Abstract
Anoplocephala perfoliata is a neglected gastro-intestinal tapeworm, commonly infecting horses worldwide. Molecular investigation of A. perfoliata is hampered by a lack of tools to better understand the host–parasite interface. This interface is likely influenced by parasite derived immune modulators released in the secretome as free proteins or components of extracellular vesicles (EVs). Therefore, adult RNA was sequenced and de novo assembled to generate the first A. perfoliata transcriptome. In addition, excretory secretory products (ESP) from adult A. perfoliata were collected and EVs isolated using size exclusion chromatography, prior to proteomic analysis of the EVs, the EV surface and EV depleted ESP. Transcriptome analysis revealed 454 sequences homologous to known helminth immune modulators including two novel Sigma class GSTs, five α-HSP90s, and three α-enolases with isoforms of all three observed within the proteomic analysis of the secretome. Furthermore, secretome proteomics identified common helminth proteins across each sample with known EV markers, such as annexins and tetraspanins, observed in EV fractions. Importantly, 49 of the 454 putative immune modulators were identified across the secretome proteomics contained within and on the surface of EVs in addition to those identified in free ESP. This work provides the molecular tools for A. perfoliata to reveal key players in the host–parasite interaction within the horse host.
1. Introduction
The gastro-intestinal tapeworm, Anoplocephala perfoliata, is one of the most prevalent tapeworm species that infects horses worldwide, with prevalence estimated between 15.8–44% of horses, yet it remains a neglected parasite with respect to management as low infection rates are often asymptomatic [1,2,3,4,5,6]. However, high burdens of A. perfoliata are linked to abdominal disturbance or pain, including spasmodic colic, ileal impaction [7], ileocaecal or caeco–caecal intussusception [8,9] and ileal or caecal rupture [10,11] due to the accumulation of adults at the ileocaecal region that attach to the caecal epithelium [2,12,13,14,15]. In such high-level infections, a localised caecal mucosal inflammatory response is activated at the site of the tapeworm attachment [2,16,17,18]. Severe caecal tissue damage and dysfunction likely predisposes horses to colic, which is often fatal during later stages without treatment [2,14]. Such documented evidence demonstrates the importance of further understanding the pathology and biology of A. perfoliata for future diagnostics and control options.
At present, there is limited mechanistic evidence of how A. perfoliata interacts with the horse intestinal environment. Therefore, an increased understanding of the underpinning molecular biology of A. perfoliata is required to delineate parasite–host interactions. Polyomic technologies, such as transcriptomics and proteomics, have generated new omic database resources to support future control for several other important helminths [19,20,21,22,23]. To date, molecular analysis of A. perfoliata has focussed on the mitochondrial genome sequence in order to simply assess the phylogenetic relationship with the closely related Anoplocephala magna [22,24]. However, at present, there is no reference genome or supporting transcriptome profiles to support discovery biology in A. perfoliata, which precludes in depth proteomic profiling. Furthermore, comprehensive nucleotide support would provide unique biological information for A. perfoliata, to facilitate the identification of genes likely involved in the pathogenesis of tapeworm infections and tapeworm-host interactions.
Molecules secreted by helminth parasites into the host environment during the course of infection are termed the excretory/secretory products (ESP) or the secretome and contain a variety of soluble proteins, glycoproteins, carbohydrates, lipids, and metabolites many of which are known to have an important role in helminth-mediated immunomodulation [16,25,26,27,28,29,30,31,32,33,34]. Investigation of the ESP from tapeworms, such as the protoscoleces of Echinococcus granulosus, have demonstrated regulation of immune cell differentiation, such as B10, B17, and Th17 cells in infected mice, accompanied by a downregulation of the inflammatory response [32]. Furthermore, A. perfoliata has been suggested to downregulate T-cell responses in the horse in live infections, which was partly attributed to ESP, which inhibited growth and induced apoptosis of Jurkat cells (human T-cell line) in vitro [16]. However, at present the active key component(s) in A. perfoliata ESP driving these potential host immunomodulatory mechanisms are yet to be determined [16].
Extracellular vesicles (EVs) are lipid membrane-bound structures released from helminths as part of the ESP. Parasite EVs are released into the host extracellular environment and are likely candidates for intercellular communication and immunomodulation [35,36,37]. To date, EVs released from tapeworms have been identified in adult Hymenolepis diminuta [38] and Taenia asiatica [39], and from metacestode stages of Taenia pisiformis [40], Echinococcus granulosus [41,42,43], Taenia crassiceps, Mesocestoides corti, and Echinococcus multilocularis [44]. However, EVs from adult equine tapeworms, including A. perfoliata, have not yet been identified.
Key secretory proteins involved in immune modulation, host interaction and parasite survival have been identified as part of the ESP as free proteins or as components of EVs for a number of helminths. Annexins, actins, cathepsin proteases, heat shock proteins (HSPs), helminth defense molecules (HDMs), glutathione transferases (GSTs), and fatty-acid binding proteins (FABP) are among a number of such proteins that have been identified from Fasciola hepatica [27,30,45,46,47,48], Calicophoron daubneyi (Rumen Fluke) [21,49], Schistosoma japonicum [50], and Schistosoma mansoni [51] representing well characterised platyhelminths. Thus, in depth characterisation of helminth secretomes has the potential to uncover host parasite interaction mechanisms.
Therefore, understanding of host–parasite interactions and subsequent pathogenesis are hampered in research neglected parasites such as A. perfoliata by a lack of fundamental molecular resources. Thus, this work generated the first transcriptome for adult A. perfoliata to support proteomic investigations. In addition, deploying this novel transcriptome revealed the A. perfoliata secretome, including EVs, EV surface expressed proteins, and EV depleted ESP via GeLC and Gel free proteomic approaches. This first comprehensive coverage of the A. perfoliata secretome has revealed the likely key players in the host–parasite interaction within the horse host.
2. Results
2.1. De Novo Assembly of the A. perfoliata Transcriptome
RNA sequencing across all six samples resulted in a total of 109,267,236 paired-end reads. The trimmed reads underwent de novo assembly individually and a total of 199,943 transcripts sequences were obtained. A total of 2353 sequences were determined to be host (E. caballus) when the peptide candidates were blasted against the E. caballus genome and related flatworm genome (Hymenolepis microstoma). Following removal of homologous sequences and host contamination, a total of 74,607 remained, with the total assembled contigs length of 56,913,324 bp and mean ± SD contig length of 763 ± 762 bp. Summary statistics for the A. perfoliata de novo transcriptome assembly are presented in Table 1. The assembled transcriptome was converted to protein sequences, using best predicted coding regions, resulting in 34,341 protein sequences (Table 1).

Table 1.
Summary statistics of the trimmed Illumina sequencing, de novo transcriptome assembly and TransDecoder protein sequences of adult A. perfoliata from naturally infected horses (n = 6).
2.2. Transcriptome Functional Annotation and Gene Ontology Terms Analysis
A total of 19,445 (56.6%) sequences were successfully annotated to the top three species hits to related tapeworm species including Hymenolepis diminuta, Echinococcus granulosus, and Hymenolepis microstoma. However, Omicsbox functional annotation revealed that 3244 (9.4%) protein sequences could not be annotated. A further 6199 (18.1%) sequences were annotated with Blast Hits and 5453 (15.9%) with GO term mapping. The majority of GO terms were classified according to the three main GO categories; biological processes (33.5%), molecular functions (39.7%), and cellular components (26.8%). The most frequent GO terms identified by level 3 are demonstrated in Supplementary Figure S1. Protein descriptions were assigned to 27,950 (81.39%) of the transcript sequences, whereas 6391 (18.61%) sequences were unnamed protein products or uncharacterised proteins; 1738 (5.06%) were of unknown function; 3244 (9.45%) did not have information provided and 1409 (4.10%) were hypothetical proteins/transcripts.
2.3. Transcripts Expression of A. perfoliata Transcriptome
The mean ± SD TPM was 12.4 ± 222, with the top 50 most abundant transcripts summarised in Table 2. Known genes of interest in other platyhelminths were noted amongst the top 50 abundance transcripts, such as, Dynein light chain (IPR037177) superfamily, EF-hand domain pair (IPR011992), Armadillo-like helical (IPR011989), Armadillo-type fold (IPR016024), and profilin superfamily (IPR036140). Protein families and domains of highly expressed genes related to these superfamilies were also found, similar to other platyhelminths, such as Dynein light chain, type 1/2 family (IPR001372), profilin family (IPR005455), EF-hand domain (IPR002048), and domain of unknown function, DUF5734 (IPR043792). Thus, demonstrating the likely validity of the transcript dataset for A. perfoliata.

Table 2.
The top 50 most abundant transcripts in A. perfoliata calculated from Salmon and expressed as a TPM value. The description of each transcript is demonstrated from Omicsbox functional annotation.
2.4. Bioinformatics of Potential Immune Modulators
An initial A. perfoliata transcriptome analysis by tBLASTn and PFAM was performed using 73 known immune modulators from helminths. This analysis demonstrated significant hits for 43 of the 73 bait immune modulators leading to a total of 454 unique contigs which were identified as potential immune modulator homologs (cutoff 1 × 10−15) (Supplementary Table S1). The top five hits, or less is fewer than five hits returned, for each of the 43 bait proteins returning hits were confirmed as homologs using Pfam domain searches with over 70% domain conservation and provided a total of 83 unique contigs confirmed as potential immune modulators.
2.4.1. Characterisation of Novel A. perfoliata Sigma Class GSTs
A total of 4 A. perfoliata sequences were identified by tBLASTn as potential Sigma class GST protein sequences, homologous to protein sequences of recognised Sigma class GSTs from mammals and helminths (cutoff 1 × 10−15). Subsequently, InterProScan followed by manual BLASTp against the NCBI (nr) database confirmed that three of these sequences were potential Sigma class GST A. perfoliata sequences (GST superfamily IPR040079) representing two distinct enzymes (ApGST-S1, ApGST-S2.1, and ApGST-S2.2 where 2.1 and 2.2 share 100% amino-acid sequence identity).
The secondary characteristic structure of multiple aligned A. perfoliata Sigma GST homolog sequences were predicted, with three β-strands and nine α-helices demonstrating the consistency of the secondary characteristic structure between ApGST-S1, ApGST-S2 (2.1 and 2.2), and recognised Sigma class GST sequences (Supplementary Figure S2). Sigma class GST homologs were investigated based on the GSH-binding sites in the N-terminal domain such as the catalytic tyrosine residue at the end of the first β-strand (Tyr8), Phe9, Arg14, Trp39, Lys43, Pro52, and Ser64) and the substrate binding sites in the C-terminal domain [52,53,54,55]. Only ApGST-S2.1 and 2.2 (212 amino acids) contained the highly conserved tyrosine residue (Tyr8) at the end of the first β-strand and also demonstrated a high homology to other GSH-binding sites (Supplementary Figure S2).
A maximum likelihood (ML) phylogenetic tree generated from the 3 A. perfoliata and 16 recognised Sigma class GST sequences demonstrated that ApGST-S1, ApGST-S2.1, and ApGST-S2.2 were clustered in a Cestode clade, which suggested that all three should be included in the Sigma class of the GST superfamily (Figure 1). ApGST-S1 clustered closest to H. microstoma Sigma like GST (accession CDS25704) with a bootstrap value of 52%, and also clustered in the Sigma group of Echinococcus multicularis Sigma like GST (accession CDS39356) and Echinococcus granulosus isozyme (accession EUB60467) with a high bootstrap value (100%) (Figure 1).
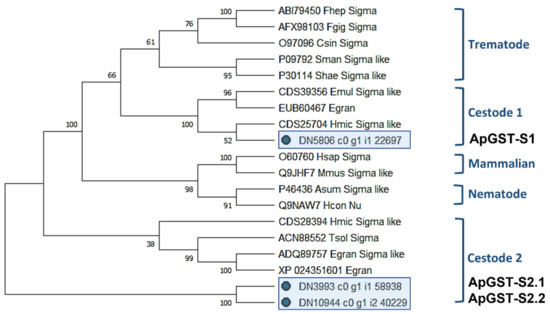
Figure 1.
Maximum likelihood (ML) tree with JTT matrix-based model inferred from A. perfoliata Sigma class GST amino acid sequences. The bootstrap consensus tree inferred from 1000 replicates. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (five categories (+G, parameter = 4.2179)). This analysis involved 19 amino acid sequences. There was a total of 279 positions in the final dataset. Evolutionary analyses were conducted in MEGA X.
2.4.2. Characterisation of Novel A. perfoliata Heat Shock Protein 90
A total of nine A. perfoliata sequences were identified by tBLASTn as potential HSP90 protein sequences, homologous to protein sequences of recognised HSP90 family including alpha and beta isoforms from mammals and helminths (cutoff 1 × 10−20). The InterProScan followed by manual BLASTp against NCBI (nr) database confirmed eight A. perfoliata sequences as part of the HSP90 protein family (IPR001404) representing five likely distinct A. perfoliata HSP90s.
The secondary characteristic structure of multiple aligned HSP90 alpha (HSP90α) A. perfoliata homologous sequences was predicted, with 17 β-strands and 25 α-helices demonstrating the consistency of the secondary characteristic structure between all eight novel A. perfoliata and recognised HSP90α sequences (Supplementary Figure S3). Among the eight novel isoforms of A. perfoliata HSP90α sequences, ApHSP90-4 (743 amino acids) contained a unique motif; the MEEVD peptide sequence in the C terminal end of cytoplasmic HSP90 isoforms, whereas ApHSP90-5.1 (777 amino acids) and ApHSP90-5.2 (575 amino acids) contained KEEL peptide sequence 75% identical to KDEL peptide sequence of unique endoplasmic reticulum isoforms (human GRP94) [56]. The cytosolic HSP90α was investigated based on a signature sequence LIP and EDD peptide sequences at residues 80–82 and 701–703, respectively [57]. However, all A. perfoliata sequences lacked this signature as did the additional included cestode sequences.
The analysis of the evolutionary relationships between the 8 A. perfoliata HSP90 and 27 recognised HSP90 protein member sequences demonstrated that there were no A. perfoliata HSP90 sequence clustered within HSP90 beta (HSP90β) clades (Figure 2). All eight potential A. perfoliata HSP90 sequences were clustered in a branch of HSP90α into a Cestode and Trematode specific clade. Of which, ApHSP90-4 was in a HSP90α Cestode specific clade containing E. granulosus (accession XP_024345770.1 and CDI70178.1) with good bootstrap support (bootstrap value 97%). The phylogenetic analysis suggests five A. perfoliata HSP90s of which ApHSP90-1.1 to 2.2 and ApHSP90-3 were clustered into a Cestode specific clade of three recognised HSP90 H. microstoma (accession CDS28179.1), E. granulosus (accession CDS25067.1), and E. multilocularis (accession CDS39694.1) with good bootstrap support (Bootstrap value 75%) (Figure 2).
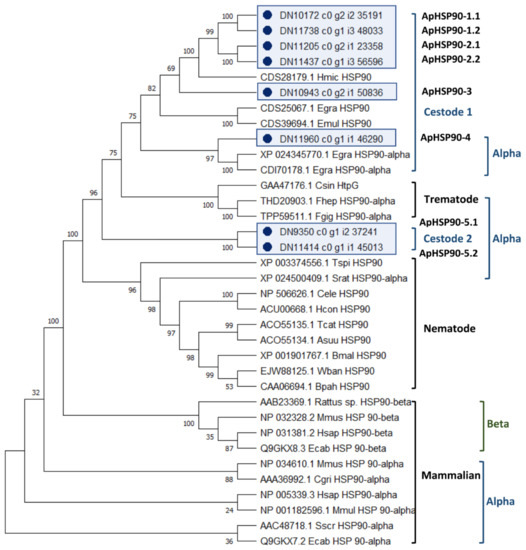
Figure 2.
The phylogenetic tree of A. perfoilata Heat Shock Protein 90 inferred by using Maximum likelihood (ML) method and JTT matrix-based model. The bootstrap consensus tree inferred from 1000 replicates. The tree with the highest log likelihood (-16485.50) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (five categories (+G, parameter = 1.4964)). This analysis involved 35 amino acid sequences. There was a total of 885 positions in the final dataset. Evolutionary analyses were conducted in MEGA X.
2.4.3. Characterisation of Novel A. perfoliata Alpha-Enolase
A total of five A. perfoliata sequences were identified by tBLASTn as potential alpha-Enolase (α-Enolase) protein sequences, homologous to protein sequences of recognised α-Enolase from mammals and helminths (cutoff 1 × 10−15). Subsequently, InterProScan followed by manual BLASTp against the NCBI (nr) database confirmed that five of these sequences were potential α-Enolase A. perfoliata sequences (Enolase superfamily IPR000941). There were three A. perfoliata α-Enolase enzymes that retained all catalytic residues and, therefore, likely to be catalytically functional, whilst the remaining two sequences represented likely incomplete fragments.
The secondary characteristic structure of the three full length α-Enolase A. perfoliata homologous sequences were predicted. All three α-Enolase A. perfoliata sequences; Apα-Enolase-1 (433 amino acids), 2 (456 amino acids) and 3 (456 amino acids) showed comparable numbers of β-strands and α-helices when compared to human α-Enolase (11 β-strands and 16 α-helices) demonstrating the consistency of the secondary characteristic structure between the three novel α-Enolase from A. perfoliata and recognised α-Enolase sequences (Supplementary Figure S4). All three α-Enolase A. perfoliata sequences and recognised α-Enolase sequences conserved amino acid residues imperative for proper catalytic function (His158, Glu167, Glu210, Lys343, Lys394, respective positions in human α-Enolase) [58].
The analysis of the evolutionary relationships between all 3 A. perfoliata α-Enolase and 14 recognised α-Enolase protein member sequences demonstrated that all 3 A. perfoliata α-Enolase sequences (Apα-Enolase-1, -2, and -3) clustered in a Cestode specific clade containing H. microstoma α-Enolase sequence (accession CDS26422 and CDS30005), T. solium (accession AHB59732), E. multilocularis (accession CDS37852), and E. granulosus (accession ACY30465) with good bootstrap support (Figure 3).
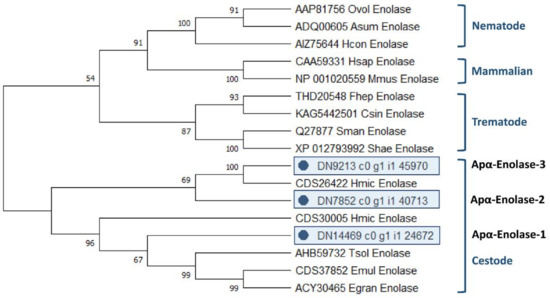
Figure 3.
Maximum likelihood (ML) tree with JTT matrix-based model inferred from A. perfoliata α-Enolase amino acid sequences. The bootstrap consensus tree inferred from 1000 replicates. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (five categories (+G, parameter = 0.4274)). This analysis involved 19 amino acid sequences. There were a total of 597 positions in the final dataset. Evolutionary analyses were conducted in MEGA X.
2.5. Morphological Characterisation and Size Distribution of A. perfoliata EVs
Both transmission electron microscopy (TEM) and nanoparticle tracking analysis (NTA) confirmed that A. perfoliata secreted EVs during in vitro maintenance (Figure 4a). TEM analysis demonstrated the morphological characteristics of size exclusion chromatography (SEC) purified A. perfoliata EVs were in a spherical shaped (cup-shaped) membrane surrounded by a phospholipid bilayer structure (Figure 4a). The proportion of the size distribution of SEC purified A. perfoliata exosome (30–100 nm) and microvesicles (100–1000 nm) determined by TEM was 86% and 14%, respectively (Figure 4b). NTA demonstrated that the majority of the EV population were found to be 67–213 nm (Figure 4c), with a mean concentration of 1.57 × 109 EV particles/mL (Table 3). The mean estimated particle size (mean ± SD) of SEC purified A. perfoliata EVs (three replicates) measured by TEM and NTA were approximately 64.16 ± 28.50 nm (n = 200) and 199.1 ± 108.7 nm in diameter, respectively. TEM showed the greatest EV size was 214.57 nm whereas and the smallest EVs size was 30.17 nm in diameter. The summary statistics of NTA of SEC purified A. perfoliata EVs at 1:600 dilution (n = 3) are shown in Table 3.
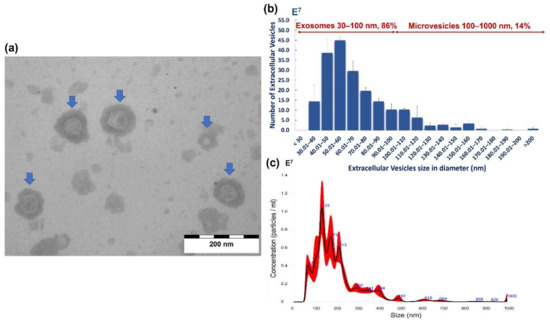
Figure 4.
Characterisation of the in vitro maintenance A. perfoliata extracellular vesicles (EVs) by transmission electron microscopy (TEM) and nanoparticle tracking analysis (NTA): (a) The TEM image from purified A. perfoliata derived EVs at 80 kV; scale bar is 200 nm; demonstrates that A. perfoliata secreted extracellular vesicles (blue arrows). A. perfoliata EV morphology is in a spherical shaped membrane surrounded by a phospholipid bilayer structure; (b) A mean size distribution of the isolated size exclusion chromatography (SEC) purified A. perfoliata EVs (mean ± SE; 200 EVs per replicates; n = 3). The number of exosomes is shown in the size range of 30–100 nm (86%) and microvesicles in the size range of 100–1000 nm (14%); (c) Representative histogram showing the EV particle size distribution and average finite track length adjustment (FTLA) concentration (EVs × 107/mL vs size in nm) of SEC purified A. perfoliata EVs at 1:600 dilution with main peaks at approximately 67–213 nm determined by nanoparticle tracking analysis. Averaged FTLA concentration, as red areas, specify the standard deviation (SD) between measurements and blue numbers indicate the maxima of individual peaks.

Table 3.
Summary statistics of nano-particle tracking analysis of size exclusion chromatography purified Anoplocephala perfoliata EVs.
2.6. Protein Profiling of A. perfoliata Proteomics Datasets
Three biological replicates of both whole EVs and EV depleted ESP produced similar patterns of protein bands for each separate sample demonstrating the similarity amongst biological replicates. Protein bands in A. perfoliata EV depleted ESP demonstrated more frequent and dense proteins on 1D SDS-PAGE gels compared to the whole EVs (Supplementary Figure S5).
The resulting three A. perfoliata mass spectrometry datasets including whole EVs, EV surface and EV depleted ESP were analysed through MASCOT via MS/MS Ion Search against the A. perfoliata transcriptome for protein identification. The full list of proteins identified in each A. perfoliata proteomics datasets is available in Supplementary Tables S2–S4. A total of 315 proteins were identified from A. perfoliata whole EVs, 301 proteins from the EV surface and 596 proteins from EV deleted ESP (Figure 5).
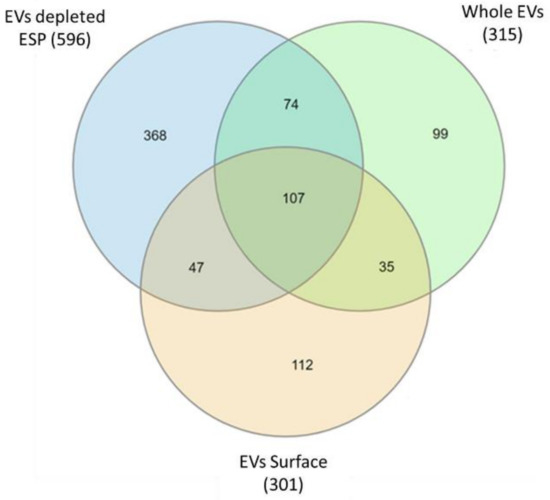
Figure 5.
Venn diagrams comparing the proteins identified in Anoplocephala perfoliata whole extracellular vesicle (EVs), extracellular vesicle (EV) surface, and extracellular vesicle depleted excretory-secretory products (EV depleted ESP) retrieved from mass spectrometry analysis and MASCOT via MS/MS Ion Search against the A. perfoliata transcriptome.
A total of 107 proteins were common between all three proteomics datasets (Figure 5), with the most abundant proteins in all datasets being WD repeat and FYVE domain-containing protein 3 (Table 4, Table 5 and Table 6). A total of 142 identified proteins were common between whole EVs and the EV surface with a total of 474 proteins identified in or on EVs. Well-known identified EV markers of interest as defined by the Exocarta database [59,60] and Vesiclepedia data [61] in A. perfoliata EV and EV surface proteomics datasets were noted amongst the top 50 most abundant proteins, such as, annexin, actin, myosin, enolase, phosphoglycerate kinase, heat shock 70 kDa protein, molecular chaperone HtpG, and programmed cell death 6-interacting protein (Table 4 and Table 5). On the surface of A. perfoliata EVs, protein pumps and transporters were identified such as ATP binding cassette subfamily B (MDR:TAP), multidrug resistance protein, V type proton ATPase 116 kDa subunit A, plasma membrane calcium-transporting ATPase 3 and solute carrier family 5 (Table 5). Key secretory proteins linked to the host–parasite interface such as enolase and calpain were identified in the top 50 most abundant of A. perfoliata EV depleted ESP (Table 6).

Table 4.
The top 50 most abundant proteins putatively identified in A. perfoliata EVs proteomics dataset by 1D SDS-PAGE, LC MS/MS (GeLC) and a MASCOT search at the significance threshold score above 48. Protein descriptions were given from Omicsbox. Protein hits shaded in grey represent known helminth released immune modulators, as also identified in the A. perfoliata transcriptome.

Table 5.
The top 50 most abundant proteins putatively identified on the A. Perfoliata EV surface proteomic dataset by gel-free LC MS/MS and a MASCOT search at the significance threshold score above 47. Protein descriptions were given from Omicsbox. Protein hits shaded in grey represent known helminth released immune modulators, as also identified in the A. perfoliata transcriptome.

Table 6.
The top 50 most abundant proteins putatively identified in A. perfoliata EV depleted ESP proteomic datasets by 1D SDS-PAGE, LC MS/MS, and a MASCOT search at the significance threshold score above 48. Protein descriptions were given from Omicsbox. Protein hits shaded in grey represent known helminth released immune modulators, as also identified in the A. perfoliata transcriptome.
The 454 putative immune modulator sequences that were identified in the transcript were also assessed in the A. perfoliata proteomics data, with a total of 49 identified as expressed proteins across all three datasets, including 22 expressed in EVs, 16 on the EV surface and 40 in the EV depleted ESP (Supplementary Tables S2–S4). Of note, only a single Sigma class GST was identified across the proteomic datasets. ApGST-S1 was identified within the EV depleted ESP proteomic dataset and relatively low abundance. When assessing the proteomics datasets for HSP90 and α-Enolase, one HSP90 (ApHSP90-4) and two α-Enolase (Apα-Enolase-1 and 2) were observed across all three proteomic datasets analysed; namely whole EVs, EV surface proteins and EV depleted ESP. ApHSP90-4 and Apα-Enolase-1 were extremely abundant in the analysis featuring in the top 30 of all three datasets (27th and 23rd in whole EVs, 30th and 24th on the EV surface and 29th and 3rd in the ESP, respectively; Table 4, Table 5 and Table 6). Additionally, Apα-Enolase-2 was identified within the EV depleted ESP proteomic dataset at very low abundance.
2.7. Gene Ontology Enrichment Analysis
A total of 173 GO terms were not propagated up the hierarchy (p < 0.05 identified significance), of which 45, 42, and 86 GO terms were enriched in whole EVs, the EV surface and EV depleted ESP, respectively. The comparison of GO term enrichment from all three proteomics datasets is presented in Supplementary Figure S6. A total of 20 GO terms were enriched across all secretome proteomic datasets, with most GO terms being in the biological processes group, of which calcium ion binding was found to be the most enriched, followed by Arp2/3 complex-mediated actin nucleation, microtubule-based process and gluconeogenesis. Seven GO terms enriched in EV samples only were all categorised in biological processes, with carbohydrate transmembrane transport being the most enriched followed by transmembrane transport, inorganic anion transport, peptidyl-lysine modification to peptidyl-hypusine, calcium-mediated signaling, protein phosphorylation, and protein processing. In the EV surface protein samples, the three most enriched GO terms were categorised in biological processes including bile acid and bile salt transport, seryl-tRNA aminoacylation and urea cycle whereas the three enriched GO terms categorised in cellular component categories included membrane, virion, and nascent polypeptide-associated complex. The top five most enriched GO terms in EV depleted ESP were categorised in biological processes included carbohydrate metabolic process, formaldehyde catabolic process, negative regulation of endopeptidase activity, proteolysis involved in cellular protein catabolic process and proteolysis.
3. Discussion
The equine tapeworm, A. perfoliata, remains a research neglected parasite with limited molecular information available and as such, a lack of understanding of the host–parasite interaction. Our study employed a polyomic approach to characterise adult A. perfoliata, generating a transcriptome of the whole worm and proteomic maps of the secretome. To our knowledge, the present study is the first to generate a de novo transcriptome assembly of this adult equine tapeworm. Moreover, we also present the first evidence that the secretome of an equine helminth parasite generates EVs, which are filled with a plethora of immune-modulatory proteins that have previously been suggested as regulators of host immune responses.
The top 50 most highly represented transcripts demonstrate the expression of common conserved genes similar to other closely related cestodes at several life stages—such as dynein light chain, tegumental protein, deoxyhypusine hydroxylase, 8 kDa glycoprotein, and expressed conserved protein—thus demonstrating the validity of the transcript assembly [62,63,64,65,66,67,68]. Moreover, the A. perfoliata transcriptome aligned well to 3 closely related tapeworm species, namely H. diminuta, E. granulosus, and H. microstoma, again confirming the validity of the dataset.
The development of the first transcriptome for A. perfoliata provides support to explore key proteins of importance linked to the host–parasite interface as has been demonstrated previously for other helminths [19,21,27,69]. At the host–parasite interface immune modulation is imperative for parasite survival [70,71] and consequently many immune modulatory proteins have been identified in platyhelminth species [40,45,49]. The current transcriptome and proteomic analysis has identified 454 transcripts as homologues of recognised immune modulators in other helminth species of which several are functionally expressed given their presence as part of the secretome. Notable immune modulators were identified in the top 50 most abundant proteins, such as sigma class GST (EV depleted ESP), enolase (EVs, EV surface and EV depleted ESP), calpain A (EVs, EV surface, and EV depleted ESP), and HSP90α (EVs, EV surface and EV depleted ESP). Thus, we have demonstrated the potential for an immune modulatory role of the A. perfoliata secretome that may have wide ranging effects on the host immune response to the parasite. The functionality of these putative immune modulators now needs further investigation.
The parasite secretome during infection is known to have an essential role in host–parasite interactions [16,26,27,28,29,30,31,32,33,34,41,72]. Our study establishes for the first time the A. perfoliata secretome; including proteomic analysis of in vitro secreted EVs purified through SEC, the EV surface and EV depleted ESP. We report identification of a total of 315 proteins from A. perfoliata whole EVs, 301 proteins from the EV surface and a further 596 proteins from EV deleted ESP. The majority of GO terms were enriched in the biological processes group, however, calcium ion binding in the molecular category was found to be the most enriched across all secretome samples, a process involved in EV biogenesis [73,74,75].
Many key secretory proteins such as GSTs, HSP90 and Enolase were secreted by A. perfoliata as free proteins (EV depleted ESP) during in vitro maintenance in the current study, which shows similarity to other cestodes [25,28,76,77]. Interestingly, enolase, which is described as a multifunctional protein and essential in the host immune system evasion through immunomodulation, was observed as the third-most-abundant protein in A. perfoliata EV depleted ESP. Likewise, enolase was found as the most abundant in E. granulosus [76] and Taenia solium ESP [25]. In addition, proteins identified in H. diminuta ESP, like peroxidasin, expressed conserved protein, NADP-dependent malic enzyme, deoxyhypusine hydroxylase, and particularly, basement membrane-specific heparan sulfate proteoglycan core protein, were also present in the 50 most abundant proteins in A. perfoliata EV depleted ESP [28].
We also confirmed for the first time that equine tapeworms, A. perfoliata, release whole EVs as part of the ESP, during in vitro maintenance, determined via TEM and NTA analysis. Both size and morphology of A. perfoliata whole EVs were similar to EVs released from other tapeworms in a spherical shaped or cup-shaped membrane surrounded by a phospholipid bilayer structure with sizes ranging from 30 to 200 nm in diameter [38,39,40,41,42,43,44]. The protein profile of A. perfoliata EVs also demonstrated a number of common EV markers in the top 50 abundant proteins, which have been reported in Exocarta, Vesiclepedia, and from other cestodes [38,43]. Furthermore, proteins such as H17 tegumental antigen, tegumental antigen, and tegumental protein, which were found in abundance in A. perfoliata, have been suggested as typical components in EVs from parasitic flatworms such as Hymenolepis diminuta [38], Echinococcus granulosus [41], and Calicophoron daubneyi [49].
The outer surface proteins of parasite derived EVs have crucial roles in establishing cell to cell communication, mediating cellular uptake, affecting immune recognition, and representing effector molecules [78]. The surface proteins of EVs from trematodes have been characterised for several species [47,49,79], but this is the first study to provide a proteome profile for the EV surface of a cestode species. Surface hydrolysis and gel free proteomics led to the identification of 301 surface proteins of A. perfoliata EVs, including many well-known EVs markers, which have been identified in other platyhelminths [47,49]. CD63 antigen, known as part of the tetraspanin family, is mainly associated with membranes of intracellular vesicles [80] and was identified on the A. perfoliata EV surface, although not in the top 50 most abundant proteins. Interestingly, Cathepsins (B, D, and L) which are commonly found on the surface of trematode EVs [47,49] were not observed on the A. perfoliata EV surface, perhaps reflecting the relative importance of parasite digestive tract secretions, that are absent from cestodes [81].
Glycolytic process and phosphorylation enzymes such as phosphoenolpyruvate carboxykinase, phosphoglycerate kinase, glyceraldehyde-3-phosphate dehydrogenase, and glucose 6 phosphate were also identified on the surface of A. perfoliata which are crucial enzyme activities providing a source of energy for helminths [82,83] and likely reflect the EV site of origin from the cestode tegument given A. perfoliata lack a digestive tract [81]. Glycogen or glucose, although sporadically available in the caecum, are likely an easy source of energy in the host’s gut which are absorbed directly through their tegument [83,84]. Additionally, membrane transport proteins on the surface of A. perfoliata EVs such as solute carrier family 5, plasma membrane calcium-transporting ATPase 3, band 3 anion transport protein particularly the pumps protein; ATP binding cassette subfamily B (MDR:TAP) and multidrug resistance protein were observed on the A. perfoliata EV surface similar to F. hepatica EVs [47]. These transport enzyme activities are likely to enhance the carbohydrate metabolism mechanisms in or between cells providing more nutrients and energy uptake to A. perfoliata.
The transcriptomics and proteomics analysis of A. perfoliata demonstrates the variety of key proteins that are relevant to the parasite–host interaction. We therefore further investigated three novel proteins using the transcriptome that have previously been identified as immunomodulators, namely Sigma class GSTs, HSP90α, and enolase to better understand the relationship of these proteins in A. perfoliata to those identified in other platyhelminths.
Sigma class GSTs have well established multi-functional roles in the host–parasite interaction, including general detoxification of xenobiotic and endogenously derived toxins and prostaglandin synthase activity and as such have been suggested as vaccine candidates within Fasciola [85,86,87] and Schistosoma [88,89,90,91,92,93,94]. We have identified two novel A. perfoliata Sigma class GSTs (ApGST-S1, GST-S2.1, and GST-S2.2) within the A. perfoliata transcriptome, confirmed by secondary structure assessment, domain analysis, and phylogenetic analysis. Furthermore, a likely functional expressed protein of ApGST-S1 was also identified in the EV depleted ESP and was initially given the protein description as AChain A, Glutathione S-transferase 28 Kda (GST class-Mu 28 kDa isozyme) following Omicsbox classification. However, domain analysis and phylogenetic analysis demonstrated that ApGST-S1 is likely be a Sigma-like GST. All A. perfoliata Sigma class GSTs were clustered well in the Sigma class GST clade, specifically as part of a cestode group. Moreover, the secondary characteristic structure prediction demonstrated the consistency and similarity with other cestodes and trematodes of the β-strand, α-helix, and random coils structures within the recognised Sigma class GST sequences, which are conserved regions of these proteins. Interestingly, the catalytic tyrosine residues (Y) positioned at the end of the first β-strand, which has been suggested as the key feature of the GSH binding site of the sigma class GST [52,53,54,55], was present in ApGST-S2.1 and S2.2 but missing in ApGST-S1 which was replaced with a histidine residue. However, ApGST-S1 demonstrates similarity to H. microstoma Sigma-like GST (Hmic; accession CDS25704) at this residue and was consequently clustered alongside this H. microstoma Sigma-like GST in the phylogenetic tree (bootstrap value 52%). Therefore, ApGST-S1 is likely be a Sigma-like GST with ApGST-S2.1 and S2.2 representing true Sigma class GSTs. However, given its secretion and alternative active site residue, ApGST-S1 may preferentially function as an immune modulator.
α-Enolase, also known as phosphopyruvate hydratase, is a glycolytic enzyme responsible for converting 2-phosphoglycerate (2-PG) into phosphoenolpyruvate (PEP) in the penultimate step of glycolysis [95]. Additionally, α-Enolase is also considered a multi-functional protein due to acting as a plasminogen receptor and concentrating proteolytic plasmin activity on the cell surface [96]. Functional characterisations of the Onchocerca volvulus α-Enolase has suggested that this enzyme possesses immunomodulatory properties due to its ability to bind to plasminogen and promote plasmin-mediated proteolysis, which subsequently leads to the degradation of the host’s extracellular matrix [37,97]. In total, three full length novel α-Enolases were identified (Apα-Enolase-1, 2, and 3) when exploring the A. perfoliata transcriptome with the potential of two further isoforms that were currently represented by small fragments. Following phylogenetic analysis, all three novel Apα-enolases were clustered well in the α-enolase clade, specifically as part of a cestode group. Assessing primary amino acid sequence of the translated contig hits showed Apα-enolase-1, -2, and -3 to conserve all five active site amino acid residues (His158, Glu167, Glu210, Lys343, Lys394, respective positions in human α-enolase), suggesting these α-enolase A. perfoliata proteins would exhibit similar enzymatic activity properties commonly associated with previously characterised α-enolase proteins [58]. Apα-enolase-1, -2, and -3 also possessed similar secondary structure positioning when compared to human α-enolase (11 β-strands and 16 α-helices), which further supports the previous hypothesis. From the proteomic datasets, only Apα-enolase-1 was expressed in EVs, on the EV surface and in EV depleted ESP, Apα-enolase-2 was expressed in EV depleted ESP, suggesting key roles in host invasion/interaction. To this end, Apα-Enolase-1, -2, and -3 are confirmed to be likely functional Apα-enolase. However, the two Apα-enolase fragments lacked the five catalytic active-site residues and comparable secondary structure patterning, which suggests enzymatic activity is not conserved in these two Apα-enolase proteins and thus further investigation is required to confirm their enzymatic activity and full transcripts.
HSP90 is a molecular chaperone and a highly conserved protein involved in signal transduction, cell cycle control, stress management and folding, degradation, and transport of proteins [98,99,100,101,102,103,104]. Furthermore, HSP90 has also been thought to be involved in host immune system modulation via platyhelminth secretomes [50,105], although information on the role of HSP90 as an immune modulator in helminth infections is less extensive than that presented for sigma class GSTs and enolase. In exploring the A. perfoliata transcriptome, five novel HSP90s were identified (ApHSP90-1 [1.1 and 1.2], -2 [2.1, 2.2], -3, -4, and -5 [5.1 and 5.2]). All putative ApHSP90s were confirmed via secondary structure assessment and phylogenetic analysis with all five identified as the HSP90 alpha (HSP90α) isoform via phylogenetics. It has been reported that only HSP90α isoforms are secreted from cells whereas HSP90β isoforms (HSP90β) primarily operate intracellularly [106]. This is of interest to the secretome dataset, which identified HSP90α proteins in EVs, on the EV surface and in EV depleted ESP, thus supporting the importance of expression of HSP90α for production of proteins to be secreted by A. perfoliata and suggesting key roles in host invasion/interaction. Further work will need to be completed to elucidate if HSP90α is involved in host immune modulation in A. perfoliata infected horses. On further assessment of the secondary structure, only ApHSP90-4 contained the cytoplasmic HSP90 sequence motif, MEEVD, whereas ApHSP90-5 (both 5.1 and 5.2) contained KEEL which is 75% conserved to the KDEL peptide sequences of the HSP90 endoplasmic reticulum (GRP94; 94-kDa glucose-regulated protein). As expected, there were no ApHSP90 sequences that contained LKID peptide sequences, which are specific to HSP90β [56,57,99]. Three ApHSP90 sequences (ApHSP90-1.1, 1.2, 2.1, 2.2, and 3) were also shown to be closely related to other recognised cytosolic HSP90 sequences within the phylogenetic tree (accession CDS28179 HmicHSP90, CDS25067 EgraHSP90, and CDS39694 EmulHSP90). Therefore, ApHSP90-1, -2, and -3 may be the cytosolic HSP90 which are not specific to alpha (inducible isoform) or beta isoforms (constitutively expressed). The LIP and EDD peptide sequences have been suggested as a signature sequences of HSP90 alpha isoforms [57], yet they were missing in ApHSP90-4, although present in the other 4 ApHSP90. Interestingly, the IIP and EDE peptide sequences were found in ApHSP90-4 instead, which are similar to that observed in E. granulosus HSP90α (accession XP_024345770 and CDI70178). To this end, ApHSP90-1, -2, -3, and -4 are most likely to be a cytosolic HSP90, which of ApHSP90-4 is most likely to be HSP90α-like, whereas ApHSP90-5 is most likely to be an endoplasmic reticulum HSP90.
In the current study we have generated the first de novo transcriptome for A. perfoliata to support functional genomics investigations into the host–parasite interaction. In addition, the first in-depth proteomic profiles of the A. perfoliata secretome has been conducted to gain insights into this important interface. In doing so, we have demonstrated that the A. perfoliata secretome contains many proteins that have previously been identified to be involved in host–parasite interactions, namely through immune modulation of the host environment, and these are found in both EVs and EV depleted ESP. We have also identified and characterised novel potential A. perfoliata immune modulators, namely sigma class GST, α-enolase, and HSP90α isoforms. However, the findings demonstrate a variety of key secretory molecules from EVs and ESP, which are not limited to those characterised within the current work, yet the wider immunomodulatory activities of the A. perfoliata secretome need to be further investigated. Importantly, our study demonstrates that A. perfoliata does have the potential to modulate the horse host immune response.
4. Materials and Methods
4.1. Collection of Adult A. perfoliata and In Vitro Maintenance
Live adult A. perfoliata were collected from the caecum at the ileocecal valve of naturally infected horses immediately post-slaughter from a commercial abattoir. Specimens were washed thrice in pre-warmed sterile phosphate-buffered saline (PBS; pH 7.4; Thermo Scientific, Loughborough, UK) at 39 °C to remove host contamination. For subsequent RNA extraction, six A. perfoliata from six separate infections were immediately snap-frozen in liquid nitrogen for 1 min and stored on dry ice for transportation to the laboratory, where they were stored at −80 °C until RNA extraction. For secretome proteomic analysis, 50 live adult A. perfoliata per replicate were maintained in vitro from three individual horse infections following the method previously described [107]. Briefly, A. perfoliata were maintained at 39 °C for 5 h in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Thermo Scientific, Loughborough, UK; supplemented with 2.2 mM Calcium acetate, 2.7 mM Magnesium sulphate, 61 mM glucose, 15 mM HEPES pH 7.0–7.6, gentamycin (5 μg/mL), and 1 μM serotonin). Following the maintenance period, adult A. perfoliata and precipitated debris were removed. The culture supernatant was collected and immediately stored at −80 °C until extracellular vesicle purification and further proteomics analysis.
4.2. Total RNA Extraction and Purification
Total RNA was extracted and purified from adult A. perfoliata (n = 6) using the Direct-zol™ RNA MiniPrep Plus Kit (Zymo Research, Cambridge, UK). A. perfoliata were removed from −80 °C and ≤50 mg of the soma, including the scolex, was removed and cut into small pieces before transferring into a 2 mL microcentrifuge tube containing 600 μL RNA Isolation Reagent, TRI Reagent® (Zymo Research, Cambridge, UK). Tissue samples were subsequently disrupted via bead beating by adding a pre-frozen (−80 °C) 5 mm stainless-steel bead (Qiagen, Manchester, UK) and samples placed in a TissueLyser LT (Qiagen, Manchester, UK) for 2 min at 50 oscillations per s. The bead beating process was repeated where tissue disruption was not complete. Samples were centrifuged at 15,000× g for 30 s to pellet any remaining debris and the supernatant extracted following the manufacturer’s protocol. RNA concentration was determined using a NanoDrop1000 spectrophotometer (Thermo Scientific, Loughborough, UK) and the integrity determined via a 2100 Bioanalyzer (Agilent Technologies, CA, USA) assessment, following the manufacturer’s instructions.
4.3. RNA-Seq Library Construction and Next Generation Sequencing
Purified RNA from all samples were sequenced at the Translation Genomics facility in IBERS, Aberystwyth University. Briefly, RNA purity was assessed using Qubit® RNA HS Assay Kits with the Qubit® Fluorometer (Invitrogen, Thermo Scientific, Loughborough, UK). cDNA libraries were then constructed by reverse transcribing 500 ng of total RNA from each sample using the TruSeq RNA Library Preparation Kit v2 according to the Low Sample (LS) Workflow (Illumina, Cambridge, UK). RNA adapter Indexes were added and ligated on both ends of cDNA to allow multiple indexing of samples pooled together. The cDNA fragments with adapters then underwent PCR amplification (Illumina, Cambridge, UK). cDNA quality was determined on a 1.2% w/v agarose gel.
Amplified cDNA libraries were quantified using an Ultrospec EPOCH (BioTek, China), at an absorbance measurement of 280 nm to normalise a pooling volume of each sample library prior to sequencing. The final concentration of pooled cDNA libraries was quantified using a Qubit® 2.0, dsDNA BR Assay Kit (Invitrogen, Life Technologies, Paisley, UK). Cluster generation and sequencing were performed according to the MiSeq Workflow using MiSeq Reagent Kit v3 (Illumina, Cambridge, UK). Briefly, cDNA libraries were adjusted in equimolar concentration to 10 nM concentration with 10 nM Tris HCl (Melford Laboratories, Suffolk, UK) and 0.05 % v/v Tween-20 solution (Sigma-Aldrich, Merck Life Sciences, Dorset, UK) followed by diluting to 2 nM with buffer EB (Qiagen, Manchester, UK). cDNA libraries were denatured to a single stranded DNA using 0.1 M sodium hydroxide (Sigma-Aldrich, Merck Life Sciences, Dorset, UK) and diluted again to final loading concentration at 6 pM in hybridisation buffer (Illumina, Cambridge, UK). Samples were clustered onto a MiSeq flow cell and paired-end sequenced on a Miseq™, according to standard protocols (Illumina, Cambridge, UK). Base pairs (bp) per read were generated in 2 × 75 bp format.
4.4. De Novo Transcriptome Sequencing Analysis Pipeline
The sequencing pipeline was performed through the Galaxy web platform hosted by IBERS, Aberystwyth University (version 17.01; [108,109,110]). Prior to assembly, all raw FASTQ sequencing data files were assessed via FastQC (Galaxy tool version 0.69; Babraham Bioinformatics; [111]). All reads with a phred quality scores <20 were discarded, although no reads were found below this cut-off. Based on the FastQC assessment, reads were trimmed via Trimmomatic (Galaxy Version 0.36.0; [112]). Illuminaclip was initially used followed by Slidingwindow to remove from the 3′ end and Minlen to remove any reads below 36 bp long. Trimmed reads were again assessed through FastQC to ensure that the read quality of the new RNA-Seq datasets had phred scores of ≥30 across more than 70% of the bases.
De novo assembly of reads was completed in Trinity (v2.11.0; [113,114]) using default parameters. To determine a common set of transcripts between all six biological replicates, all six assemblies were clustered together with Cluster Database at High Identity with Tolerance (cd-hit) software (version 4.8.1; [115,116,117,118,119]). All coding regions within transcript sequences were identified using the assembled unitranscripts as input through Transdecoder (part of the Trinity package; [114]). To detect open reading frames (ORF), parameter settings used at least 100 amino acid long and the ORFs retention of 3000.
Host (horse) contamination was removed from the transcriptome by comparing to protein and CDS files from the Equus caballus genome from Ensemble (version 3.0; [120]), and Hymenolepis microstoma as the closest relative genome sequenced cestode (PRJEB124; [64]), using BLASTp or BLASTx with default options and a minimum e-value of 0.1. Transcripts which were more similar to the host (horse) rather than to Hymenolepis microstoma were deemed to be likely host contaminants and were subsequently removed from the transcriptome.
4.5. Functional Annotation and Gene Ontology (GO) Terms Analysis
The resulting assembly was functionally annotated using Omicsbox [121] to predict the functional description (DE) and GO functional classification of the unigenes. The expression level of transcripts using RNA-seq data was quantified by Salmon [122]. The top 50 abundant transcripts were searched against the Omicsbox output to obtain protein descriptions. Transcripts not found in the output were subsequently translated into protein sequences using ExPASy Translate tools [123] followed by manually BLASTp against the NCBI (nr) protein database using a protein query (BLASTp; [124]) and cut-off set at 1.0 × 10−03.
4.6. Bioinformatic Analysis of Potential Immune Modulators
The transcriptome was analysed for the presence of characterised immune modulators, previously identified in helminths by performing tBLASTn searched against the A. perfoliata transcriptome through BioEdit Sequence Alignment Editor (Version 7.2.6.1; [125]). The number of expected hits of similar quality (e-value) cutoff was set at 1.0 × 10−15 using various immunomodulator bait peptide sequences retrieved from Genbank and NCBI Reference Sequence (see immunomodulators listed in Supplementary Table S1). Subsequently, the top 5 hits from each bait sequence were searched and translated with ExPASy Translate tools [123] to identify the best opening reading frames (ORFs). The peptide sequence of the bait proteins were submitted to Pfam database (version 34.0; [126]) to confirm for protein domain conservation.
Three potential immune modulators in the transcript database, sigma class glutathione transferase (Sigma class GST), cytoplasmic heat shock protein 90 (HSP90), and alpha-enolase (α-Enolase), were selected for further bioinformatic analysis. Protein sequences of recognised Sigma class GST, HSP90 family (alpha and beta isoforms) and α-Enolase from 13, 22, 14 different species, respectively, covering mammalians, nematodes, trematodes, and cestodes were retrieved from Genbank and NCBI Reference Sequence (see recognised Sigma class GST, HSP90 and α-Enolase proteins sequences in Supplementary Table S5, [52,56,64,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142]. Recognised sequences were blasted against the A. perfoliata transcriptome (cutoff set at 1.0 × 10−15, 1.0 × 10−20, and 1.0 × 10−15 for Sigma GST, HSP90, and α-Enolase, respectively) and representative A. perfoliata sequences translated into protein sequences, as previously described. To ensure that selected protein sequences were either Sigma class GST, HSP90 or α-Enolase, each protein sequence was searched against the NCBI (nr) protein database using a protein query (BLASTp; [124]). Subsequently, all representative protein sequences were classified into protein super-families, domain prediction and functional site analysis through InterProScan databases (version 77.0; [143,144]). The resulting InterPro domains classified as Sigma class GST, HSP90, and α-enolase with N-terminal domain (NTD), C-terminal domain (CTD) were kept as a unique sigma class GST, HSP90, and α-enolase protein sequence. All unique classified sigma class GST, HSP90, and α-enolase sequences, or one representative if isoforms were presented, were taken as a final sigma class GST, HSP90, and α-enolase representative protein sequence in A. perfoliata for subsequent phylogenetic analysis.
4.7. Sequence Alignment and Phylogenetic Analysis of Potential Immune Modulators
All multiple sequence alignments of the resulting final sigma class GST, HSP90α, and α-enolase representative protein sequences of A. perfoliata and recognised sigma class GST, HSP90α, and α-enolase protein sequences were completed using ClustalW through BioEdit Sequence Alignment Editor. The secondary characteristic structure including beta sheets and alpha helixes of novel A. perfoliata sigma class GST, HSP90α, and α-enolase sequences were then predicted using the Predict Secondary Structure (PSIPRED) Protein Analysis Workbench (PSIPRED 4.0; [145,146]) followed by protein domain identification and architecture analysis using Simple Modular Architecture Research (SMART) tools [147,148] to obtain the novel A. perfoliata Sigma class GST, HSP90α, and α-enolase sequences.
Subsequently, phylogenetic trees were constructed and visualised in MEGA X (version 10.1.7; [149,150]). Reliability of the phylogenetic tree was estimated with 1000 bootstrap replicates, using both a neighbour-joining (NJ) method and a maximum likelihood (ML) method. For NJ method, the parameters were set as the correction of the amino acid data based on the gamma distribution of rates at 1.0 with a Poisson correction method, pairwise deletions and number of treat at 3. For ML method the parameters were set with a likelihood of amino acid data determined based upon five discrete gamma rate categories. An initial tree for the heuristic search was obtained automatically by applying NJ and BIONJ algorithms to a matrix of pairwise distances estimated using a Jones–Taylor–Thornton (JTT) substitution model, ML heuristic method, with nearest-neighbor-interchange (NNI), and number of threads at 3.
4.8. Extracellular Vesicles Purification by Size Exclusion Chromatography
EVs were purified from A. perfoliata culture media following the protocol described [45]. Briefly, media was centrifuged at 4 °C at 300× g for 10 min and then 700× g for 30 min. Subsequently, residual cells and debris were removed by filtering supernatant through a 0.45 μm PES syringe membrane filter (STARLAB, Milton Keynes, UK). A. perfoliata supernatant was concentrated using 10 KDaMWCO Amicon® Ultra-15 Centrifugal Filter Units (MerckMillipore, Merck Life Sciences, Dorset, UK), following the manufacturer’s guidelines. Briefly, samples were centrifuged at 3000× g for 20 min at 4 °C, until approximately 500 μL of sample was retained in the filter unit. Filtration flow-through was stored at −80 °C for further analysis.
A. perfoliata EVs were then purified using a qEV original size exclusion chromatography (SEC) column (iZON Science, Oxford, UK), according to the manufacturer’s protocol. Briefly, 10 mL of filtered (0.22 μm, STARLAB) PBS (pH 7.4) was loaded through the qEVoriginal SEC column, followed by 500 μL of the concentrated supernatant. The first 3 mL of the filtration flow-through was discarded and SEC purified EVs collected from the next 1.5 mL of the filtration flow-through. Subsequently, 10 mL of filtered PBS was added to the qEVoriginal SEC column and the next 7.5 mL of the filtration flow-through collected EV depleted SEC ESP. Both EVs and EV depleted SEC ESP were stored at −80 °C until further proteomics analysis.
4.9. Characterization of Extracellular Vesicles Released from A. perfoliata
4.9.1. Transmission Electron Microscopy (TEM) Analysis
A. perfoliata EV sample (n = 3) preparation for TEM was performed following the protocol described [151]. Briefly, 10 μL of EVs in 2% paraformaldehyde (PFA) were fixed onto formvar/Carbon coated Copper TEM grids (400 Mesh, Agar Scientific, Stansted, UK). Following fixation, each grid was then washed in 100 μL of PBS (pH 7.4) for 1 min followed by fixing in a 1% (v/v) glutaraldehyde solution (Sigma-Aldrich, Merck Life Sciences, Dorset, UK) for 5 min. Each TEM grid was washed with distilled water for 2 min for a total of eight times. Grids were then contrast-stained in 50 μL of uranyl-oxalate solution (pH 7) for 5 min. Finally, TEM grids were embedded in 50 μL of methyl cellulose uranyl-oxalate, for 10 min on ice, and stored at the room temperature before imaging via transmission electron microscope (JEM1010 Transmission Electron Microscope, Jeol, Tokyo, Japan) at 80 kV as previously described [45]. The size (dimension) of 200 EVs per purification sample imaged by TEM were measured using ImageJ (version 1.52a; [152].
4.9.2. Nanoparticle Tracking Analysis (NTA)
A. perfoliata EVs (n = 3) underwent nanoparticle tracking with size distribution and number of particles in each replicate determined using a Nanosight NS500 system (Malvern Instruments, Malvern, UK) equipped with a green 532 nm laser and a high sensitivity electron multiplying charge-coupled device (EMCCD) camera (Andor Technology, Belfast, UK), following the manufacturer’s instruction. Samples were diluted in PBS (pH 7.4) to obtain a concentration of particles ranging between 106 and 109 particles/mL (Malvern Instruments, Malvern, UK). For each sample, videos of the particles moving under Brownian motion were captured, with a camera level of 15. Subsequently, the captured video data were analysed using the NanoSight software (NTA version 3.2 Dev Build 3.2.16) to assess the particle size and concentration of EVs, with the analysis setting set at a detection threshold of 5.
4.9.3. Extracellular Vesicle Surface Protein Hydrolysis
Surface proteins of SEC purified EVs of A. perfoliata were removed through hydrolysis with trypsin as previously described [49]. Briefly, SEC purified EVs were diluted with PBS to a final concentration of 200 μg in 250 μL total volume. Sequencing grade modified trypsin (100 μg/mL; Roche, U.K) was added to the EVs obtained a final concentration of 50 μg/mL and incubated for 5 min at 37 °C. The treated EVs were then centrifuged for 1 h at 100,000× g at 4 °C. The resulting supernatant was stored at −20 °C prior to gel free mass spectrometry analysis.
4.10. Secretome Proteomics Analysis
4.10.1. One Dimensional Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis
EV depleted SEC ESP were concentrated by precipitation with ice-cold 20% (w/v) trichloroacetic acid (Thermo Scientific, Loughborough, UK) in 100% acetone as previously described [107]. Precipitated pellets were re-suspended in Isoelectric Focusing (IEF) Buffer Z (8 M urea, 2% w/v CHAPS (C32H58N2O7S), 33 mM dithiothreitol, 0.5% carrier ampholytes v/v BioLyte® 3/10) prior to protein quantification. EVs depleted ESP samples were quantified using Bradford assay [153] according to the manufacturer’s protocol, through an UV–visible spectrophotometer (Cary 50, Agilent Technologies, Cheshire, UK) at an absorbance measurement of A595 nm. The Qubit® Protein Assay Kits along with the Qubit® 2.0 Fluorometer (Invitrogen, Thermo Scientific, Loughborough, UK) was employed to quantify the concentration of EV samples according to the manufacturer’s protocol. Samples were evaporated under a vacuum centrifugation for approximately 1 h to concentrate the samples, which was repeated until an acceptable concentration was reached and quantified. All EVs and EV depleted ESP samples were stored at −20 °C until further 1D SDS-PAGE electrophoresis.
Both EVs and EV depleted ESP samples were run on a mini 1D SDS-PAGE gel, 7cm 12.5 % resolving acrylamide gels. All samples were mixed with 4x SDS loading buffer, heated to 95 °C for 10 min and centrifuged at 21,000× g for 10 min. All samples were run at a constant voltage of 70V (BioRad) for approximately 20 min until the bromophenol blue moved through the stacking gel, and then increased to 150 V until completion. Gels were fixed in 40% (v/v) ethanol and 10% (v/v) acetic acid (Thermo Scientific) for 1 h and stained overnight with colloidal Coomassie™ Brilliant Blue (80% (v/v) dye stock solution and 20% (v/v) methanol). Gels were de-stained with 1% v/v acetic acid and then imaged with a GS-800™ Calibrated Densitometer (BioRad).
4.10.2. Trypsin In-Gel Digestion and Liquid Chromatography-Tandem Mass Spectrometry
SDS PAGE lanes containing either EVs or EV depleted ESP were divided into 9 and 12 sections respectively. Each of these sections were excised for in-gel digestion with trypsin, as previously described [31]. Briefly, excised gel bands were de-stained with 50% (v/v) acetonitrile (and 50% (v/v) 50 mM ammonium bicarbonate (Thermo Scientific) at 37 °C for 15 min). The supernatant was discarded and the process repeated until gel pieces were de-stained. Gel bands were then dehydrated in 100% acetonitrile at 37 °C for 15 min, and the gel dried at 50 °C. Following drying, 10 mM dithiothreitol in 50 mM ammonium bicarbonate was added to gel pieces and incubated at 80 °C for 30 min before then incubating with 55 mM iodoacetamide in in 50 mM ammonium bicarbonate (Sigma-Aldrich) at room temperature for 20 min. Gel pieces were washed with 50% (v/v) acetonitrile and 50% (v/v) 50 mM ammonium bicarbonate at room temperature for 15 min, dehydrated with 100% acetonitrile at room temperature for 15 min and then dried at 50 °C. Gel pieces were rehydrated and digested with 50 mM ammonium bicarbonate containing trypsin at 10 ng/μL at 37 °C for approximately 16 h. Gel pieces were then centrifuged at 10,000× g for 10 min and then MilliQ water added before placing on a shaker at room temperature for 10 min, and the supernatant retained. 50% (v/v) acetonitrile and 5% (v/v) formic acid were again added to the gel pieces at room temperature for 60 min. Gel pieces were centrifuged as previously and the supernatant retained and added to the previously retained extraction. All retained supernatants containing peptides were dried until pelleted ready for mass spectrometry analysis.
Liquid chromatography tandem mass spectrometry was performed at the Advanced Mass Spectrometry Facility, School of Biosciences, University of Birmingham as a commercial service. Briefly, dried peptide pellets were re-suspended in 0.1% v/v formic acid and then loaded with an autosampler to be analysed by liquid chromatography tandem mass spectrometry (Q Exactive™ HF Hybrid Quadrupole-Orbitrap™ Mass Spectrometer, Thermo Scientific) equipped with a TriVersa Nanomate (Advion, Harlow, UK) and nanoflow liquid chromatography system (Dionex, Thermo Scientific).
4.10.3. Protein Identification and Gene Ontology Terms Enrichment Analysis
Protein identification of A. perfoliata proteomics profiles were performed through MASCOT [154] hosted by IBERS, Aberystwyth University, according to the method described [107]. Briefly, the acquired MS/MS spectra (Mascot Generic Files) were submitted to a MASCOT MS/MS ions search (Matrix Science, v2.6; [154]) against the A. perfoliata transcriptome and Equus caballus genome version 3.0. Search parameters used the following: trypsin enzymatic cleavage with up to one missed cleavage allowed, fixed modifications to carboxymethyl cysteine with variable modifications set for oxidation of methionine, fixing fragment monoisotopic mass error with peptide tolerances of ±1.2 Da and MS/MS of ±0.6 Da, peptide charge 1+, 2+, and 3+, monoisotopic, data format with mascot generic, electrospray ionization (ESI) TRAP. The resulting identified proteins that indicated the identity or extensive homology (p < 0.05) were selected according to the individual MASCOT ions score with scores set at greater than 48 for EVs and EVs depleted ESP samples and 47 for EVs surface samples. Subsequently, unique peptides presented in at least two replicates were then used for searching against the A. perfoliata annotation database (obtained from the Omicsbox) to assign the protein description and Gene Ontology (GO) terms. The resulting number of proteins identified from MS/MS analysis within A. perfoliata proteomics datasets were then visualised in Venn-diagrams using InteractiVenn [155]. Gene Ontology terms enrichment analysis (GOEA) on gene sets of all proteomics datasets was performed using GOATOOLS python package (v0.5.9, [156]) whether the GO terms were propagated up the hierarchy (prop) or were not propagated up the hierarchy (nop) (p < 0.05 identified significance).
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/pathogens10070912/s1, Figure S1: Summary of the distribution of GO terms in the novel adult A. perfoliata transcriptome at level 3 representing the relative abundance of GO terms in each three main categories, Table S1: Immune modulators sequences and Immune modulators identified from A. perfoliata transcriptome; Figure S2: Multiple sequence alignment of the two novel Sigma class GSTs identified in A. perfoliata and secondary protein structure prediction; Figure S3: Multiple sequence alignment of the five novel alpha Heat shock protein 90s identified in A. perfoliata and secondary protein structure prediction; Figure S4: Multiple sequence alignment of the three novel α-Enolase from A. perfoliata and secondary protein structure prediction; Figure S5: Protein profiles of the A. perfoliata secretome fractions. (A) whole extracellular vesicles (EVs) and (B) EV depleted excretory/secretory protein (ESP) from 3 biological replicates; Table S2: The full list of proteins identified in A. perfoliata whole extracellular vesicles (EVs) proteomic dataset; Table S3: The full list of proteins identified on A. perfoliata extracellular vesicles (EVs) surface proteomic dataset; Table S4: The full list of proteins identified in A. perfoliata extracellular vesicles (EVs) depleted excretory/secretory protein (ESP) proteomic dataset; Figure S6: Gene enrichment analysis of A perfoliata proteomics datasets; Table S5: Protein sequences of recognised Sigma class GST, HSP90, and α-Enolase.
Author Contributions
Conceptualization, R.E.W. and R.M.M.; Methodology, B.W., J.J.T., N.R.A., M.J.H., M.T.S., R.E.W., and R.M.M.; Formal analysis, B.W., B.J.H., C.N.D., S.D.D., A.R.C., and H.C.P.; Investigation, B.W. and B.J.H.; Writing—original draft preparation, B.W. and B.J.H.; Writing—review and editing, P.M.B., R.E.W., and R.M.M.; Supervision, R.E.W. and R.M.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Rajamangala University of Technology Srivijaya, Thailand. The Institute of Biology, Environmental and Rural Sciences (Aberystwyth University, Wales) receives strategic funding from Biotechnology and Biological Sciences Research that also supported the current work.
Institutional Review Board Statement
The study was approved by Aberystwyth University Animal Welfare and Ethical Review Board. As the study used naturally infected horses that were being sacrificed as part of commercial processes at an abattoir, approval under the Animal (Scientific Procedures) Act 1986 was not required.
Informed Consent Statement
Not applicable.
Data Availability Statement
The Transcriptome Shotgun Assembly project has been deposited at DDBJ/EMBL/GenBank under the accession GJFT00000000. The version described in this paper is the first version, GJFT01000000. The A. perfoliata transcriptome is available for BLAST analysis at https://sequenceserver.ibers.aber.ac.uk. Proteomics data from LC-MSMS analysis has been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD027105 and 10.6019/PXD027105.
Acknowledgments
We would like to thank Ravin Jugdaohsingh (Department of Veterinary Medicine, University of Cambridge) for nanoparticle tracking analysis of A. perfoliata extracellular vesicles.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Gasser, R.B.; Williamson, R.M.C.; Beveridge, I. Anoplocephala perfoliata of horses—Significant scope for further research, improved diagnosis and control. Parasitology 2005, 131, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lyons, E.T.; Bolin, D.C.; Bryant, U.K.; Cassone, L.M.; Jackson, C.B.; Janes, J.G.; Kennedy, L.A.; Loynachan, A.T.; Boll, K.R.; Burkhardt, A.S.; et al. Postmortem examination (2016–2017) of weanling and older horses for the presence of select species of endoparasites: Gasterophilus spp., Anoplocephala spp. and Strongylus spp. in specific anatomical sites. Vet. Parasitol. Reg. Stud. Rep. 2018, 13, 98–104. [Google Scholar] [CrossRef]
- Mathewos, M.; Girma, D.; Fesseha, H.; Yirgalem, M.; Eshetu, E. Prevalence of Gastrointestinal Helminthiasis in Horses and Donkeys of Hawassa District, Southern Ethiopia. Vet. Med. Int. 2021, 2021, 6686688. [Google Scholar] [CrossRef]
- Nielsen, M.K. Equine tapeworm infections: Disease, diagnosis and control. Equine Vet. Educ. 2016, 28, 388–395. [Google Scholar] [CrossRef]
- Rehbein, S.; Visser, M.; Winter, R. Prevalence, intensity and seasonality of gastrointestinal parasites in abattoir horses in Germany. Parasitol. Res. 2013, 112, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Tomczuk, K.; Grzybek, M.; Szczepaniak, K.; Studzińska, M.; Demkowska-Kutrzepa, M.; Roczeń-Karczmarz, M.; Abbass, Z.A.; Kostro, K.; Junkuszew, A. Factors affecting prevalence and abundance of A. perfoliata infections in horses from south-eastern Poland. Vet. Parasitol. 2017, 246, 19–24. [Google Scholar] [CrossRef]
- Proudman, C.J.; French, N.P.; Trees, A.J. Tapeworm infection is a significant risk factor for spasmodic colic and ileal impaction colic in the horse. Equine Vet. J. 1998, 30, 194–199. [Google Scholar] [CrossRef]
- Owen, R.A.; Jagger, D.W.; Quan-Taylor, R. Caecal intussusceptions in horses and the significance of Anoplocephala perfoliata. Vet. Rec. 1989, 124, 34–37. [Google Scholar] [CrossRef]
- Barclay, W.P.; Phillips, T.N.; Foerner, J.J. Intussusception associated with Anoplocephala perfoliata infection in five horses. J. Am. Vet. Med. Assoc. 1982, 180, 752–753. [Google Scholar]
- Ryu, S.H.; Bak, U.B.; Kim, J.G.; Yoon, H.J.; Seo, H.S.; Kim, J.T.; Park, J.Y.; Lee, C.W. Cecal rupture by Anoplocephala perfoliata infection in a thoroughbred horse in Seoul Race Park, South Korea. J. Vet. Sci. 2001, 2, 189–193. [Google Scholar] [CrossRef]
- Proudman, C.; Trees, A. Tapeworms as a Cause of Intestinal Disease in Horses. Parasitol. Today 1999, 15, 156–159. [Google Scholar] [CrossRef]
- Fogarty, U.; Del Piero, F.; Purnell, R.E.; Mosurski, K.R. Incidence of Anoplocephala perfoliata in horses examined at an Irish abattoir. Vet. Rec. 1994, 134, 515–518. [Google Scholar] [CrossRef] [PubMed]
- Proudman, C.J.; Edwards, G.B. Are tapeworms associated with equine colic? A case control study. Equine Vet. J. 1993, 25, 224–226. [Google Scholar] [CrossRef]
- Sallé, G.; Guillot, J.; Tapprest, J.; Foucher, N.; Sevin, C.; Laugier, C. Compilation of 29 years of postmortem examinations identifies major shifts in equine parasite prevalence from 2000 onwards. Int. J. Parasitol. 2020, 50, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Williamson, R.M.C.; Gasser, R.B.; Middleton, D.; Beveridge, I. The distribution of Anoplocephala perfoliata in the intestine of the horse and associated pathological changes. Vet. Parasitol. 1997, 73, 225–241. [Google Scholar] [CrossRef]
- Lawson, A.L.; Pittaway, C.E.; Sparrow, R.M.; Balkwill, E.C.; Coles, G.C.; Tilley, A.; Wilson, A.D. Analysis of caecal mucosal inflammation and immune modulation during Anoplocephala perfoliata infection of horses. Parasite Immunol. 2019, 41. [Google Scholar] [CrossRef]
- Pearson, G.R.; Davies, L.W.; White, A.L.; O’Brien, J.K. Pathological lesions associated with Anoplocephala perfoliata at the ileo-caecal junction of horses. Vet. Rec. 1993, 132, 179–182. [Google Scholar] [CrossRef]
- Pavone, S.; Veronesi, F.; Genchi, C.; Fioretti, D.P.; Brianti, E.; Mandara, M.T. Pathological changes caused by Anoplocephala perfoliata in the mucosa/submucosa and in the enteric nervous system of equine ileocecal junction. Vet. Parasitol. 2011, 176, 43–52. [Google Scholar] [CrossRef]
- Cantacessi, C.; Mulvenna, J.; Young, N.D.; Kasny, M.; Horak, P.; Aziz, A.; Hofmann, A.; Loukas, A.; Gasser, R.B. A deep exploration of the transcriptome and “excretory/secretory” proteome of adult Fascioloides magna. Mol. Cell. Proteom. 2012, 11, 1340–1353. [Google Scholar] [CrossRef]
- Choudhary, V.; Garg, S.; Chourasia, R.; Hasnani, J.J.; Patel, P.V.; Shah, T.M.; Bhatt, V.D.; Mohapatra, A.; Blake, D.P.; Joshi, C.G. Transcriptome analysis of the adult rumen fluke Paramphistomum cervi following next generation sequencing. Gene 2015, 570, 64–70. [Google Scholar] [CrossRef]
- Huson, K.M.; Morphew, R.M.; Allen, N.R.; Hegarty, M.J.; Worgan, H.J.; Girdwood, S.E.; Jones, E.L.; Phillips, H.C.; Vickers, M.; Swain, M.; et al. Polyomic tools for an emerging livestock parasite, the rumen fluke Calicophoron daubneyi; identifying shifts in rumen functionality. Parasit. Vectors 2018, 11, 617. [Google Scholar] [CrossRef]
- Liu, G.-H.; Xu, M.-J.; Song, H.-Q.; Wang, C.-R.; Zhu, X.-Q. De novo assembly and characterization of the transcriptome of the pancreatic fluke Eurytrema pancreaticum (trematoda: Dicrocoeliidae) using Illumina paired-end sequencing. Gene 2016, 576, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-X.; Cong, W.; Elsheikha, H.M.; Liu, G.-H.; Ma, J.-G.; Huang, W.-Y.; Zhao, Q.; Zhu, X.-Q. De novo transcriptome sequencing and analysis of the juvenile and adult stages of Fasciola gigantica. Infect. Genet. Evol. 2017, 51, 33–40. [Google Scholar] [CrossRef]
- Guo, A. The complete mitochondrial genome of Anoplocephala perfoliata, the first representative for the family Anoplocephalidae. Parasites Vectors 2015, 8, 549. [Google Scholar] [CrossRef] [PubMed]
- Victor, B.; Kanobana, K.; Gabriël, S.; Polman, K.; Deckers, N.; Dorny, P.; Deelder, A.M.; Palmblad, M. Proteomic analysis of Taenia solium metacestode excretion-secretion proteins. Proteomics 2012, 12, 1860–1869. [Google Scholar] [CrossRef] [PubMed]
- Wangchuk, P.; Kouremenos, K.; Eichenberger, R.M.; Pearson, M.; Susianto, A.; Wishart, D.S.; McConville, M.J.; Loukas, A. Metabolomic profiling of the excretory–secretory products of hookworm and whipworm. Metabolomics 2019, 15, 101. [Google Scholar] [CrossRef]
- Robinson, M.W.; Menon, R.; Donnelly, S.M.; Dalton, J.P.; Ranganathan, S. An integrated transcriptomics and proteomics analysis of the secretome of the helminth pathogen Fasciola hepatica: Proteins associated with invasion and infection of the mammalian host. Mol. Cell. Proteom. 2009, 8, 1891–1907. [Google Scholar] [CrossRef] [PubMed]
- Bień, J.; Sałamatin, R.; Sulima, A.; Savijoki, K.; Bruce Conn, D.; Näreaho, A.; Młocicki, D. Mass spectrometry analysis of the excretory-secretory (E-S) products of the model cestode Hymenolepis diminut a reveals their immunogenic properties and the presence of new E-S proteins in cestodes. Acta Parasitol. 2016, 61, 429–442. [Google Scholar] [CrossRef]
- Han, C.; Yu, J.; Zhang, Z.; Zhai, P.; Zhang, Y.; Meng, S.; Yu, Y.; Li, X.; Song, M. Immunomodulatory effects of Trichinella spiralis excretory-secretory antigens on macrophages. Exp. Parasitol. 2019, 196, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Marcilla, A.; Trelis, M.; Cortés, A.; Sotillo, J.; Cantalapiedra, F.; Minguez, M.T.; Valero, M.L.; Sánchez del Pino, M.M.; Muñoz-Antoli, C.; Toledo, R.; et al. Extracellular Vesicles from Parasitic Helminths Contain Specific Excretory/Secretory Proteins and Are Internalized in Intestinal Host Cells. PLoS ONE 2012, 7, e45974. [Google Scholar] [CrossRef]
- Morphew, R.M.; Wright, H.A.; LaCourse, E.J.; Porter, J.; Barrett, J.; Woods, D.J.; Brophy, P.M. Towards Delineating Functions within the Fasciola Secreted Cathepsin L Protease Family by Integrating In Vivo Based Sub-Proteomics and Phylogenetics. PLoS Negl. Trop. Dis. 2011, 5, e937. [Google Scholar] [CrossRef]
- Pan, W.; Hao, W.T.; Shen, Y.J.; Li, X.Y.; Wang, Y.J.; Sun, F.F.; Yin, J.H.; Zhang, J.; Tang, R.X.; Cao, J.P.; et al. The excretory-secretory products of Echinococcus granulosus protoscoleces directly regulate the differentiation of B10, B17 and Th17 cells. Parasites Vectors 2017, 10, 1–11. [Google Scholar] [CrossRef]
- Vendelova, E.; Camargo de Lima, J.; Lorenzatto, K.R.; Monteiro, K.M.; Mueller, T.; Veepaschit, J.; Grimm, C.; Brehm, K.; Hrčková, G.; Lutz, M.B.; et al. Proteomic Analysis of Excretory-Secretory Products of Mesocestoides corti Metacestodes Reveals Potential Suppressors of Dendritic Cell Functions. PLoS Negl. Trop. Dis. 2016, 10, 5061. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Bai, X.; Zhu, H.; Wang, X.; Shi, H.; Tang, B.; Boireau, P.; Cai, X.; Luo, X.; Liu, M.; et al. Immunoproteomic analysis of the excretory-secretory products of Trichinella pseudospiralis adult worms and newborn larvae. Parasites Vectors 2017, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Eichenberger, R.M.; Sotillo, J.; Loukas, A. Immunobiology of parasitic worm extracellular vesicles. Immunol. Cell Biol. 2018, 96, 704–713. [Google Scholar] [CrossRef] [PubMed]
- Zakeri, A.; Hansen, E.P.; Andersen, S.D.; Williams, A.R.; Nejsum, P. Immunomodulation by helminths: Intracellular pathways and extracellular vesicles. Front. Immunol. 2018, 9, 2349. [Google Scholar] [CrossRef]
- Maizels, R.M.; Smits, H.H.; McSorley, H.J. Modulation of Host Immunity by Helminths: The Expanding Repertoire of Parasite Effector Molecules. Immunity 2018, 49, 801–818. [Google Scholar] [CrossRef]
- Mazanec, H.; Koník, P.; Gardian, Z.; Kuchta, R. Extracellular vesicles secreted by model tapeworm Hymenolepis diminuta: Biogenesis, ultrastructure and protein composition. Int. J. Parasitol. 2021, 51, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.; Mao, L.; Zhang, S.; Guo, X.; Liu, G.; Wang, L.; Hou, J.; Zheng, Y.; Luo, X. Identification and molecular characterization of exosome-like vesicles derived from the Taenia asiatica adult worm. Acta Trop. 2019, 198, 105036. [Google Scholar] [CrossRef]
- Wang, L.Q.; Liu, T.L.; Liang, P.H.; Zhang, S.H.; Li, T.S.; Li, Y.P.; Liu, G.X.; Mao, L.; Luo, X.N. Characterization of exosome-like vesicles derived from Taenia pisiformis cysticercus and their immunoregulatory role on macrophages. Parasites Vectors 2020, 13, 1–16. [Google Scholar] [CrossRef]
- Nicolao, M.C.; Rodriguez Rodrigues, C.; Cumino, A.C. Extracellular vesicles from Echinococcus granulosus larval stage: Isolation, characterization and uptake by dendritic cells. PLoS Negl. Trop. Dis. 2019, 13, e0007032. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, W.; Cui, F.; Shi, C.; Ma, Y.; Yu, Y.; Zhao, W.; Zhao, J. Extracellular vesicles derived from Echinococcus granulosus hydatid cyst fluid from patients: Isolation, characterization and evaluation of immunomodulatory functions on T cells. Int. J. Parasitol. 2019, 49, 1029–1037. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Cai, M.; Yang, J.; Li, Y.; Ding, J.; Kandil, O.M.; Kutyrev, I.; Ayaz, M.; Zheng, Y. Comparative analysis of different extracellular vesicles secreted by Echinococcus granulosus protoscoleces. Acta Trop. 2021, 213, 105756. [Google Scholar] [CrossRef] [PubMed]
- Ancarola, M.E.; Marcilla, A.; Herz, M.; Macchiaroli, N.; Pérez, M.; Asurmendi, S.; Brehm, K.; Poncini, C.; Rosenzvit, M.; Cucher, M. Cestode parasites release extracellular vesicles with microRNAs and immunodiagnostic protein cargo. Int. J. Parasitol. 2017, 47, 675–686. [Google Scholar] [CrossRef]
- Davis, C.N.; Phillips, H.; Tomes, J.J.; Swain, M.T.; Wilkinson, T.J.; Brophy, P.M.; Morphew, R.M. The importance of extracellular vesicle purification for downstream analysis: A comparison of differential centrifugation and size exclusion chromatography for helminth pathogens. PLoS Negl. Trop. Dis. 2019, 13, e0007191. [Google Scholar] [CrossRef] [PubMed]
- Cwiklinski, K.; De La Torre-Escudero, E.; Trelis, M.; Bernal, D.; Dufresne, P.J.; Brennan, G.P.; O’Neill, S.; Tort, J.; Paterson, S.; Marcilla, A.; et al. The extracellular vesicles of the helminth pathogen, Fasciola hepatica: Biogenesis pathways and cargo molecules involved in parasite pathogenesis. Mol. Cell. Proteom. 2015, 14, 3258–3273. [Google Scholar] [CrossRef] [PubMed]
- De la Torre-Escudero, E.; Gerlach, J.Q.; Bennett, A.P.S.; Cwiklinski, K.; Jewhurst, H.L.; Huson, K.M.; Joshi, L.; Kilcoyne, M.; O’Neill, S.; Dalton, J.P.; et al. Surface molecules of extracellular vesicles secreted by the helminth pathogen Fasciola hepatica direct their internalisation by host cells. PLoS Negl. Trop. Dis. 2019, 13, e0007087. [Google Scholar] [CrossRef]
- Bennett, A.P.S.; de la Torre-Escudero, E.; Oliver, N.A.M.; Huson, K.M.; Robinson, M.W. The cellular and molecular origins of extracellular vesicles released by the helminth pathogen, Fasciola hepatica. Int. J. Parasitol. 2020, 50, 671–683. [Google Scholar] [CrossRef]
- Allen, N.R.; Taylor-Mew, A.R.; Wilkinson, T.J.; Huws, S.; Phillips, H.; Morphew, R.M.; Brophy, P.M. Modulation of Rumen Microbes Through Extracellular Vesicle Released by the Rumen Fluke Calicophoron daubneyi. Front. Cell. Infect. Microbiol. 2021, 11, 263. [Google Scholar] [CrossRef]
- Liu, F.; Cui, S.J.; Hu, W.; Feng, Z.; Wang, Z.Q.; Han, Z.G. Excretory/secretory proteome of the adult developmental stage of human blood fluke, Schistosoma japonicum. Mol. Cell. Proteom. 2009, 8, 1236–1251. [Google Scholar] [CrossRef]
- Kifle, D.W.; Pearson, M.S.; Becker, L.; Pickering, D.; Loukas, A.; Sotillo, J. Proteomic analysis of two populations of Schistosoma mansoni-derived extracellular vesicles: 15k pellet and 120k pellet vesicles. Mol. Biochem. Parasitol. 2020, 236, 111264. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.A.; Bae, Y.A.; Lee, E.G.; Kim, S.H.; Diaz-Camacho, S.P.; Nawa, Y.; Kang, I.; Kong, Y. A novel sigma-like glutathione transferase of Taenia solium metacestode. Int. J. Parasitol. 2010, 40, 1097–1106. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zhou, X.; Chen, L.; Zhang, Z.; Wang, C.; Gu, X.; Wang, T.; Peng, X.; Yang, G. Cloning and characterization of a novel sigma-like glutathione S-transferase from the giant panda parasitic nematode, Baylisascaris schroederi. Parasites Vectors 2015, 8, 44. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.C.; Yang, Y.J.; Ma, K.X.; Shi, C.Y.; Chen, G.W.; Liu, D.Z. A novel sigma class glutathione S-transferase gene in freshwater planarian Dugesia japonica: Cloning, characterization and protective effects in herbicide glyphosate stress. Ecotoxicology 2020, 29, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Fujii, H.; Aso, Y.; Banno, Y.; Koga, K. Expression and characterization of a sigma-class glutathione S-transferase of the fall webworm, Hyphantria cunea. Biosci. Biotechnol. Biochem. 2007, 71, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Skantar, A.M.; Carta, L.K. Molecular characterization and phylogenetic evaluation of the Hsp90 gene from selected nematodes. J. Nematol. 2004, 36, 466–480. [Google Scholar]
- Chen, B.; Zhong, D.; Monteiro, A. Comparative genomics and evolution of the HSP90 family of genes across all kingdoms of organisms. BMC Genom. 2006, 7, 156. [Google Scholar] [CrossRef]
- Pancholi, V. Multifunctional α-enolase: Its role in diseases. Cell. Mol. Life Sci. 2001, 58, 902–920. [Google Scholar] [CrossRef]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef]
- Mathivanan, S.; Fahner, C.J.; Reid, G.E.; Simpson, R.J. ExoCarta 2012: Database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012, 40, D1241–D1244. [Google Scholar] [CrossRef]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borràs, F.E.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A Compendium for Extracellular Vesicles with Continuous Community Annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef]
- Parkinson, J.; Wasmuth, J.D.; Salinas, G.; Bizarro, C.V.; Sanford, C.; Berriman, M.; Ferreira, H.B.; Zaha, A.; Blaxter, M.L.; Maizels, R.M.; et al. A Transcriptomic Analysis of Echinococcus granulosus Larval Stages: Implications for Parasite Biology and Host Adaptation. PLoS Negl. Trop. Dis. 2012, 6, e1897. [Google Scholar] [CrossRef]
- Debarba, J.A.; Sehabiague, M.P.C.; Monteiro, K.M.; Gerber, A.L.; Vasconcelos, A.T.R.; Ferreira, H.B.; Zaha, A. Transcriptomic analysis of the early strobilar development of echinococcus granulosus. Pathogens 2020, 9, 465. [Google Scholar] [CrossRef]
- Tsai, I.J.; Zarowiecki, M.; Holroyd, N.; Garciarrubio, A.; Sanchez-Flores, A.; Brooks, K.L.; Tracey, A.; Bobes, R.J.; Fragoso, G.; Sciutto, E.; et al. The genomes of four tapeworm species reveal adaptations to parasitism. Nature 2013, 496, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Preza, M.; Calvelo, J.; Langleib, M.; Hoffmann, F.; Castillo, E.; Koziol, U.; Iriarte, A. Stage-specific transcriptomic analysis of the model cestode Hymenolepis microstoma. Genomics 2021, 113, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Olson, P.D.; Zarowiecki, M.; James, K.; Baillie, A.; Bartl, G.; Burchell, P.; Chellappoo, A.; Jarero, F.; Tan, L.Y.; Holroyd, N.; et al. Genome-wide transcriptome profiling and spatial expression analyses identify signals and switches of development in tapeworms. Evodevo 2018, 9, 1–29. [Google Scholar] [CrossRef]
- Yang, D.; Fu, Y.; Wu, X.; Xie, Y.; Nie, H.; Chen, L.; Nong, X.; Gu, X.; Wang, S.; Peng, X.; et al. Annotation of the transcriptome from taenia pisiformis and its comparative analysis with three taeniidae species. PLoS ONE 2012, 7, e32283. [Google Scholar] [CrossRef]
- García-Montoya, G.M.; Mesa-Arango, J.A.; Isaza-Agudelo, J.P.; Agudelo-Lopez, S.P.; Cabarcas, F.; Barrera, L.F.; Alzate, J.F. Transcriptome profiling of the cysticercus stage of the laboratory model Taenia crassiceps, strain ORF. Acta Trop. 2016, 154, 50–62. [Google Scholar] [CrossRef]
- Pan, W.; Shen, Y.; Han, X.; Wang, Y.; Liu, H.; Jiang, Y.; Zhang, Y.; Wang, Y.; Xu, Y.; Cao, J. Transcriptome Profiles of the Protoscoleces of Echinococcus granulosus Reveal that Excretory-Secretory Products Are Essential to Metabolic Adaptation. PLoS Negl. Trop. Dis. 2014, 8, e3392. [Google Scholar] [CrossRef]
- Hewitson, J.P.; Grainger, J.R.; Maizels, R.M. Helminth immunoregulation: The role of parasite secreted proteins in modulating host immunity. Mol. Biochem. Parasitol. 2009, 167, 1–11. [Google Scholar] [CrossRef]
- Gazzinelli-Guimaraes, P.H.; Nutman, T.B. Helminth parasites and immune regulation. F1000Research 2018, 7. [Google Scholar] [CrossRef]
- Nono, J.K.; Pletinckx, K.; Lutz, M.B.; Brehm, K. Excretory/secretory-products of echinococcus multilocularis larvae induce apoptosis and tolerogenic properties in dendritic cells in vitro. PLoS Negl. Trop. Dis. 2012, 6, e1516. [Google Scholar] [CrossRef]
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell. Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Savina, A.; Furlán, M.; Vidal, M.; Colombo, M.I. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J. Biol. Chem. 2003, 278, 20083–20090. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.; Azimi, I.; Monteith, G.; Bebawy, M. Ca2+ mediates extracellular vesicle biogenesis through alternate pathways in malignancy. J. Extracell. Vesicles 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiao, D.; Shen, Y.; Han, X.; Zhao, F.; Li, X.; Wu, W.; Zhou, H.; Zhang, J.; Cao, J. Proteomic analysis of the excretory/secretory products and antigenic proteins of Echinococcus granulosus adult worms from infected dogs. BMC Vet. Res. 2015, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Virginio, V.G.; Monteiro, K.M.; Drumond, F.; De Carvalho, M.O.; Vargas, D.M.; Zaha, A.; Ferreira, H.B. Excretory/secretory products from in vitro-cultured Echinococcus granulosus protoscoleces. Mol. Biochem. Parasitol. 2012, 183, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Buzás, E.I.; Tóth, E.; Sódar, B.W.; Szabó-Taylor, K. Molecular interactions at the surface of extracellular vesicles. Semin. Immunopathol. 2018, 40, 453–464. [Google Scholar] [CrossRef]
- Murphy, A.; Cwiklinski, K.; Lalor, R.; O’Connell, B.; Robinson, M.W.; Gerlach, J.; Joshi, L.; Kilcoyne, M.; Dalton, J.P.; O’Neill, S.M. Fasciola hepatica extracellular vesicles isolated from excretory-secretory products using a gravity flow method modulate dendritic cell phenotype and activity. PLoS Negl. Trop. Dis. 2020, 14, e0008626. [Google Scholar] [CrossRef]
- Andreu, Z.; Yáñez-Mó, M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014, 5, 442. [Google Scholar] [CrossRef]
- Margulis, L.; Chapman, M.J. Kingdoms and Domains: An Illustrated Guide to the Phyla of Life on Earth; Academic Press: Cambridge, MA, USA, 2009. [Google Scholar]
- Mansour, T.E.; Mansour, J.M. Energy Metabolism in Parasitic Helminths: Targets for Antiparasitic Agents. In Chemotherapeutic Targets in Parasites: Contemporary Strategies; Mansour, T.E., Ed.; Cambridge University Press: Cambridge, UK, 2002; pp. 33–57. ISBN 9780521620659. [Google Scholar]
- Barrett, J. Forty years of helminth biochemistry. Parasitology 2009, 136, 1633–1642. [Google Scholar] [CrossRef]
- Chappell, L.H. The biology of the external surfaces of helminth parasites. Proc. R. Soc. Edinburgh. Sect. B Biol. Sci. 1980, 79, 145–172. [Google Scholar] [CrossRef]
- LaCourse, E.J.; Perally, S.; Morphew, R.M.; Moxon, J.V.; Prescott, M.; Dowling, D.J.; O’Neill, S.M.; Kipar, A.; Hetzel, U.; Hoey, E.; et al. The Sigma Class Glutathione Transferase from the Liver Fluke Fasciola hepatica. PLoS Negl. Trop. Dis. 2012, 6, e1666. [Google Scholar] [CrossRef] [PubMed]
- Dowling, D.J.; Hamilton, C.M.; Donnelly, S.; La Course, J.; Brophy, P.M.; Dalton, J.; O’Neill, S.M. Major secretory antigens of the Helminth Fasciola hepatica activate a suppressive dendritic cell phenotype that attenuates Th17 cells but fails to activate Th2 immune responses. Infect. Immun. 2010, 78, 793–801. [Google Scholar] [CrossRef]
- Zafra, R.; Pérez-écija, R.A.; Buffoni, L.; Pacheco, I.L.; Martínez-Moreno, A.; LaCourse, E.J.; Perally, S.; Brophy, P.M.; Pérez, J. Early hepatic and peritoneal changes and immune response in goats vaccinated with a recombinant glutathione transferase sigma class and challenged with Fasciola hepatica. Res. Vet. Sci. 2013, 94, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Grezel, D.; Capron, M.; Grzych, J.-M.; Fontaine, J.; Lecocq, J.-P.; Capron, A. Protective immunity induced in rat schistosomiasis by a single dose of the Sm28GST recombinant antigen: Effector mechanisms involving IgE and IgA antibodies. Eur. J. Immunol. 1993, 23, 454–460. [Google Scholar] [CrossRef]
- Fonseca, C.T.; Oliveira, S.C.; Alves, C.C. Eliminating schistosomes through vaccination: What are the best immune weapons? Front. Immunol. 2015, 6, 95. [Google Scholar] [CrossRef] [PubMed]
- Ivanoff, N.; Phillips, N.; Schacht, A.M.; Heydari, C.; Capron, A.; Riveau, G. Mucosal vaccination against schistosomiasis using liposome-associated Sm 28 kDa glutathione S-transferase. Vaccine 1996, 14, 1123–1131. [Google Scholar] [CrossRef]
- Boulanger, D.; Reid, G.D.F.; Sturrock, R.F.; Wolowczuk, I.; Balloul, J.M.; Grezel, D.; Pierce, R.J.; Otieno, M.F.; Guerret, S.; Grimaud, A.; et al. Immunization of mice and baboons with the recombinant Sm28GST affects both worm viability and fecundity after experimental infection with Schistosoma mansoni. Parasite Immunol. 1991, 13, 473–490. [Google Scholar] [CrossRef]
- Riveau, G.; Schacht, A.M.; Dompnier, J.P.; Deplanque, D.; Seck, M.; Waucquier, N.; Senghor, S.; Delcroix-Genete, D.; Hermann, E.; Idris-Khodja, N.; et al. Safety and efficacy of the rSh28GST urinary schistosomiasis vaccine: A phase 3 randomized, controlled trial in Senegalese children. PLoS Negl. Trop. Dis. 2018, 12, e0006968. [Google Scholar] [CrossRef]
- Molehin, A.J. Schistosomiasis vaccine development: Update on human clinical trials. J. Biomed. Sci. 2020, 27, 1–7. [Google Scholar] [CrossRef]
- Al-Naseri, A.; Al-Absi, S.; El Ridi, R.; Mahana, N. A comprehensive and critical overview of schistosomiasis vaccine candidates. J. Parasit. Dis. 2021, 45, 557–580. [Google Scholar] [CrossRef] [PubMed]
- Fukano, K.; Kimura, K. Measurement of enolase activity in cell lysates. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2014; Volume 542, pp. 115–124. ISBN 9780124166189. [Google Scholar]
- Díaz-Ramos, À.; Roig-Borrellas, A.; García-Melero, A.; López-Alemany, R. α-enolase, a multifunctional protein: Its role on pathophysiological situations. J. Biomed. Biotechnol. 2012, 2012, 156795. [Google Scholar] [CrossRef] [PubMed]
- Jolodar, A.; Fischer, P.; Bergmann, S.; Büttner, D.W.; Hammerschmidt, S.; Brattig, N.W. Molecular cloning of an α-enolase from the human filarial parasite Onchocerca volvulus that binds human plasminogen. Biochim. Biophys. Acta Gene Struct. Expr. 2003, 1627, 111–120. [Google Scholar] [CrossRef]
- Johnson, J.L. Evolution and function of diverse Hsp90 homologs and cochaperone proteins. Biochim. Biophys. Acta Mol. Cell Res. 2012, 1823, 607–613. [Google Scholar] [CrossRef]
- Hoter, A.; El-Sabban, M.; Naim, H. The HSP90 Family: Structure, Regulation, Function, and Implications in Health and Disease. Int. J. Mol. Sci. 2018, 19, 2560. [Google Scholar] [CrossRef] [PubMed]
- Backe, S.J.; Sager, R.A.; Woodford, M.R.; Makedon, A.M.; Mollapour, M. Post-translational modifications of Hsp90 and translating the chaperone code. J. Biol. Chem. 2020, 295, 11099–11117. [Google Scholar] [CrossRef] [PubMed]
- Biebl, M.M.; Buchner, J. Structure, function, and regulation of the hsp90 machinery. Cold Spring Harb. Perspect. Biol. 2019, 11, a034017. [Google Scholar] [CrossRef]
- Roy, N.; Nageshan, R.K.; Ranade, S.; Tatu, U. Heat shock protein 90 from neglected protozoan parasites. Biochim. Biophys. Acta Mol. Cell Res. 2012, 1823, 707–711. [Google Scholar] [CrossRef]
- Zininga, T.; Ramatsui, L.; Shonhai, A. Heat shock proteins as immunomodulants. Molecules 2018, 23, 2846. [Google Scholar] [CrossRef]
- Gillan, V.; Devaney, E. Nematode Hsp90: Highly conserved but functionally diverse. Parasitology 2014, 141, 1203–1215. [Google Scholar] [CrossRef]
- Xu, Z.; Ji, M.; Li, C.; Du, X.; Hu, W.; McManus, D.P.; You, H. A biological and immunological characterization of schistosoma japonicum heat shock proteins 40 and 90α. Int. J. Mol. Sci. 2020, 21, 4034. [Google Scholar] [CrossRef]
- Jayaprakash, P.; Dong, H.; Zou, M.; Bhatia, A.; O’Brien, K.; Chen, M.; Woodley, D.T.; Li, W. Hsp90α and Hsp90β together operate a hypoxia and nutrient paucity stress-response mechanism during wound healing. J. Cell Sci. 2015, 128, 1475–1480. [Google Scholar] [CrossRef]
- Morphew, R.M.; MacKintosh, N.; Hart, E.H.; Prescott, M.; LaCourse, E.J.; Brophy, P.M. In vitro biomarker discovery in the parasitic flatworm Fasciola hepatica for monitoring chemotherapeutic treatment. EuPA Open Proteom. 2014, 3, 85–99. [Google Scholar] [CrossRef]
- Goecks, J.; Nekrutenko, A.; Taylor, J.; Afgan, E.; Ananda, G.; Baker, D.; Blankenberg, D.; Chakrabarty, R.; Coraor, N.; Goecks, J.; et al. Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Baker, D.; Batut, B.; Van Den Beek, M.; Bouvier, D.; Ech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [PubMed]
- Jalili, V.; Afgan, E.; Gu, Q.; Clements, D.; Blankenberg, D.; Goecks, J.; Taylor, J.; Nekrutenko, A. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucleic Acids Res. 2020, 48, W395–W402, Erratum in 2020, 48, 8205–8207. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 5 August 2019).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Li, W.; Jaroszewski, L.; Godzik, A. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics 2001, 17, 282–283. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Jaroszewski, L.; Godzik, A. Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics 2002, 18, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Niu, B.; Gao, Y.; Fu, L.; Li, W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics 2010, 26, 680–682. [Google Scholar] [CrossRef] [PubMed]
- Howe, K.L.; Achuthan, P.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2021. Nucleic Acids Res. 2021, 49, D884–D891. [Google Scholar] [CrossRef] [PubMed]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Morphew, R.M.; Eccleston, N.; Wilkinson, T.J.; McGarry, J.; Perally, S.; Prescott, M.; Ward, D.; Williams, D.; Paterson, S.; Raman, M.; et al. Proteomics and in Silico Approaches To Extend Understanding of the Glutathione Transferase Superfamily of the Tropical Liver Fluke Fasciola gigantica. J. Proteome Res. 2012, 11, 5876–5889. [Google Scholar] [CrossRef]
- Trottein, F.; Godin, C.; Pierce, R.J.; Sellin, B.; Taylor, M.G.; Gorillot, I.; Silva, M.S.; Lecocq, J.P.; Capron, A. Inter-species variation of schistosome 28-kDa glutathione S-transferases. Mol. Biochem. Parasitol. 1992, 54, 63–72. [Google Scholar] [CrossRef]
- Balloul, J.M.; Sondermeyer, P.; Dreyer, D.; Capron, M.; Grzych, J.M.; Pierce, R.J.; Carvallo, D.; Lecocq, J.P.; Capron, A. Molecular cloning of a protective antigen of schistosomes. Nature 1987, 326, 149–153. [Google Scholar] [CrossRef]
- Faya, N.; Penkler, D.L.; Tastan Bishop, Ö. Human, vector and parasite Hsp90 proteins: A comparative bioinformatics analysis. FEBS Open Bio 2015, 5, 916–927. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, Z.; Li, D.; Chen, Q. Identification and characterization of a Bursaphelenchus xylophilus (aphelenchida: Aphelenchoididae) thermotolerance-related gene: Bx-HSP90. Int. J. Mol. Sci. 2012, 13, 8819–8833. [Google Scholar] [CrossRef] [PubMed]
- Pantzartzi, C.N.; Drosopoulou, E.; Scouras, Z.G. Assessment and Reconstruction of Novel HSP90 Genes: Duplications, Gains and Losses in Fungal and Animal Lineages. PLoS ONE 2013, 8, 73217. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Cheng, Y.S.; Shih, H.H. Expression patterns and structural modelling of Hsp70 and Hsp90 in a fish-borne zoonotic nematode Anisakis pegreffii. Vet. Parasitol. 2015, 212, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Pepin, K.; Momose, F.; Ishida, N.; Nagata, K. Molecular Cloning of Horse Hsp90 cDNA and Its Comparative Analysis with Other Vertebrate Hsp90 Sequences. J. Vet. Med. Sci. 2001, 63, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Wang, L.; Liu, Y.; He, L.; Li, M.; Lu, W.; Xue, C. Molecular characterization and expression of a heat shock protein gene (HSP90) from the carmine spider mite, Tetranychus cinnabarinus (Boisduval). J. Insect Sci. 2010, 10, 1536–2442. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Zhang, W.; Zhang, L.; Zhang, Z.; Li, J.; Lu, G.; Zhu, Y.; Wang, Y.; Huang, Y.; Liu, J.; et al. The genome of the hydatid tapeworm Echinococcus granulosus. Nat. Genet. 2013, 45, 1168–1175. [Google Scholar] [CrossRef]
- Iriarte, A.; Arbildi, P.; La-Rocca, S.; Musto, H.; Fernández, V. Identification of novel glutathione transferases in Echinococcus granulosus. An evolutionary perspective. Acta Trop. 2012, 123, 208–216. [Google Scholar] [CrossRef]
- Kanaoka, Y.; Fujimori, K.; Kikuno, R.; Sakaguchi, Y.; Urade, Y.; Hayaishi, O. Structure and chromosomal localization of human and mouse genes for hematopoietic prostaglandin D synthase. Eur. J. Biochem. 2000, 267, 3315–3322. [Google Scholar] [CrossRef]
- Liebau, E.; Schönberger, Ö.L.; Walter, R.D.; Henkle-Dührsen, K.J. Molecular cloning and expression of a cDNA encoding glutathione S-transferase from Ascaris suum. Mol. Biochem. Parasitol. 1994, 63, 167–170. [Google Scholar] [CrossRef]
- Liebau, E.; Eckelt, V.H.O.; Wildenburg, G.; Teesdale-Spittle, P.; Brophy, P.M.; Walter, R.D.; Henkle-Dührsen, K. Structural and functional analysis of a glutathione S-transferase from Ascaris suum. Biochem. J. 1997, 324, 659–666. [Google Scholar] [CrossRef]
- Sharp, P.J.; Smith, D.R.J.; Bach, W.; Wagland, B.M.; Cobon, G.S. Purified glutathione s-transferases from parasites as candidate protective antigens. Int. J. Parasitol. 1991, 21, 839–846. [Google Scholar] [CrossRef]
- Kang, S.Y.; Ahn, I.Y.; Park, C.Y.; Chung, Y.B.; Hong, S.T.; Kong, Y.; Cho, S.Y.; Hong, S.J. Clonorchis sinensis: Molecular cloning and characterization of 28-kDa glutathione S-transferase. Exp. Parasitol. 2001, 97, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Blum, M.; Chang, H.Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Jones, D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Buchan, D.W.A.; Jones, D.T. The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res. 2019, 47, W402–W407. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Hall, B.G. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Perkins, D.N.; Pappin, D.J.C.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Heberle, H.; Meirelles, V.G.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
- Klopfenstein, D.V.; Zhang, L.; Pedersen, B.S.; Ramírez, F.; Vesztrocy, A.W.; Naldi, A.; Mungall, C.J.; Yunes, J.M.; Botvinnik, O.; Weigel, M.; et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 2018, 8, 1–17. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).