Extracellular Proteome Analysis Shows the Abundance of Histidine Kinase Sensor Protein, DNA Helicase, Putative Lipoprotein Containing Peptidase M75 Domain and Peptidase C39 Domain Protein in Leptospira interrogans Grown in EMJH Medium
Abstract
1. Introduction
2. Results
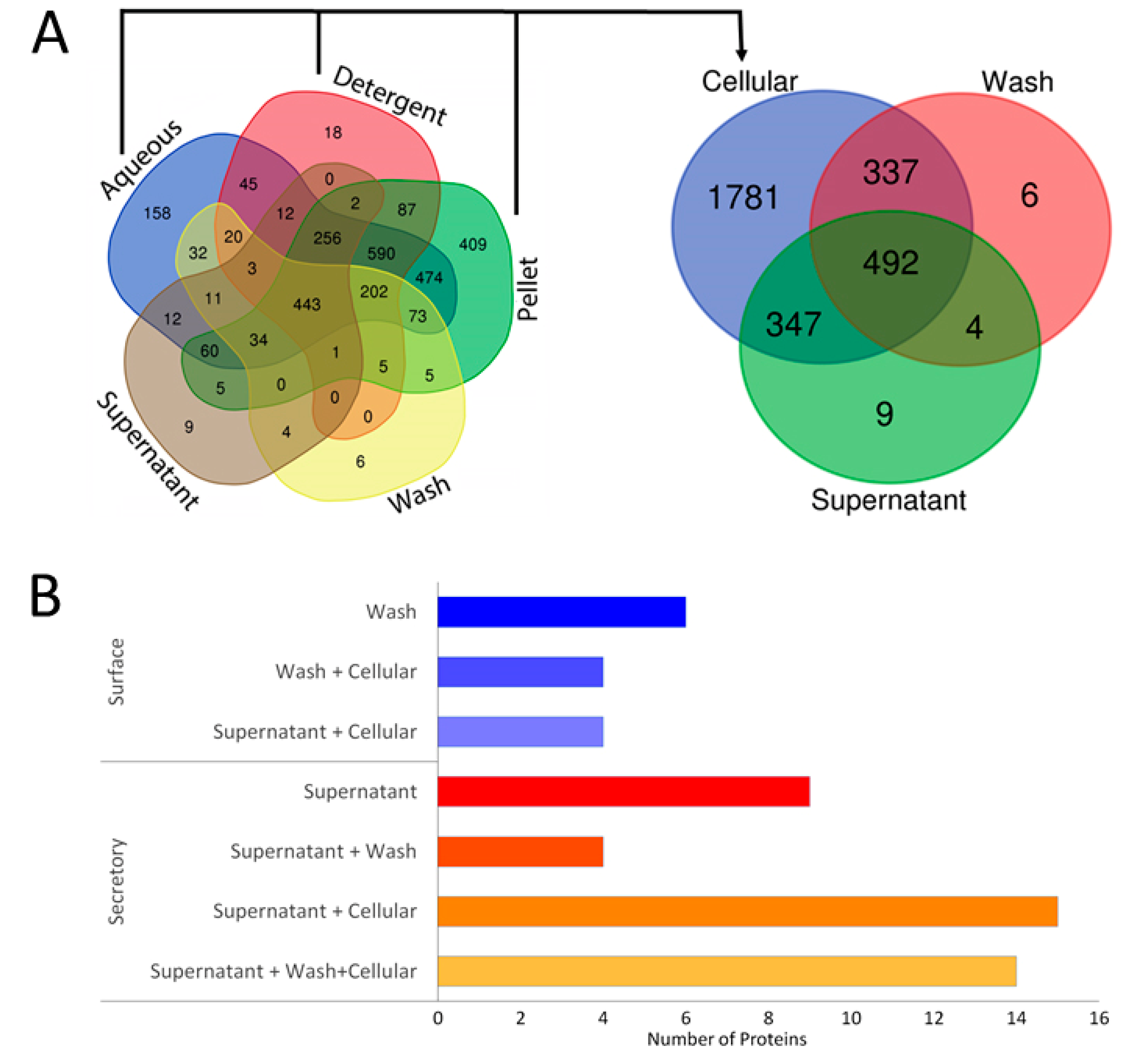
2.1. Bioinformatics Analysis of Secretory Proteins
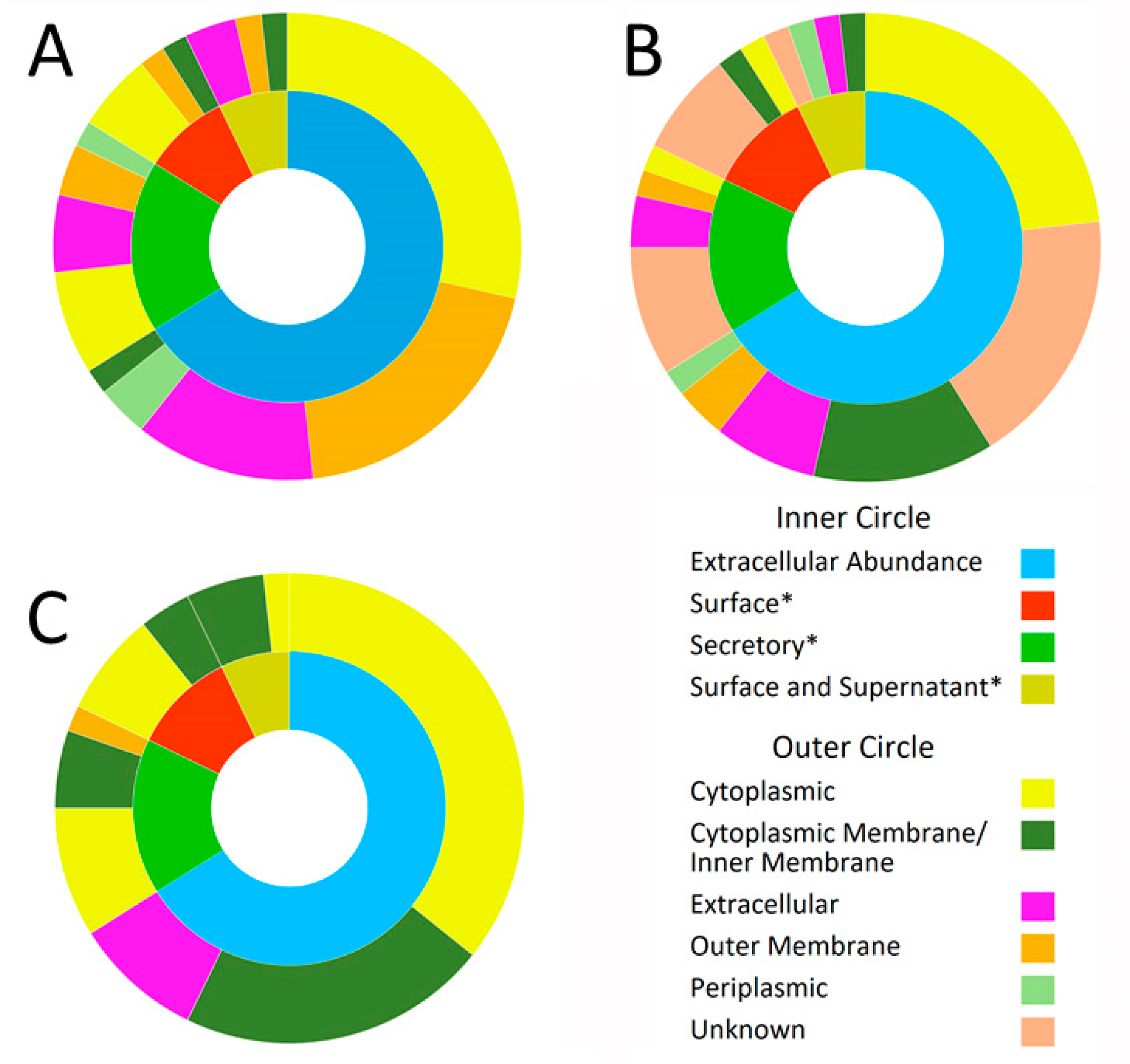
2.1.1. Prediction of Subcellular Location of Extracellular Proteins Identified in Leptospira
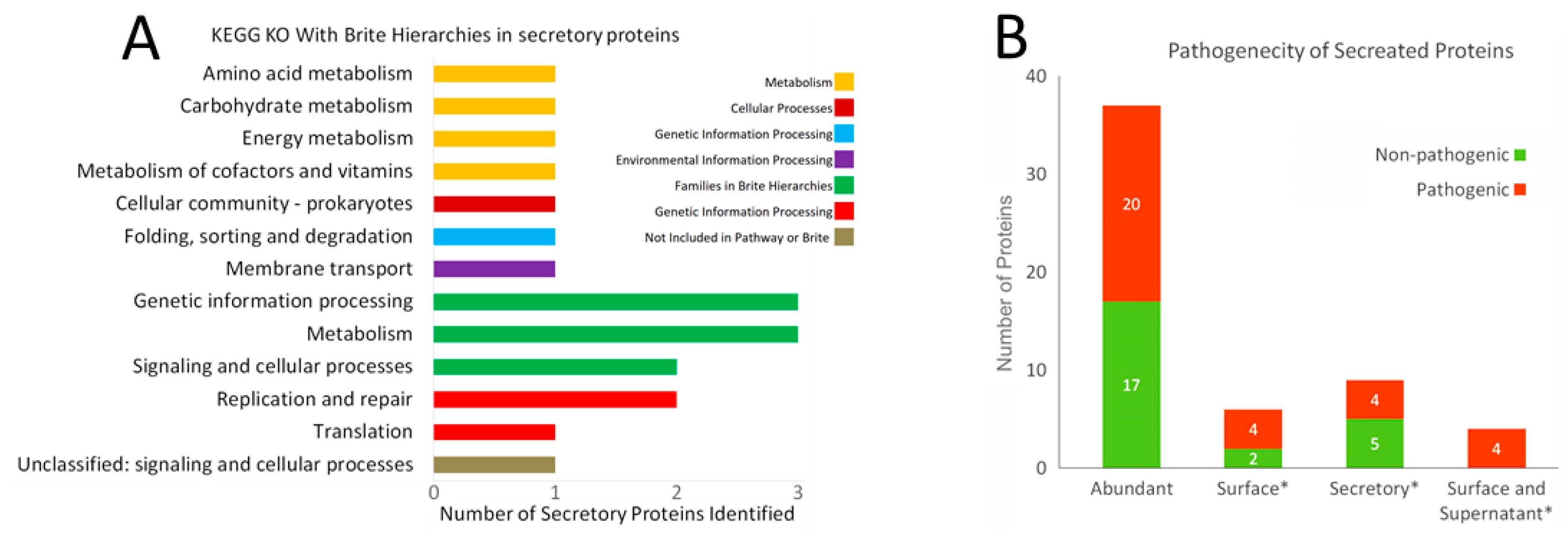
2.1.2. Prediction of Protein Function and Pathogenic Nature
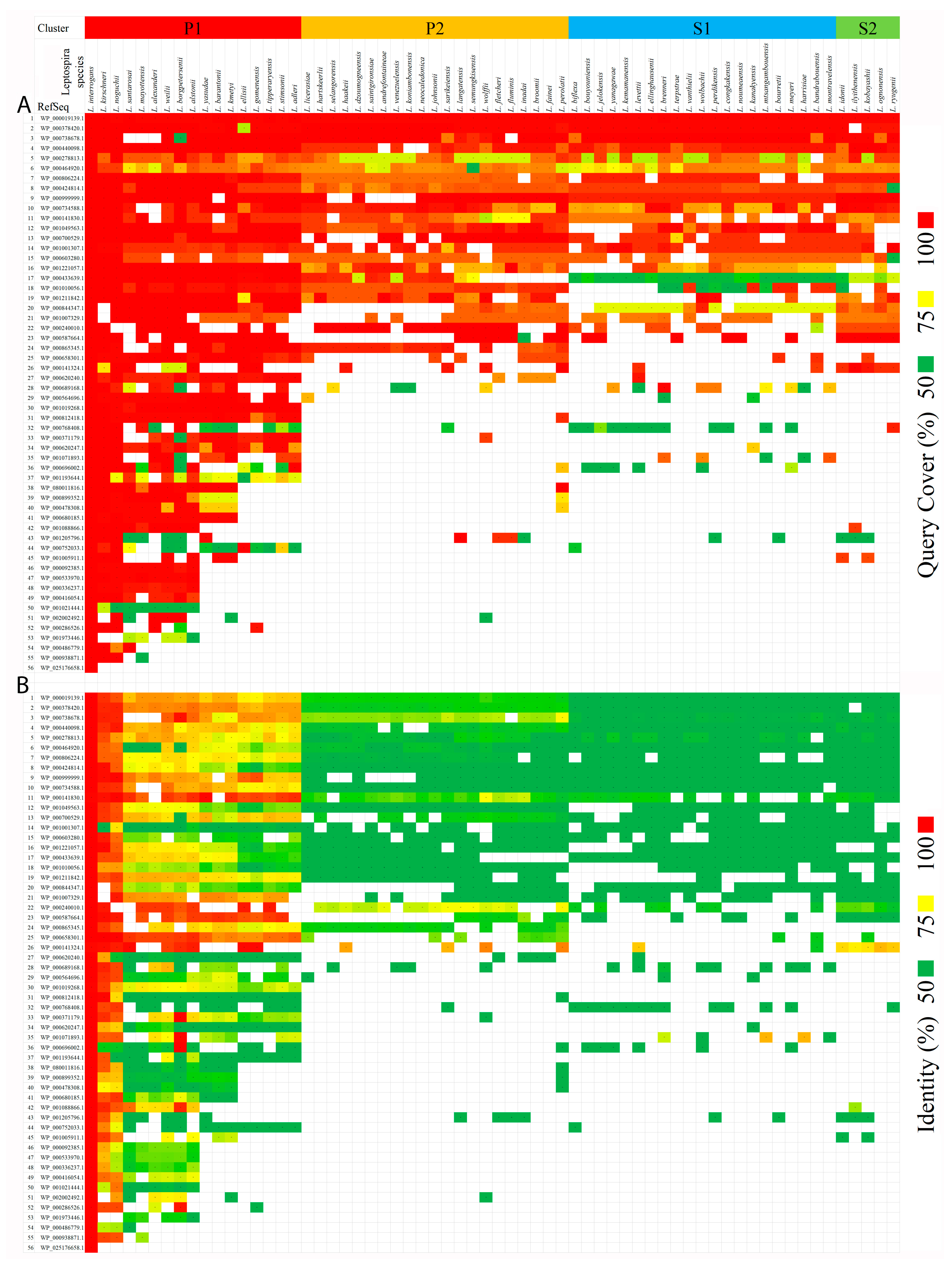
2.1.3. Identification of Orthologous Proteins
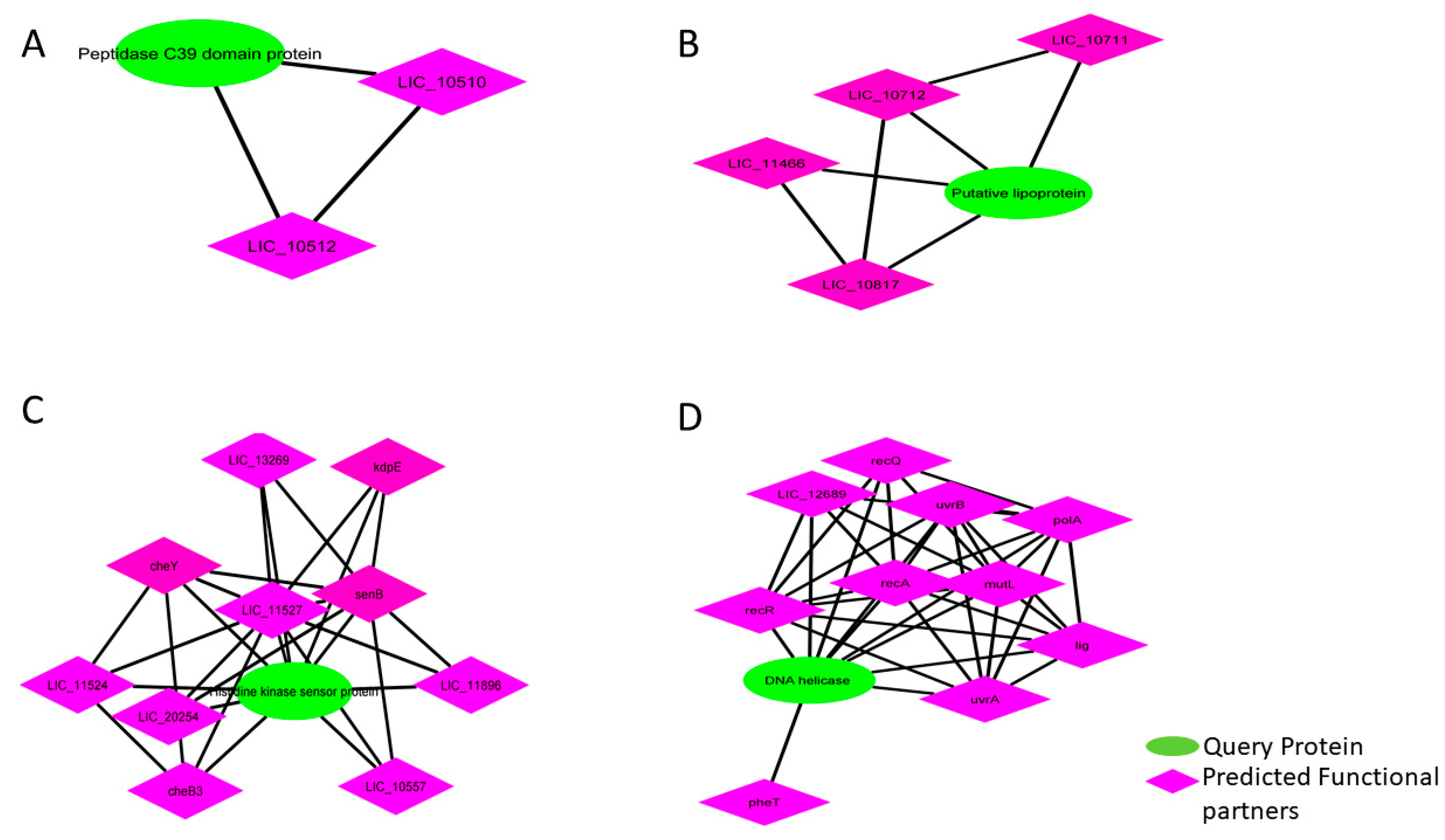
2.1.4. Determination of Protein Interactions
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Leptospira Strain and Culture
4.3. Enrichment of Surface and Extracellular/Secreted Protein
4.4. Extraction of Extracellular/Secreted Proteins
4.5. Extraction of Surface Proteins
4.6. Triton X-114 Extraction
4.7. Mass Spectrometry Analysis
4.7.1. In-Solution Digestion
4.7.2. Basic pH RPLC Based Fractionation
4.7.3. LC-MS/MS Analysis
4.7.4. MS/MS Data Analysis
4.7.5. Data Availability
4.7.6. Bioinformatics Analysis
Sub-Cellular Localization
Prediction of Protein Function and Pathogenic Nature
Identification of Orthologous Proteins
Analysis of Interacting Proteins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Karpagam, K.B.; Ganesh, B. Leptospirosis: A Neglected Tropical Zoonotic Infection of Public Health Importance—An Updated Review. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 835–846. [Google Scholar] [CrossRef] [PubMed]
- Samrot, A.V.; Sean, T.C.; Bhavya, K.S.; Sahithya, C.S.; Chandrasekaran, S.; Palanisamy, R.; Robinson, E.R.; Subbiah, S.K.; Mok, P.L. Leptospiral Infection, Pathogenesis and Its Diagnosis—A Review. Pathogens 2021, 10, 145. [Google Scholar] [CrossRef]
- Nogueira, S.V.; Backstedt, B.T.; Smith, A.A.; Qin, J.H.; Wunder, E.A.; Ko, A.; Pal, U. Leptospira Interrogans Enolase Is Secreted Extracellularly and Interacts with Plasminogen. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Asrat, S.; Davis, K.M.; Isberg, R.R. Modulation of the Host Innate Immune and Inflammatory Response by Translocated Bacterial Proteins. Cell. Microbiol. 2015, 17, 785–795. [Google Scholar] [CrossRef] [PubMed]
- Depluverez, S.; Devos, S.; Devreese, B. The Role of Bacterial Secretion Systems in the Virulence of Gram-Negative Airway Pathogens Associated with Cystic Fibrosis. Front. Microbiol. 2016, 7, 1336. [Google Scholar] [CrossRef]
- Lebrun, I.; Marques-Porto, R.; Pereira, A.; Pereira, A.; Perpetuo, E. Bacterial Toxins: An Overview on Bacterial Proteases and Their Action as Virulence Factors. Mini-Rev. Med. Chem. 2009, 9, 820–828. [Google Scholar] [CrossRef]
- da Silva, L.B.; Menezes, M.C.; Kitano, E.S.; Oliveira, A.K.; Abreu, A.G.; Souza, G.O.; Heinemann, M.B.; Isaac, L.; Fraga, T.R.; Serrano, S.M.T.; et al. Leptospira Interrogans Secreted Proteases Degrade Extracellular Matrix and Plasma Proteins From the Host. Front. Cell. Infect. Microbiol. 2018, 8, 92. [Google Scholar] [CrossRef]
- Zuerner, R.L.; Knudtson, W.; Bolin, C.A.; Trueba, G. Characterization of Outer Membrane and Secreted Proteins of Leptospira Interrogans Serovar Pomona. Microb. Pathog. 1991, 10, 311–322. [Google Scholar] [CrossRef]
- Wang, H.; Wu, Y.; Ojcius, D.M.; Yang, X.F.; Zhang, C.; Ding, S.; Lin, X.; Yan, J. Leptospiral Hemolysins Induce Proinflammatory Cytokines through Toll-like Receptor 2-and 4-Mediated JNK and NF-κB Signaling Pathways. PLoS ONE 2012. [Google Scholar] [CrossRef]
- Bernheimer, A.W.; Bey, R.F. Copurification of Leptospira Interrogans Serovar Pomona Hemolysin and Sphingomyelinase C. Infect. Immun. 1986, 54, 262–264. [Google Scholar] [CrossRef]
- Carvalho, E.; Barbosa, A.S.; Gómez, R.M.; Oliveira, M.L.S.; Romero, E.C.; Gonçales, A.P.; Morais, Z.M.; Vasconcellos, S.A.; Ho, P.L. Evaluation of the Expression and Protective Potential of Leptospiral Sphingomyelinases. Curr. Microbiol. 2010, 60, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, J.; Medeiros, M.A.; Sanchez, Y.; Werneid, K.F.; Ko, A.I. Osmotic Regulation of Expression of Two Extracellular Matrix-Binding Proteins and a Haemolysin of Leptospira Interrogans: Differential Effects on LigA and Sph2 Extracellular Release. Microbiology 2007. [Google Scholar] [CrossRef] [PubMed]
- Biswas, D.; Roy, S.; Vijayachari, P.; Sugunan, A.P.; Natarajaseenivasan, K.; Sehgal, S.C. Comparison of Immunoreactive Proteins of Commonly Circulating Serogroups of Leptospira in Andaman Islands, India. Indian J. Med. Res. 2005, 121, 151–158. [Google Scholar] [PubMed]
- Natarajaseenivasan, K.; Vijayachari, P.; Sugunan, A.P.; Sharma, S.; Sehgal, S.C. Leptospiral Proteins Expressed during Acute & Convalescent Phases of Human Leptospirosis. Indian J. Med. Res. 2004, 120, 151–159. [Google Scholar]
- Pietrocola, G.; Nobile, G.; Rindi, S.; Speziale, P. Staphylococcus Aureus Manipulates Innate Immunity through Own and Host-Expressed Proteases. Front. Cell. Infect. Microbiol. 2017, 7, 166. [Google Scholar] [CrossRef]
- Galyov, E.E.; Håkansson, S.; Forsberg, Å.; Wolf-Watz, H. A Secreted Protein Kinase of Yersinia Pseudotuberculosis Is an Indispensable Virulence Determinant. Nature 1993, 361, 730–732. [Google Scholar] [CrossRef]
- Martinez, E.; Huc-Brandt, S.; Brelle, S.; Allombert, J.; Cantet, F.; Gannoun-Zaki, L.; Burette, M.; Martin, M.; Letourneur, F.; Bonazzi, M.; et al. The Secreted Protein Kinase CstK from Coxiella Burnetii Influences Vacuole Development and Interacts with the GTPase-Activating Host Protein TBC1D5. J. Biol. Chem. 2020, 295, 7391–7403. [Google Scholar] [CrossRef]
- Zhang, Y.; Lai, B.S.; Juhas, M.; Zhang, Y. Toxoplasma Gondii Secretory Proteins and Their Role in Invasion and Pathogenesis. Microbiol. Res. 2019, 227, 126293. [Google Scholar] [CrossRef]
- Lima, T.S.; Lodoen, M.B. Mechanisms of Human Innate Immune Evasion by Toxoplasma Gondii. Front. Cell. Infect. Microbiol. 2019, 9. [Google Scholar] [CrossRef]
- Gomez, S.; Adalid-Peralta, L.; Palafox-Fonseca, H.; Cantu-Robles, V.A.; Soberón, X.; Sciutto, E.; Fragoso, G.; Bobes, R.J.; Laclette, J.P.; Del Pozo Yauner, L.; et al. Genome Analysis of Excretory/secretory Proteins in Taenia Solium Reveals Their Abundance of Antigenic Regions (AAR). Sci. Rep. 2015, 5. [Google Scholar] [CrossRef]
- Rana, A.; Thakur, S.; Bhardwaj, N.; Kumar, D.; Akhter, Y. Excavating the Surface-Associated and Secretory Proteome of Mycobacterium Leprae for Identifying Vaccines and Diagnostic Markers Relevant Immunodominant Epitopes. Pathog. Dis. 2016, 74. [Google Scholar] [CrossRef]
- Thoduvayil, S.; Dhandapani, G.; Brahma, R.; Devasahayam Arokia Balaya, R.; Mangalaparthi, K.K.; Patel, K.; Kumar, M.; Tennyson, J.; Satheeshkumar, P.K.; Kulkarni, M.J.; et al. Triton X-114 Fractionated Subcellular Proteome of Leptospira Interrogans Shows Selective Enrichment of Pathogenic and Outer Membrane Proteins in the Detergent Fraction. Proteomics 2020, 20, e2000170. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.T.; Schiettekatte, O.; Goarant, C.; Neela, V.K.; Bernet, E.; Thibeaux, R.; Ismail, N.; Mohd Khalid, M.K.N.; Amran, F.; Masuzawa, T.; et al. Revisiting the Taxonomy and Evolution of Pathogenicity of the Genus Leptospira through the Prism of Genomics. PLoS Negl. Trop. Dis. 2019, 13, e0007270. [Google Scholar] [CrossRef] [PubMed]
- Passos da Silva, D.; Schofield, M.; Parsek, M.; Tseng, B. An Update on the Sociomicrobiology of Quorum Sensing in Gram-Negative Biofilm Development. Pathogens 2017, 6, 51. [Google Scholar] [CrossRef]
- Viratyosin, W.; Ingsriswang, S.; Pacharawongsakda, E.; Palittapongarnpim, P. Genome-Wide Subcellular Localization of Putative Outer Membrane and Extracellular Proteins in Leptospira Interrogans Serovar Lai Genome Using Bioinformatics Approaches. BMC Genom. 2008. [Google Scholar] [CrossRef] [PubMed]
- Vemulapalli, R.; Duncan, A.J.; Boyle, S.M.; Sriranganathan, N.; Toth, T.E.; Schurig, G.G. Cloning and Sequencing of yajC and secDHomologs of Brucella Abortus and Demonstration of Immune Responses to YajC in Mice Vaccinated with B. Abortus RB51. Infect. Immun. 1998, 66, 5684–5691. [Google Scholar] [CrossRef] [PubMed]
- Stock, A.M.; Robinson, V.L.; Goudreau, P.N. Two-Component Signal Transduction. Annu. Rev. Biochem. 2000, 69, 183–215. [Google Scholar] [CrossRef] [PubMed]
- Falke, J.J.; Bass, R.B.; Butler, S.L.; Chervitz, S.A.; Danielson, M.A. The Two-Component Signaling Pathway of Bacterial Chemotaxis: A Molecular View of Signal Transduction by Receptors, Kinases, and Adaptation Enzymes. Annu. Rev. Cell Dev. Biol. 1997, 13, 457–512. [Google Scholar] [CrossRef]
- Wolanin, P.M.; Thomason, P.A.; Stock, J.B. Histidine Protein Kinases: Key Signal Transducers Outside the Animal Kingdom. Genome Biol. 2002, 3. [Google Scholar] [CrossRef]
- Welch, M.; Chinardet, N.; Mourey, L.; Birck, C.; Samana, J.P. Structure of the CheY-Binding Domain of Histidine Kinase CheA in Complex with CheY. Nat. Struct. Biol. 1998, 5, 25–29. [Google Scholar] [CrossRef]
- Lambert, A.; Wong Ng, J.; Picardeau, M. Gene Inactivation of a Chemotaxis Operon in the Pathogen Leptospira Interrogans. FEMS Microbiol. Lett. 2015, 362, 1–8. [Google Scholar] [CrossRef]
- Matilla, M.A.; Krell, T. The Effect of Bacterial Chemotaxis on Host Infection and Pathogenicity. FEMS Microbiol. Rev. 2018, 42, 40–67. [Google Scholar] [CrossRef]
- Truglio, J.J.; Croteau, D.L.; Skorvaga, M.; DellaVecchia, M.J.; Theis, K.; Mandavilli, B.S.; Van Houten, B.; Kisker, C. Interactions between UvrA and UvrB: The Role of UvrB’s Domain 2 in Nucleotide Excision Repair. EMBO J. 2004, 23, 2498–2509. [Google Scholar] [CrossRef]
- Crowley, D.; Boubriak, I.; Berquist, B.; Clark, M.; Richard, E.; Sullivan, L.; DasSarma, S.; McCready, S. The uvrA, uvrB and uvrC Genes Are Required for Repair of Ultraviolet Light Induced DNA Photoproducts in Halobacterium Sp. NRC-1. Saline Syst. 2006, 2, 1–13. [Google Scholar] [CrossRef][Green Version]
- Yamaguchi, M.; Dao, V.; Modrich, P. MutS and MutL Activate DNA Helicase II in a Mismatch-Dependent Manner. J. Biol. Chem. 1998, 273, 9197–9201. [Google Scholar] [CrossRef]
- Bernstein, K.A.; Gangloff, S.; Rothstein, R. The RecQ DNA Helicases in DNA Repair. Annu. Rev. Genet. 2010, 44, 393–417. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Rawlings, N.D.; Farr, C.L.; Chiu, H.-J.; Grant, J.C.; Jaroszewski, L.; Klock, H.E.; Knuth, M.W.; Miller, M.D.; Weekes, D.; et al. Structural and Sequence Analysis of Imelysin-like Proteins Implicated in Bacterial Iron Uptake. PLoS ONE 2011, 6, e21875. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Rathinam, S.R.; Priya, C.G.; Muthukkaruppan, V.R.; Stevenson, B.; Timoney, J.F. LruA and LruB Antibodies in Sera of Humans with Leptospiral Uveitis. Clin. Vaccine Immunol. 2008, 15, 1019–1023. [Google Scholar] [CrossRef]
- Verma, A.; Kumar, P.; Babb, K.; Timoney, J.F.; Stevenson, B. Cross-Reactivity of Antibodies against Leptospiral Recurrent Uveitis-Associated Proteins A and B (LruA and LruB) with Eye Proteins. PLoS Negl. Trop. Dis. 2010, 4, e778. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Artiushin, S.; Matsunaga, J.; Haake, D.A.; Timoney, J.F. LruA and LruB, Novel Lipoproteins of Pathogenic Leptospira Interrogans Associated with Equine Recurrent Uveitis. Infect. Immun. 2005, 73, 7259–7266. [Google Scholar] [CrossRef] [PubMed]
- Eshghi, A.; Pappalardo, E.; Hester, S.; Thomas, B.; Pretre, G.; Picardeau, M. Pathogenic Leptospira Interrogans Exoproteins Are Primarily Involved in Heterotrophic Processes. Infect. Immun. 2015, 83, 3061–3073. [Google Scholar] [CrossRef]
- Havarstein, L.S.; Diep, D.B.; Nes, I.F. A Family of Bacteriocin ABC Transporters Carry out Proteolytic Processing of Their Substrates Concomitant with Export. Mol. Microbiol. 1995, 16, 229–240. [Google Scholar] [CrossRef]
- Wang, J.; Yang, B.; An, Y.; Marquez-Lago, T.; Leier, A.; Wilksch, J.; Hong, Q.; Zhang, Y.; Hayashida, M.; Akutsu, T.; et al. Systematic Analysis and Prediction of Type IV Secreted Effector Proteins by Machine Learning Approaches. Brief. Bioinform. 2019, 20, 931–951. [Google Scholar] [CrossRef]
- Wang, J.; Yang, B.; Leier, A.; Marquez-Lago, T.T.; Hayashida, M.; Rocker, A.; Zhang, Y.; Akutsu, T.; Chou, K.-C.; Strugnell, R.A.; et al. Bastion6: A Bioinformatics Approach for Accurate Prediction of Type VI Secreted Effectors. Bioinformatics 2018, 34, 2546–2555. [Google Scholar] [CrossRef]
- Wang, J.; Li, J.; Yang, B.; Xie, R.; Marquez-Lago, T.T.; Leier, A.; Hayashida, M.; Akutsu, T.; Zhang, Y.; Chou, K.-C.; et al. Bastion3: A Two-Layer Ensemble Predictor of Type III Secreted Effectors. Bioinformatics 2019, 35, 2017–2028. [Google Scholar] [CrossRef] [PubMed]
- Gallique, M.; Bouteiller, M.; Merieau, A. The Type VI Secretion System: A Dynamic System for Bacterial Communication? Front. Microbiol. 2017, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Garcia, F.; Ruiz-Perez, F.; Cataldi, Á.; Larzábal, M. Type VI Secretion System in Pathogenic Escherichia Coli: Structure, Role in Virulence, and Acquisition. Front. Microbiol. 2019, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Mariano, G.; Trunk, K.; Williams, D.J.; Monlezun, L.; Strahl, H.; Pitt, S.J.; Coulthurst, S.J. A Family of Type VI Secretion System Effector Proteins That Form Ion-Selective Pores. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Diallo, A.; Foster, H.R.; Gromek, K.A.; Perry, T.N.; Dujeancourt, A.; Krasteva, P.V.; Gubellini, F.; Falbel, T.G.; Burton, B.M.; Fronzes, R. Bacterial Transformation: ComFA Is a DNA-Dependent ATPase That Forms Complexes with ComFC and DprA. Mol. Microbiol. 2017, 105, 741–754. [Google Scholar] [CrossRef] [PubMed]
- Knizewski, L.; Steczkiewicz, K.; Kuchta, K.; Wyrwicz, L.; Plewczynski, D.; Kolinski, A.; Rychlewski, L.; Ginalski, K. Uncharacterized DUF1574 Leptospira Proteins Are SGNH Hydrolases. Cell Cycle 2008, 7, 542–544. [Google Scholar] [CrossRef]
- Benaroudj, N.; Saul, F.; Bellalou, J.; Miras, I.; Weber, P.; Bondet, V.; Murray, G.L.; Adler, B.; Ristow, P.; Louvel, H.; et al. Structural and Functional Characterization of an Orphan ATP-Binding Cassette ATPase Involved in Manganese Utilization and Tolerance in Leptospira Spp. J. Bacteriol. 2013, 195, 5583–5591. [Google Scholar] [CrossRef] [PubMed]
- Thulasiraman, V.; Lin, S.; Gheorghiu, L.; Lathrop, J.; Lomas, L.; Hammond, D.; Boschetti, E. Reduction of the Concentration Difference of Proteins in Biological Liquids Using a Library of Combinatorial Ligands. Electrophoresis 2005, 26, 3561–3571. [Google Scholar] [CrossRef]
- Guerrier, L.; Thulasiraman, V.; Castagna, A.; Fortis, F.; Lin, S.; Lomas, L.; Righetti, P.G.; Boschetti, E. Reducing Protein Concentration Range of Biological Samples Using Solid-Phase Ligand Libraries☆. J. Chromatogr. B 2006, 833, 33–40. [Google Scholar] [CrossRef]
- Patil, A.H.; Datta, K.K.; Behera, S.K.; Kasaragod, S.; Pinto, S.M.; Koyangana, S.G.; Mathur, P.P.; Gowda, H.; Pandey, A.; Prasad, T.S.K. Dissecting Candida Pathobiology: Post-Translational Modifications on the Candida Tropicalis Proteome. Omi. A J. Integr. Biol. 2018, 22, 544–552. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.-S.; Lin, C.-J.; Hwang, J.-K. Predicting Subcellular Localization of Proteins for Gram-Negative Bacteria by Support Vector Machines Based on N -Peptide Compositions. Protein Sci. 2004, 13, 1402–1406. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.-S.; Chen, Y.-C.; Lu, C.-H.; Hwang, J.-K. Prediction of Protein Subcellular Localization. Proteins Struct. Funct. Bioinforma. 2006, 64, 643–651. [Google Scholar] [CrossRef]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved Protein Subcellular Localization Prediction with Refined Localization Subcategories and Predictive Capabilities for All Prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Profiti, G.; Casadio, R. BUSCA: An Integrative Web Server to Predict Subcellular Localization of Proteins. Nucleic Acids Res. 2018, 46, W459–W466. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New Perspectives on Genomes, Pathways, Diseases and Drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Kapil, R.; Dhakan, D.B.; Sharma, V.K. MP3: A Software Tool for the Prediction of Pathogenic Proteins in Genomic and Metagenomic Data. PLoS ONE 2014, 9, e93907. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Lata, K.S.; Sharma, P.; Bhairappanavar, S.B.; Soni, S.; Das, J. Inferring Pathogen-Host Interactions between Leptospira Interrogans and Homo Sapiens Using Network Theory. Sci. Rep. 2019, 9, 1434. [Google Scholar] [CrossRef] [PubMed]

| Sl. No. | Accession | Gene Names (Ordered Locus) | Description | Total Value of Triton X-114 Fractions (C) | Wash (W) (Surface) | Supernatant (S) (Secretory) | Abundance of Extracellular Proteins (EC) = (W+S)/C |
|---|---|---|---|---|---|---|---|
| 1 | WP_000286526.1 | hypothetical protein | - | 0.1 | - | Ex. W | |
| 2 | WP_000424814.1 | ComF family protein | - | 8.4 | - | Ex. W | |
| 3 | WP_000486779.1 | LIC_10697 | hypothetical protein | - | 2.2 | - | Ex. W |
| 4 | WP_000700529.1 | hypothetical protein | - | 0.5 | - | Ex. W | |
| 5 | WP_000865345.1 | LIC_13053 | fatty acid desaturase | - | 0.3 | - | Ex. W |
| 6 | WP_001973446.1 | hypothetical protein | - | 0.7 | - | Ex. W | |
| 7 | WP_000371179.1 | LIC_13177 | hypothetical protein | - | - | 5.9 | Ex. S |
| 8 | WP_000416054.1 | LIC_11345 | TonB-dependent siderophore receptor | - | - | 0.04 | Ex. S |
| 9 | WP_000738678.1 | LIC_12988 | lipase | - | - | 14.3 | Ex. S |
| 10 | WP_000812418.1 | LIC_10645 | hypothetical protein | - | - | 1.4 | Ex. S |
| 11 | WP_000844347.1 | LIC_10346 | SGNH/GDSL hydrolase family protein | - | - | 1.2 | Ex. S |
| 12 | WP_000899352.1 | sphingomyelin phosphodiesterase | - | - | 1.7 | Ex. S | |
| 13 | WP_001021444.1 | DUF1563 domain-containing protein | - | - | 18.2 | Ex. S | |
| 14 | WP_001088866.1 | LIC_12191 | NUDIX hydrolase | - | - | 2.3 | Ex. S |
| 15 | WP_025176658.1 | hypothetical protein | - | - | 17.3 | Ex. S | |
| 16 | WP_000336237.1 | LIC_12715 | DUF1561 domain-containing protein | - | 0.7 | 3.2 | Ex. EC |
| 17 | WP_000533970.1 | LIC_12986 | DUF1561 domain-containing protein | - | 0.5 | 3.9 | Ex. EC |
| 18 | WP_001193644.1 | hypothetical protein | - | 2.1 | 4.7 | Ex. EC | |
| 19 | WP_001211842.1 | LIC_11088 | di-heme enzyme | - | 0.4 | 4.8 | Ex. EC |
| 20 | WP_000433639.1 | LIC_11528 | PAS domain S-box protein | 0.5 | 941.1 | 7.0 | 1771.4 |
| 21 | WP_000378420.1 | LIC_11624 | ATPase AAA | 21.5 | 813.4 | 1871.9 | 124.9 |
| 22 | WP_001049563.1 | LIC_10713 | peptidase M75 | 23.5 | 11.8 | 1833.1 | 78.5 |
| 23 | WP_001205796.1 | LIC_10511 | hypothetical protein | 0.1 | - | 4.0 | 75.6 |
| 24 | WP_000938871.1 | LIC_20255 | hypothetical protein | 1.7 | 42.3 | 29.6 | 42.5 |
| 25 | WP_000752033.1 | LIC_10370 | hypothetical protein | 2.1 | 10.8 | 72.9 | 38.9 |
| 26 | WP_001071893.1 | LIC_10704 | hypothetical protein | 2.9 | - | 101.8 | 35.2 |
| 27 | WP_000689168.1 | SGNH/GDSL hydrolase family protein | 2.0 | - | 55.3 | 28.0 | |
| 28 | WP_002002492.1 | LIC_11904 | hypothetical protein | 3.3 | - | 86.1 | 26.3 |
| 29 | WP_001010056.1 | hypothetical protein | 3.6 | - | 94.0 | 26.2 | |
| 30 | WP_000141324.1 | LIC_11265 | MULTISPECIES: DUF1858 domain-containing protein | 5.4 | - | 139.8 | 25.9 |
| 31 | WP_000620240.1 | LIC_10371 | hypothetical protein | 12.1 | 2.4 | 246.8 | 20.7 |
| 32 | WP_080011816.1 | sphingomyelin phosphodiesterase | 0.4 | - | 7.0 | 19.4 | |
| 33 | WP_000806224.1 | LIC_13164 | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase | 11.1 | 1.0 | 206.8 | 18.6 |
| 34 | WP_000768408.1 | LIC_12891 | hypothetical protein | 2.2 | 1.6 | 35.9 | 16.7 |
| 35 | WP_000680185.1 | LIC_10365 | DUF1565 domain-containing protein | 0.6 | 1.3 | 7.6 | 14.7 |
| 36 | WP_000620247.1 | LIC_10373 | hypothetical protein | 4.6 | 2.5 | 65.8 | 14.7 |
| 37 | WP_000564696.1 | LIC_10183 | hypothetical protein | 0.8 | 11.0 | - | 14.4 |
| 38 | WP_000999999.1 | LIC_11240 | ATP synthase subunit delta | 205.7 | 0.7 | 2731.9 | 13.3 |
| 39 | WP_000696002.1 | LIC_13248 | DUF1554 domain-containing protein | 0.7 | - | 8.6 | 12.6 |
| 40 | WP_000092385.1 | LIC_12791 | DUF1561 domain-containing protein | 0.6 | - | 6.7 | 11.7 |
| 41 | WP_000019139.1 | LIC_12413 | DUF115 domain-containing protein | 3.4 | - | 38.3 | 11.2 |
| 42 | WP_000603280.1 | LIC_20247 | lytic transglycosylase domain-containing protein | 0.7 | - | 6.9 | 10.3 |
| 43 | WP_000734588.1 | hypothetical protein | 11.2 | - | 92.3 | 8.2 | |
| 44 | WP_000440098.1 | DUF2203 domain-containing protein | 3.1 | 20.5 | - | 6.6 | |
| 45 | WP_000141830.1 | LIC_12540 | MULTISPECIES: preprotein translocase subunit YajC | 1067.4 | 1534.6 | 4361.7 | 5.5 |
| 46 | WP_001001307.1 | hypothetical protein | 0.5 | 2.4 | - | 5.2 | |
| 47 | WP_001019268.1 | LIC_12755 | MULTISPECIES: hypothetical protein | 94.8 | 21.7 | 289.9 | 3.3 |
| 48 | WP_001005911.1 | LIC_10552 | hybrid sensor histidine kinase/response regulator | 0.8 | - | 2.7 | 3.2 |
| 49 | WP_000587664.1 | VOC family protein | 198.7 | 35.6 | 428.8 | 2.3 | |
| 50 | WP_000658301.1 | DNA starvation/stationary phase protection protein | 33.3 | 54.1 | 22.1 | 2.3 | |
| 51 | WP_000278813.1 | DUF192 domain-containing protein | 2.0 | 2.9 | 1.2 | 2.1 | |
| 52 | WP_001221057.1 | LIC_10711 | hypothetical protein | 5.1 | 8.7 | - | 1.7 |
| 53 | WP_001007329.1 | LIC_12437 | hypothetical protein | 94.4 | 8.0 | 151.1 | 1.7 |
| 54 | WP_000464920.1 | stage II sporulation protein E | 2.6 | - | 4.1 | 1.6 | |
| 55 | WP_000478308.1 | sphingomyelin phosphodiesterase | 1.8 | 0.7 | 2.1 | 1.6 | |
| 56 | WP_000240010.1 | LIC_11555 | 30S ribosomal protein S16 | 27.9 | - | 42.3 | 1.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarma, A.; Gunasekaran, D.; Rex, D.A.B.; Sikha, T.; Phukan, H.; Kiran, K.M.; Pinto, S.M.; Prasad, T.S.K.; Madanan, M.G. Extracellular Proteome Analysis Shows the Abundance of Histidine Kinase Sensor Protein, DNA Helicase, Putative Lipoprotein Containing Peptidase M75 Domain and Peptidase C39 Domain Protein in Leptospira interrogans Grown in EMJH Medium. Pathogens 2021, 10, 852. https://doi.org/10.3390/pathogens10070852
Sarma A, Gunasekaran D, Rex DAB, Sikha T, Phukan H, Kiran KM, Pinto SM, Prasad TSK, Madanan MG. Extracellular Proteome Analysis Shows the Abundance of Histidine Kinase Sensor Protein, DNA Helicase, Putative Lipoprotein Containing Peptidase M75 Domain and Peptidase C39 Domain Protein in Leptospira interrogans Grown in EMJH Medium. Pathogens. 2021; 10(7):852. https://doi.org/10.3390/pathogens10070852
Chicago/Turabian StyleSarma, Abhijit, Dhandapani Gunasekaran, Devasahayam Arokia Balaya Rex, Thoduvayil Sikha, Homen Phukan, Kumar Mangalaparthi Kiran, Sneha M. Pinto, Thottethodi Subrahmanya Keshava Prasad, and Madathiparambil G. Madanan. 2021. "Extracellular Proteome Analysis Shows the Abundance of Histidine Kinase Sensor Protein, DNA Helicase, Putative Lipoprotein Containing Peptidase M75 Domain and Peptidase C39 Domain Protein in Leptospira interrogans Grown in EMJH Medium" Pathogens 10, no. 7: 852. https://doi.org/10.3390/pathogens10070852
APA StyleSarma, A., Gunasekaran, D., Rex, D. A. B., Sikha, T., Phukan, H., Kiran, K. M., Pinto, S. M., Prasad, T. S. K., & Madanan, M. G. (2021). Extracellular Proteome Analysis Shows the Abundance of Histidine Kinase Sensor Protein, DNA Helicase, Putative Lipoprotein Containing Peptidase M75 Domain and Peptidase C39 Domain Protein in Leptospira interrogans Grown in EMJH Medium. Pathogens, 10(7), 852. https://doi.org/10.3390/pathogens10070852






