ. 2020, 726, 138149. [Google Scholar] [CrossRef] [PubMed]
- Daughton, C.G. Wastewater surveillance for population-wide Covid-19: The present and future. Sci. Total. Environ. 2020, 139631. [Google Scholar] [CrossRef] [PubMed]
- Andrés-Costa, M.J.; Andreu, V.; Pico, Y. Liquid chromatography–mass spectrometry as a tool for wastewater-based epidemiology: Assessing new psychoactive substances and other human biomarkers. TrAC Trends Anal. Chem. 2017, 94, 21–38. [Google Scholar] [CrossRef]
- Hellmér, M.; Paxéus, N.; Magnius, L.; Enache, L.; Arnholm, B.; Johansson, A.; Bergström, T.; Norder, H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl. Environ. Microbiol. 2014, 80, 6771–6781. [Google Scholar] [CrossRef] [Green Version]
- Sinclair, R.G.; Choi, C.Y.; Riley, M.R.; Gerba, C.P. Pathogen surveillance through monitoring of sewer systems. Adv. Appl. Microbiol. 2008, 65, 249. [Google Scholar]
- Weidhaas, J.; Aanderud, Z.T.; Roper, D.K.; VanDerslice, J.; Gaddis, E.B.; Ostermiller, J.; Hoffman, K.; Jamal, R.; Heck, P.; Zhang, Y. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci. Total. Environ. 2021, 775, 145790. [Google Scholar] [CrossRef]
- Hamouda, M.; Mustafa, F.; Maraqa, M.; Rizvi, T.; Hassan, A.A. Wastewater surveillance for SARS-CoV-2: Lessons learnt from recent studies to define future applications. Sci. Total. Environ. 2020, 143493. [Google Scholar] [CrossRef]
- Mao, K.; Zhang, H.; Yang, Z. Can a paper-based device trace COVID-19 sources with wastewater-based epidemiology? Environ. Sci. Technol. 2020, 54, 3733–3735. [Google Scholar] [CrossRef]
- Barcelo, D. An environmental and health perspective for COVID-19 outbreak: Meteorology and air quality influence, sewage epidemiology indicator, hospitals disinfection, drug therapies and recommendations. J. Environ. Chem. Eng. 2020, 8, 104006. [Google Scholar] [CrossRef]
- Miura, F.; Kitajima, M.; Omori, R. Duration of SARS-CoV-2 viral shedding in faeces as a parameter for wastewater-based epidemiology: Re-analysis of patient data using a shedding dynamics model. Sci. Total. Environ. 2021, 769, 144549. [Google Scholar] [CrossRef]
- Aguiar-Oliveira, M.d.L.; Campos, A.; Matos, A.R.; Rigotto, C.; Sotero-Martins, A.; Teixeira, P.F.; Siqueira, M.M. Wastewater-Based Epidemiology (WBE) and Viral Detection in Polluted Surface Water: A Valuable Tool for COVID-19 Surveillance—A Brief Review. Int. J. Environ. Res. Public Health 2020, 17, 9251. [Google Scholar] [CrossRef]
- Ahmed, F.; Islam, M.A.; Kumar, M.; Hossain, M.; Bhattacharya, P.; Islam, M.T.; Hossen, F.; Hossain, M.S.; Islam, M.S.; Uddin, M.M. First detection of SARS-CoV-2 genetic material in the vicinity of COVID-19 isolation Centre in Bangladesh: Variation along the sewer network. Sci. Total. Environ. 2021, 776, 145724. [Google Scholar] [CrossRef]
- Randazzo, W.; Truchado, P.; Cuevas-Ferrando, E.; Simón, P.; Allende, A.; Sánchez, G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020, 181, 115942. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Feng, H.; Zhang, S.; Ni, Z.; Ni, L.; Chen, Y.; Zhuo, L.; Zhong, Z.; Qu, T. SARS-CoV-2 RNA detection of hospital isolation wards hygiene monitoring during the Coronavirus Disease 2019 outbreak in a Chinese hospital. Int. J. Infect. Dis. 2020, 94, 103–106. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Water, Sanitation, Hygiene, and Waste Management for the COVID-19 Virus: Interim Guidance; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Medema, G.; Heijnen, L.; Elsinga, G.; Italiaander, R.; Brouwer, A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020, 7, 511–516. [Google Scholar] [CrossRef]
- Annalaura, C.; Ileana, F.; Dasheng, L.; Marco, V. Making waves: Coronavirus detection, presence and persistence in the water environment: State of the art and knowledge needs for public health. Water Res. 2020, 115907. [Google Scholar]
- Ihsanullah, I.; Bilal, M.; Naushad, M. Coronavirus 2 (SARS-CoV-2) in water environments: Current status, challenges and research opportunities. J. Water Process. Eng. 2021, 39, 101735. [Google Scholar] [CrossRef]
- Kitajima, M.; Ahmed, W.; Bibby, K.; Carducci, A.; Gerba, C.P.; Hamilton, K.A.; Haramoto, E.; Rose, J.B. SARS-CoV-2 in wastewater: State of the knowledge and research needs. Sci. Total. Environ. 2020, 739, 139076. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, G.; Bonadonna, L.; Lucentini, L.; Kenmoe, S.; Suffredini, E. Coronavirus in water environments: Occurrence, persistence and concentration methods-A scoping review. Water Res. 2020, 179, 115899. [Google Scholar] [CrossRef]
- Saawarn, B.; Hait, S. Occurrence, fate and removal of SARS-CoV-2 in wastewater: Current knowledge and future perspectives. J. Environ. Chem. Eng. 2020, 104870. [Google Scholar]
- Chen, C.; Kostakis, C.; Gerber, J.P.; Tscharke, B.J.; Irvine, R.J.; White, J.M. Towards finding a population biomarker for wastewater epidemiology studies. Sci. Total. Environ. 2014, 487, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, M.; Picó, Y. Wastewater-based epidemiology: Current status and future prospects. Curr. Opin. Environ. Sci. Health 2019, 9, 77–84. [Google Scholar] [CrossRef]
- Kumar, M.; Patel, A.K.; Shah, A.V.; Raval, J.; Rajpara, N.; Joshi, M.; Joshi, C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total. Environ. 2020, 746, 141326. [Google Scholar] [CrossRef]
- Barceló, D. Wastewater-Based Epidemiology to monitor COVID-19 outbreak: Present and future diagnostic methods to be in your radar. Case Stud. Chem. Environ. Eng. 2020, 2, 100042. [Google Scholar] [CrossRef]
- Collivignarelli, M.C.; Collivignarelli, C.; Miino, M.C.; Abbà, A.; Pedrazzani, R.; Bertanza, G. SARS-CoV-2 in sewer systems and connected facilities. Process. Saf. Environ. Prot. 2020, 143, 196–203. [Google Scholar] [CrossRef]
- Foladori, P.; Cutrupi, F.; Segata, N.; Manara, S.; Pinto, F.; Malpei, F.; Bruni, L.; La Rosa, G. SARS-CoV-2 from faeces to wastewater treatment: What do we know? A review. Sci. Total. Environ. 2020, 743, 140444. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Zhang, H.; Pan, Y.; Yang, Z. Biosensors for wastewater-based epidemiology for monitoring public health. Water Res. 2020, 116787. [Google Scholar] [CrossRef]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Guo, C.; Tang, L.; Hong, Z.; Zhou, J.; Dong, X.; Yin, H.; Xiao, Q.; Tang, Y.; Qu, X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020, 5, 434–435. [Google Scholar] [CrossRef]
- Zhang, T.; Cui, X.; Zhao, X.; Wang, J.; Zheng, J.; Zheng, G.; Guo, W.; Cai, C.; He, S.; Xu, Y. Detectable SARS-CoV-2 viral RNA in feces of three children during recovery period of COVID-19 pneumonia. J. Med Virol. 2020, 92, 909–914. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, S.; Shi, J.; Luby, S.P.; Jiang, G. Uncertainties in estimating SARS-CoV-2 prevalence by wastewater-based epidemiology. Chem. Eng. J. 2021, 415, 129039. [Google Scholar] [CrossRef]
- Carrillo-Reyes, J.; Barragán-Trinidad, M.; Buitrón, G. Surveillance of SARS-CoV-2 in sewage and wastewater treatment plants in Mexico. J. Water Process. Eng. 2021, 40, 101815. [Google Scholar] [CrossRef]
- Zhang, D.; Ling, H.; Huang, X.; Li, J.; Li, W.; Yi, C.; Zhang, T.; Jiang, Y.; He, Y.; Deng, S. Potential spreading risks and disinfection challenges of medical wastewater by the presence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of Fangcang Hospital. Sci. Total. Environ. 2020, 741, 140445. [Google Scholar] [CrossRef] [PubMed]
- EU. Commission Recommendation (EU) 2021/472, Official Journal of the European Union. 17 March 2021. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32021H0472&from=EN (accessed on 1 July 2021).
- Lotfi, M.; Hamblin, M.R.; Rezaei, N. COVID-19: Transmission, prevention, and potential therapeutic opportunities. Clin. Chim. Acta 2020, 508, 254–266. [Google Scholar] [CrossRef]
- Paital, B. Nurture to nature via COVID-19, a self-regenerating environmental strategy of environment in global context. Sci. Total. Environ. 2020, 729, 139088. [Google Scholar] [CrossRef] [PubMed]
- Paital, B.; Agrawal, P.K. Air pollution by NO2 and PM2.5 explains COVID-19 infection severity by overexpression of angiotensin-converting enzyme 2 in respiratory cells: A review. Environ. Chem. Lett. 2020, 18, 1–18. [Google Scholar] [CrossRef]
- Paital, B.; Das, K.; Parida, S.K. Inter nation social lockdown versus medical care against COVID-19, a mild environmental insight with special reference to India. Sci. Total. Environ. 2020, 728, 138914. [Google Scholar] [CrossRef]
- Zhang, N.; Gong, Y.; Meng, F.; Bi, Y.; Yang, P.; Wang, F. Virus shedding patterns in nasopharyngeal and fecal specimens of COVID-19 patients. MedRxiv 2020. [Google Scholar] [CrossRef]
- Hart, O.E.; Halden, R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: Feasibility, economy, opportunities and challenges. Sci. Total. Environ. 2020, 730, 138875. [Google Scholar] [CrossRef]
- Ahn, D.-G.; Shin, H.-J.; Kim, M.-H.; Lee, S.; Kim, H.-S.; Myoung, J.; Kim, B.-T.; Kim, S.-J. Current status of epidemiology, diagnosis, therapeutics, and vaccines for novel coronavirus disease 2019 (COVID-19). J. Microbiol. Biotechnol. 2020, 30, 313–324. [Google Scholar] [CrossRef]
- Das, K.; Paital, B. First week of social lockdown versus medical care against COVID-19-with special reference to India. Curr. Trend Biotechnol. Pharm. 2020, 14, 190–210. [Google Scholar] [CrossRef]
- Van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef]
- Rooney, C.M.; Moura, I.B.; Wilcox, M.H. Tracking COVID-19 via sewage. Curr. Opin. Gastroenterol. 2021, 37, 4–8. [Google Scholar] [CrossRef] [PubMed]
- Saguti, F.; Magnil, E.; Enache, L.; Churqui, M.P.; Johansson, A.; Lumley, D.; Davidsson, F.; Dotevall, L.; Mattsson, A.; Trybala, E. Surveillance of wastewater revealed peaks of SARS-CoV-2 preceding those of hospitalized patients with COVID-19. Water Res. 2021, 189, 116620. [Google Scholar] [CrossRef] [PubMed]
- Urban, R.C.; Nakada, L.Y.K. COVID-19 pandemic: Solid waste and environmental impacts in Brazil. Sci. Total. Environ. 2021, 755, 142471. [Google Scholar] [CrossRef]
- Westhaus, S.; Weber, F.-A.; Schiwy, S.; Linnemann, V.; Brinkmann, M.; Widera, M.; Greve, C.; Janke, A.; Hollert, H.; Wintgens, T. Detection of SARS-CoV-2 in raw and treated wastewater in Germany–suitability for COVID-19 surveillance and potential transmission risks. Sci. Total. Environ. 2021, 751, 141750. [Google Scholar] [CrossRef]
- Das, K.; Behera, T.R.; Paital, B. Heat Shock Protein, Corona Virus COVID-19. In Outbreak Challenges in Indian Migrant Pregnant and Lactating Mothers: Learnt Lesson Demands Hierarchical Strategy for Such Future Situation; Kaur, P., Asea, A., Eds.; Springer: New York, NY, USA, 2021. [Google Scholar]
- Paital, B.; Das, K.; Behera, T.R. Social lockdown and ecological intervention for the prevention of the community spread of COVID-19. Cancer Res. Stat. Treat. 2020, 3, 667. [Google Scholar]
- Al Huraimel, K.; Alhosani, M.; Kunhabdulla, S.; Stietiya, M.H. SARS-CoV-2 in the environment: Modes of transmission, early detection and potential role of pollutions. Sci. Total. Environ. 2020, 140946. [Google Scholar] [CrossRef]
- Nicastri, E.; D’Abramo, A.; Faggioni, G.; De Santis, R.; Mariano, A.; Lepore, L.; Molinari, F.; Petralito, G.; Fillo, S.; Munzi, D. Coronavirus disease (COVID-19) in a paucisymptomatic patient: Epidemiological and clinical challenge in settings with limited community transmission, Italy, February 2020. Eurosurveillance 2020, 25, 2000230. [Google Scholar] [CrossRef] [Green Version]
- Urban, R.C.; Nakada, L.Y.K. GIS-based spatial modelling of COVID-19 death incidence in São Paulo, Brazil. Environ. Urban. 2020, 33, 229–238. [Google Scholar] [CrossRef]
- Fuschi, C.; Pu, H.; Negri, M.; Colwell, R.; Chen, J. Wastewater-Based Epidemiology for Managing the COVID-19 Pandemic. ACS Est. Water 2021. [Google Scholar] [CrossRef]
- Rimoldi, S.G.; Stefani, F.; Gigantiello, A.; Polesello, S.; Comandatore, F.; Mileto, D.; Maresca, M.; Longobardi, C.; Mancon, A.; Romeri, F. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total. Environ. 2020, 744, 140911. [Google Scholar] [CrossRef]
- Döhla, M.; Wilbring, G.; Schulte, B.; Kümmerer, B.M.; Diegmann, C.; Sib, E.; Richter, E.; Haag, A.; Engelhart, S.; Eis-Hübinger, A.M. SARS-CoV-2 in environmental samples of quarantined households. medRxiv 2020. [Google Scholar] [CrossRef]
- La Rosa, G.; Mancini, P.; Ferraro, G.B.; Veneri, C.; Iaconelli, M.; Bonadonna, L.; Lucentini, L.; Suffredini, E. SARS-CoV-2 has been circulating in northern Italy since December 2019: Evidence from environmental monitoring. Sci. Total. Environ. 2021, 750, 141711. [Google Scholar] [CrossRef] [PubMed]
- Chavarria-Miró, G.; Anfruns-Estrada, E.; Guix, S.; Paraira, M.; Galofré, B.; Sáanchez, G.; Pintó, R.; Bosch, A. Sentinel surveillance of SARS-CoV-2 in wastewater anticipates the occurrence of COVID-19 cases. MedRxiv 2020. [Google Scholar] [CrossRef]
- Or, I.B.; Yaniv, K.; Shagan, M.; Ozer, E.; Erster, O.; Mendelson, E.; Mannasse, B.; Shirazi, R.; Kramarsky-Winter, E.; Nir, O. Regressing SARS-CoV-2 sewage measurements onto COVID-19 burden in the population: A proof-of-concept for quantitative environmental surveillance. MedRxiv 2020. [Google Scholar] [CrossRef]
- Wurtzer, S.; Marechal, V.; Mouchel, J.; Maday, Y.; Teyssou, R.; Richard, E.; Almayrac, J.; Moulin, L. Evaluation of lockdown effect on SARS-CoV-2 dynamics through viral genome quantification in waste water, Greater Paris, France, 5 March to 23 April 2020. Eurosurveillance 2020, 25, 2000776. [Google Scholar] [CrossRef]
- Wu, F.; Zhang, J.; Xiao, A.; Gu, X.; Lee, W.L.; Armas, F.; Kauffman, K.; Hanage, W.; Matus, M.; Ghaeli, N. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. Msystems 2020, 5, 9. [Google Scholar] [CrossRef] [PubMed]
- Sherchan, S.P.; Shahin, S.; Ward, L.M.; Tandukar, S.; Aw, T.G.; Schmitz, B.; Ahmed, W.; Kitajima, M. First detection of SARS-CoV-2 RNA in wastewater in North America: A study in Louisiana, USA. Sci. Total. Environ. 2020, 743, 140621. [Google Scholar] [CrossRef] [PubMed]
- D’Aoust, P.M.; Mercier, E.; Montpetit, D.; Jia, J.-J.; Alexandrov, I.; Neault, N.; Baig, A.T.; Mayne, J.; Zhang, X.; Alain, T. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021, 188, 116560. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, G.; Iaconelli, M.; Mancini, P.; Ferraro, G.B.; Veneri, C.; Bonadonna, L.; Lucentini, L.; Suffredini, E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total. Environ. 2020, 736, 139652. [Google Scholar] [CrossRef]
- Kocamemi, B.A.; Kurt, H.; Hacioglu, S.; Yarali, C.; Saatci, A.M.; Pakdemirli, B. First data-set on SARS-CoV-2 detection for Istanbul wastewaters in Turkey. MedRxiv 2020. [Google Scholar] [CrossRef]
- Mlejnkova, H.; Sovova, K.; Vasickova, P.; Ocenaskova, V.; Jasikova, L.; Juranova, E. Preliminary study of Sars-Cov-2 occurrence in wastewater in the Czech Republic. Int. J. Environ. Res. Public Health 2020, 17, 5508. [Google Scholar] [CrossRef]
- Gonçalves, J.; Koritnik, T.; Mioč, V.; Trkov, M.; Bolješič, M.; Berginc, N.; Prosenc, K.; Kotar, T.; Paragi, M. Detection of SARS-CoV-2 RNA in hospital wastewater from a low COVID-19 disease prevalence area. Sci. Total. 2021, 755, 143226. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, E.; Xagoraraki, I. A water-focused one-health approach for early detection and prevention of viral outbreaks. One Heal. 2019, 7, 100094. [Google Scholar] [CrossRef] [PubMed]
- Prado, T.; Fumian, T.M.; Mannarino, C.F.; Maranhão, A.G.; Siqueira, M.M.; Miagostovich, M.P. Preliminary results of SARS-CoV-2 detection in sewerage system in Niterói municipality, Rio de Janeiro, Brazil. Memórias Do Inst. Oswaldo Cruz 2020, 115. [Google Scholar] [CrossRef]
- Haramoto, E.; Malla, B.; Thakali, O.; Kitajima, M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total. Environ. 2020, 737, 140405. [Google Scholar] [CrossRef]
- Guerrero-Latorre, L.; Ballesteros, I.; Villacrés-Granda, I.; Granda, M.G.; Freire-Paspuel, B.; Ríos-Touma, B. SARS-CoV-2 in river water: Implications in low sanitation countries. Sci. Total. Environ. 2020, 743, 140832. [Google Scholar] [CrossRef]
- Iglesias, N.G.; Gebhard, L.G.; Carballeda, J.M.; Aiello, I.; Recalde, E.; Terny, G.; Ambrosolio, S.; L’Arco, G.; Konfino, J.; Brardinelli, J.I. SARS-CoV-2 surveillance in untreated wastewater: First detection in a low-resource community in Buenos Aires, Argentina. medRxiv 2020. [Google Scholar] [CrossRef]
- Ampuero, M.; Valenzuela, S.; Valiente-Echeverria, F.; Soto-Rifo, R.; Barriga, G.P.; Chnaiderman, J.; Rojas, C.; Guajardo-Leiva, S.; Diez, B.; Gaggero, A. SARS-CoV-2 Detection in Sewage in Santiago, Chile-Preliminary results. MedRxiv 2020. [Google Scholar] [CrossRef]
- Khan, R.; Saxena, A.; Shukla, S.; Sekar, S.; Goel, P. Effect of COVID-19 lockdown on the water quality index of River Gomti, India, with potential hazard of faecal-oral transmission. Environ. Sci. Pollut. Res. 2021, 28, 33021–33029. [Google Scholar] [CrossRef]
- Baldovin, T.; Amoruso, I.; Fonzo, M.; Buja, A.; Baldo, V.; Cocchio, S.; Bertoncello, C. SARS-CoV-2 RNA detection and persistence in wastewater samples: An experimental network for COVID-19 environmental surveillance in Padua, Veneto Region (NE Italy). Sci. Total. Environ. 2021, 760, 143329. [Google Scholar] [CrossRef] [PubMed]
- Hokajärvi, A.-M.; Rytkönen, A.; Tiwari, A.; Kauppinen, A.; Oikarinen, S.; Lehto, K.-M.; Kankaanpää, A.; Gunnar, T.; Al-Hello, H.; Blomqvist, S. The detection and stability of the SARS-CoV-2 RNA biomarkers in wastewater influent in Helsinki, Finland. Sci. Total. Environ. 2021, 770, 145274. [Google Scholar] [CrossRef] [PubMed]
- Sharif, S.; Ikram, A.; Khurshid, A.; Salman, M.; Mehmood, N.; Arshad, Y.; Ahmad, J.; Angez, M.; Alam, M.M.; Rehman, L. Detection of SARS-Coronavirus-2 in wastewater, using the existing environmental surveillance network: An epidemiological gateway to an early warning for COVID-19 in communities. MedRxiv 2020. [Google Scholar] [CrossRef]
- Ahmed, W.; Tscharke, B.; Bertsch, P.M.; Bibby, K.; Bivins, A.; Choi, P.; Clarke, L.; Dwyer, J.; Edson, J.; Nguyen, T.M.H. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: A temporal case study. Sci. Total. Environ. 2021, 761, 144216. [Google Scholar] [CrossRef]
- Johnson, R.; Muller, C.; Ghoor, S.; Louw, J.; Archer, E.; Surujlal-Naicker, S.; Berkowitz, N.; Volschenk, M.; Bröcker, L.; Wolfaardt, G. Qualitative and quantitative detection of SARS-CoV-2 RNA in untreated wastewater in Western Cape Province, South Africa. South Afr. Med. J. 2021. [Google Scholar] [CrossRef]
- Mousazadeh, M.; Paital, B.; Naghdali, Z.; Mortezania, Z.; Hashemi, M.; Niaragh, E.K.; Aghababaei, M.; Ghorbankhani, M.; Lichtfouse, E.; Sillanpää, M. Positive environmental effects of the coronavirus 2020 episode: A review. Environ. Dev. Sustain. 2021, 23, 12738–12760. [Google Scholar] [CrossRef]
- Ahmed, W.; Angel, N.; Edson, J.; Bibby, K.; Bivins, A.; O’Brien, J.W.; Choi, P.M.; Kitajima, M.; Simpson, S.L.; Li, J. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total. Environ. 2020, 728, 138764. [Google Scholar] [CrossRef]
- Ahmed, W.; Bertsch, P.M.; Bivins, A.; Bibby, K.; Farkas, K.; Gathercole, A.; Haramoto, E.; Gyawali, P.; Korajkic, A.; McMinn, B.R. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total. Environ. 2020, 739, 139960. [Google Scholar] [CrossRef]
- Jafferali, M.H.; Khatami, K.; Atasoy, M.; Birgersson, M.; Williams, C.; Cetecioglu, Z. Benchmarking virus concentration methods for quantification of SARS-CoV-2 in raw wastewater. Sci. Total. Environ. 2021, 755, 142939. [Google Scholar] [CrossRef]
- Kitamura, K.; Sadamasu, K.; Muramatsu, M.; Yoshida, H. Efficient detection of SARS-CoV-2 RNA in the solid fraction of wastewater. Sci. Total. Environ. 2021, 763, 144587. [Google Scholar] [CrossRef]
- Hasan, S.W.; Ibrahim, Y.; Daou, M.; Kannout, H.; Jan, N.; Lopes, A.; Alsafar, H.; Yousef, A.F. Detection and quantification of SARS-CoV-2 RNA in wastewater and treated effluents: Surveillance of COVID-19 epidemic in the United Arab Emirates. Sci. Total. Environ. 2021, 764, 142929. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, I.; Challant, J.; Jeulin, H.; Hartard, C.; Mathieu, L.; Lopez, S.; Obépine, S.I.G.; Schvoerer, E.; Courtois, S.; Gantzer, C. Epidemiological surveillance of SARS-CoV-2 by genome quantification in wastewater applied to a city in the northeast of France: Comparison of ultrafiltration-and protein precipitation-based methods. Int. J. Hyg. Environ. Health 2021, 233, 113692. [Google Scholar] [CrossRef] [PubMed]
- Torii, S.; Furumai, H.; Katayama, H. Applicability of polyethylene glycol precipitation followed by acid guanidinium thiocyanate-phenol-chloroform extraction for the detection of SARS-CoV-2 RNA from municipal wastewater. Sci. Total. Environ. 2021, 756, 143067. [Google Scholar] [CrossRef]
- Kumar, M.; Kuroda, K.; Patel, A.K.; Patel, N.; Bhattacharya, P.; Joshi, M.; Joshi, C.G. Decay of SARS-CoV-2 RNA along the wastewater treatment outfitted with Upflow Anaerobic Sludge Blanket (UASB) system evaluated through two sample concentration techniques. Sci. Total. Environ. 2021, 754, 142329. [Google Scholar] [CrossRef] [PubMed]
- Balboa, S.; Mauricio-Iglesias, M.; Rodríguez, S.; Martínez-Lamas, L.; Vasallo, F.J.; Regueiro, B.; Lema, J.M. The fate of SARS-CoV-2 in wastewater treatment plants points out the sludge line as a suitable spot for incidence monitoring. medRxiv 2020. [Google Scholar] [CrossRef]
- Martin, J.; Klapsa, D.; Wilton, T.; Zambon, M.; Bentley, E.; Bujaki, E.; Fritzsche, M.; Mate, R.; Majumdar, M. Tracking SARS-CoV-2 in sewage: Evidence of changes in virus variant predominance during COVID-19 pandemic. Viruses 2020, 12, 1144. [Google Scholar] [CrossRef] [PubMed]
- Jahn, K.; Dreifuss, D.; Topolsky, I.; Kull, A.; Ganesanandamoorthy, P.; Fernandez-Cassi, X.; Bänziger, C.; Stachler, E.; Fuhrmann, L.; Jablonski, K.P. Detection of SARS-CoV-2 variants in Switzerland by genomic analysis of wastewater samples. medRxiv 2021. [Google Scholar] [CrossRef]
- Forés, E.; Bofill-Mas, S.; Itarte, M.; Martínez-Puchol, S.; Hundesa, A.; Calvo, M.; Borrego, C.M.; Corominas, L.; Girones, R.; Rusiñol, M. Evaluation of two rapid ultrafiltration-based methods for SARS-CoV-2 concentration from wastewater. Sci. Total. Environ. 2021, 768, 144786. [Google Scholar] [CrossRef]
- Thurston-Enriquez, J.A.; Haas, C.N.; Jacangelo, J.; Riley, K.; Gerba, C.P. Inactivation of feline calicivirus and adenovirus type 40 by UV radiation. Appl. Environ. Microbiol. 2003, 69, 577–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerrity, D.; Papp, K.; Stoker, M.; Sims, A.; Frehner, W. Early-pandemic wastewater surveillance of SARS-CoV-2 in Southern Nevada: Methodology, occurrence, and incidence/prevalence considerations. Water Res. 2021, 10, 100086. [Google Scholar]
- Barril, P.A.; Pianciola, L.A.; Mazzeo, M.; Ousset, M.J.; Jaureguiberry, M.V.; Alessandrello, M.; Sánchez, G.; Oteiza, J.M. Evaluation of viral concentration methods for SARS-CoV-2 recovery from wastewaters. Sci. Total. Environ. 2021, 756, 144105. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, H.; Yoshida, H.; Usuku, S. Environmental surveillance can dynamically track ecological changes in enteroviruses. Appl. Environ. Microbiol. 2019, 85, e01604–e01619. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-step method of rna isolation by acid guanidinium thiocyanate phenol chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Kocamemi, B.; Kurt, H.; Sait, A.; Sarac, F.; Saatci, A.; Pakdemirli, B. SARS-CoV-2 Detection in Istanbul Wastewater Treatment Plant Sludges. medRxiv 2020. [Google Scholar] [CrossRef]
- Hata, A.; Hara-Yamamura, H.; Meuchi, Y.; Imai, S.; Honda, R. Detection of SARS-CoV-2 in wastewater in Japan during a COVID-19 outbreak. Sci. Total. Environ. 2021, 758, 143578. [Google Scholar] [CrossRef]
- Calgua, B.; Rodriguez-Manzano, J.; Hundesa, A.; Sunen, E.; Calvo, M.; Bofill-Mas, S.; Girones, R. New methods for the concentration of viruses from urban sewage using quantitative PCR. J. Virol. Methods 2013, 187, 215–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, N.A.; Tharpe, C.; Meschke, J.S.; Ferguson, C.M. Survey of rapid development of environmental surveillance methods for SARS-CoV-2 detection in wastewater. Sci. Total. Environ. 2021, 769, 144852. [Google Scholar] [CrossRef] [PubMed]
- Pastorino, B.; Touret, F.; Gilles, M.; de Lamballerie, X.; Charrel, R.N. Prolonged infectivity of SARS-CoV-2 in fomites. Emerg. Infect. Dis. 2020, 26, 2256. [Google Scholar] [CrossRef]
- Graham, K.E.; Loeb, S.K.; Wolfe, M.K.; Catoe, D.; Sinnott-Armstrong, N.; Kim, S.; Yamahara, K.M.; Sassoubre, L.M.; Mendoza Grijalva, L.M.; Roldan-Hernandez, L. SARS-CoV-2 RNA in Wastewater Settled Solids Is Associated with COVID-19 Cases in a Large Urban Sewershed. Environ. Sci. Technol. 2020, 55, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Ikner, L.A.; Gerba, C.P.; Bright, K.R. Concentration and recovery of viruses from water: A comprehensive review. Food Environ. Virol. 2012, 4, 41–67. [Google Scholar] [CrossRef]
- Farkas, K.; Hillary, L.S.; Malham, S.K.; McDonald, J.E.; Jones, D.L. Wastewater and public health: The potential of wastewater surveillance for monitoring COVID-19. Curr. Opin. Environ. Sci. Health 2020, 17, 14–20. [Google Scholar] [CrossRef]
- Katayama, H.; Shimasaki, A.; Ohgaki, S. Development of a virus concentration method and its application to detection of enterovirus and Norwalk virus from coastal seawater. Appl. Environ. Microbiol. 2002, 68, 1033–1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petala, M.; Dafou, D.; Kostoglou, M.; Karapantsios, T.; Kanata, E.; Chatziefstathiou, A.; Sakaveli, F.; Kotoulas, K.; Arsenakis, M.; Roilides, E. A physicochemical model for rationalizing SARS-CoV-2 concentration in sewage. Case study: The city of Thessaloniki in Greece. Sci. Total. Environ. 2021, 755, 142855. [Google Scholar] [CrossRef]
- Trottier, J.; Darques, R.; Mouheb, N.A.; Partiot, E.; Bakhache, W.; Deffieu, M.S.; Gaudin, R. Post-lockdown detection of SARS-CoV-2 RNA in the wastewater of Montpellier, France. One Health 2020, 10, 100157. [Google Scholar] [CrossRef] [PubMed]
- Albertsson, P.-å.; Frick, G. Partition of virus particles in a liquid two-phase system. Biochim. Et Biophys. Acta 1960, 37, 230–237. [Google Scholar] [CrossRef]
- Lu, D.; Huang, Z.; Luo, J.; Zhang, X.; Sha, S. Primary concentration–The critical step in implementing the wastewater based epidemiology for the COVID-19 pandemic: A mini-review. Sci. Total. Environ. 2020, 747, 141245. [Google Scholar] [CrossRef]
- Shuval, H.I.; Cymbalista, B.F.S.; Goldblum, N. The phase-separation method for the concentration and detection of viruses in water. Water Res. 1969, 3, 225–240. [Google Scholar] [CrossRef]
- Wallis, C.; Grinstein, S.; Melnick, J.L.; Fields, J.E. Concentration of viruses from sewage and excreta on insoluble polyelectrolytes. Appl. Microbiol. 1969, 18, 1007–1014. [Google Scholar] [CrossRef]
- Word Health Organization. Guidelines for Environmental Surveillance of Poliovirus Circulation; WHO: Geneva, Swithzerland, 2003. [Google Scholar]
- Rothman, J.A.; Loveless, T.B.; Griffith, M.L.; Steele, J.A.; Griffith, J.F.; Whiteson, K.L. Metagenomics of Wastewater Influent from Southern California Wastewater Treatment Facilities in the Era of COVID-19. Microbiol. Resour. Announc. 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, T.H.; Johns, M.W. Improved detection of F-specific RNA coliphages in fecal material by extraction and polyethylene glycol precipitation. Appl. Environ. Microbiol. 2009, 75, 6142–6146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- United States Environmental Protection Agency. Method 1623: Cryptosporidium and Giardia in Water by Filtration/IMS/FAEPA 821-R-01–025; EPA US Environmental Protection Agency: Washington, DC, USA, 2005.
- Miyani, B.; Fonoll, X.; Norton, J.; Mehrotra, A.; Xagoraraki, I. SARS-CoV-2 in Detroit wastewater. J. Environ. Eng. 2020, 146, 06020004. [Google Scholar] [CrossRef]
- Philo, S.E.; Keim, E.K.; Swanstrom, R.; Ong, A.Q.; Burnor, E.A.; Kossik, A.L.; Harrison, J.C.; Demeke, B.A.; Zhou, N.A.; Beck, N.K. A comparison of SARS-CoV-2 wastewater concentration methods for environmental surveillance. Sci. Total. Environ. 2021, 760, 144215. [Google Scholar] [CrossRef]
- Michael-Kordatou, I.; Karaolia, P.; Fatta-Kassinos, D. Sewage analysis as a tool for the COVID-19 pandemic response and management: The urgent need for optimised protocols for SARS-CoV-2 detection and quantification. J. Environ. Chem. Eng. 2020, 8, 104306. [Google Scholar] [CrossRef]
- Tavares, L.; Alves, P.M.; Ferreira, R.B.; Santos, C.N. Comparison of different methods for DNA-free RNA isolation from SK-N-MC neuroblastoma. BMC Res. Notes 2011, 4, 1–5. [Google Scholar] [CrossRef]
- CDC. 2019-nCoV Real-Time RT-PCR Diagnostic Panel. Instructions for Use; (Effective 4 February 2020); CDC: Atlanta, GA, USA, 2020. [Google Scholar]
- Bhatt, A.; Arora, P.; Prajapati, S.K. Occurrence, fates and potential treatment approaches for removal of viruses from wastewater: A review with emphasis on SARS-CoV-2. J. Environ. Chem. Eng. 2020, 104429. [Google Scholar] [CrossRef]
- Lodder, W.; de Roda Husman, A.M. SARS-CoV-2 in wastewater: Potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020, 5, 533–534. [Google Scholar] [CrossRef]
- Mallapaty, S. How sewage could reveal true scale of coronavirus outbreak. Nature 2020, 580, 176–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, R.; Curtis, K.; Bivins, A.; Bibby, K.; Weir, M.H.; Yetka, K.; Thompson, H.; Keeling, D.; Mitchell, J.; Gonzalez, D. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020, 186, 116296. [Google Scholar] [CrossRef] [PubMed]
- Langone, M.; Petta, L.; Cellamare, C.; Ferraris, M.; Guzzinati, R.; Mattioli, D.; Sabia, G. SARS-CoV-2 in water services: Presence and impacts. Environ. Pollut. 2020, 268, 115806. [Google Scholar] [CrossRef] [PubMed]
- Nemudryi, A.; Nemudraia, A.; Wiegand, T.; Surya, K.; Buyukyoruk, M.; Cicha, C.; Vanderwood, K.K.; Wilkinson, R.; Wiedenheft, B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020, 1, 100098. [Google Scholar] [CrossRef]
- Jung, Y.; Park, G.-S.; Moon, J.H.; Ku, K.; Beak, S.-H.; Lee, C.-S.; Kim, S.; Park, E.C.; Park, D.; Lee, J.-H. Comparative analysis of primer–probe sets for RT-qPCR of COVID-19 causative virus (SARS-CoV-2). ACS Infect. Dis. 2020, 6, 2513–2523. [Google Scholar] [CrossRef]
- Nalla, A.K.; Casto, A.M.; Huang, M.-L.W.; Perchetti, G.A.; Sampoleo, R.; Shrestha, L.; Wei, Y.; Zhu, H.; Jerome, K.R.; Greninger, A.L. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J. Clin. Microbiol. 2020, 58, e00557-20. [Google Scholar] [CrossRef] [Green Version]
- Vogels, C.B.; Brito, A.F.; Wyllie, A.L.; Fauver, J.R.; Ott, I.M.; Kalinich, C.C.; Petrone, M.E.; Casanovas-Massana, A.; Muenker, M.C.; Moore, A.J. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat. Microbiol. 2020, 5, 1299–1305. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.K.; Pan, Y.; Cheng, S.M.; Hui, K.P.; Krishnan, P.; Liu, Y.; Ng, D.Y.; Wan, C.K.; Yang, P.; Wang, Q. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020, 66, 549–555. [Google Scholar] [CrossRef] [Green Version]
- Shirato, K.; Nao, N.; Katano, H.; Takayama, I.; Saito, S.; Kato, F.; Katoh, H.; Sakata, M.; Nakatsu, Y.; Mori, Y. Development of genetic diagnostic methods for novel coronavirus 2019 (nCoV-2019) in Japan. Jpn. J. Infect. Dis. 2020, 73. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).