Multi-Epitope Vaccine Design Using an Immunoinformatic Approach for SARS-CoV-2
Abstract
1. Introduction
2. Results
2.1. Prediction of B-Cell Epitopes
2.2. Prediction of T-Cell Epitopes
2.3. Multi-Epitope Vaccine Design
2.4. Interaction of Predicted Peptides with HLA Alleles
2.5. Humoral Immune Responses to SARS-CoV-2 S Protein
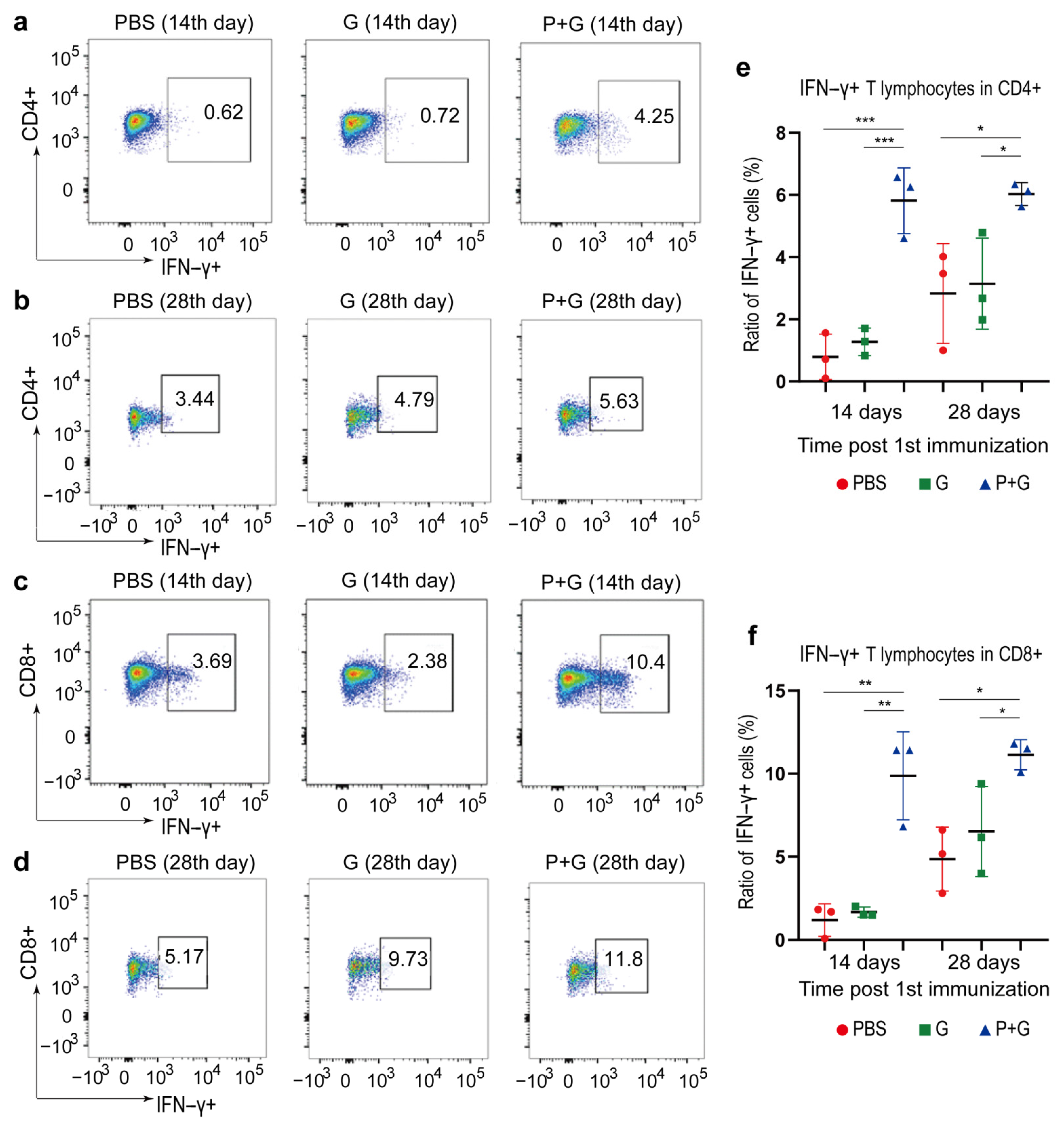
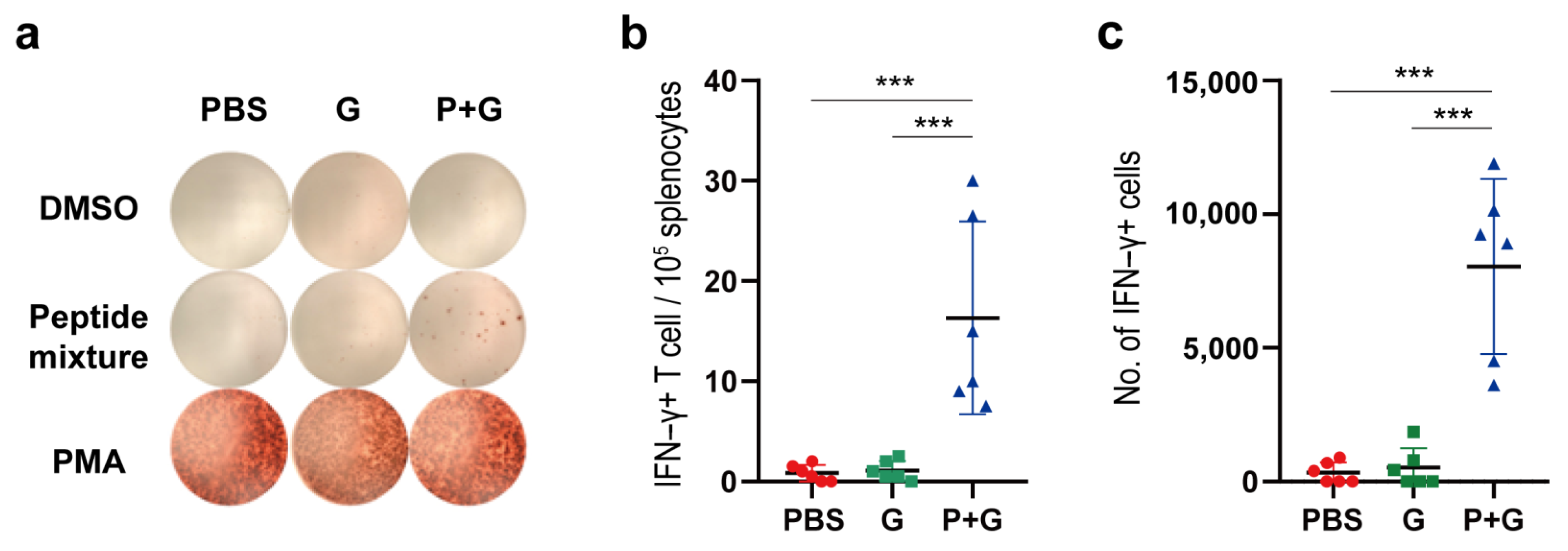
2.6. Cellular Immune Responses to SARS-CoV-2 S Proteins
3. Discussion
4. Methods
4.1. Data Retrieval
4.2. B-Cell Epitope Prediction
4.3. T-Cell Epitope Prediction
4.4. Vaccine Peptide Design
4.5. Structural Analysis
4.6. Immuno-Stimulation of B Lymphocytes
4.7. Immuno-Stimulation of T-Cells
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Surveillance Case Definitions for Human Infection with Novel Coronavirus (Ncov): Interim Guidance v1, January 2020; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Perlman, S. Another decade, another coronavirus. N. Engl. J. Med. 2020, 382, 760–762. [Google Scholar] [CrossRef]
- Wu, J.T.; Leung, K.; Leung, G. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: A modelling study. Lancet 2020, 395, 689–697. [Google Scholar] [CrossRef]
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Layber, C.; Leontovich, A.M.; Neuman, B.W. Severe acute respiratory syndrome-related coronavirus: The species and its viruses—A statement of the Coronavirus Study Group. bioRxiv 2020. [Google Scholar] [CrossRef]
- Coronavirus COVID-19 Global Cases by the Center for Systems Sciences and Engineering (CSSE) at Johns Hopkins University (JHU). Available online: https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html#/bda7594740fd40299423467b48e9ecf6 (accessed on 31 December 2020).
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L. A pneumonia out-break associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Benvenuto, D.; Giovannetti, M.; Ciccozzi, A.; Spoto, S.; Angeletti, S.; Ciccozzi, M. The 2019-new coronavirus epidemic: Evidence for virus evolution. J. Med. Virol. 2020, 92, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Forni, G.; Mantovani, A. COVID-19 vaccines: Where we stand and challenges ahead. Cell Death Differ. 2021, 28, 626–639. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef]
- Chen, J.; Shi, J.; Yau, T.; Liu, C.; Li, X.; Zhao, Q.; Ruan, J.; Gao, S. Bioinformatics analysis of the 2019 novel coronavirus genome. Chin. J. Bioinform. 2020, 18, 96–102. (In Chinese) [Google Scholar]
- Rahimi, A.; Mirzazadeh, A.; Tavakolpour, S. Genetics and genomics of SARS-CoV-2: A review of the literature with the special focus on genetic diversity and SARS-CoV-2 genome detection. Genomics 2021, 113, 1221–1232. [Google Scholar] [CrossRef]
- Tian, X.; Li, C.; Huang, A.; Xia, S.; Lu, S.; Shi, Z.; Lu, L.; Jiang, S.; Yang, Z.; Wu, Y.; et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020, 9, 382–385. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.; Yang, X.; Ye, L.; Chen, K.; Chan, E.W.-C.; Chen, S. Genomic and protein structure modelling analysis depicts the origin and infectivity of 2019-nCoV, a new coronavirus which caused a pneumonia outbreak in Wuhan, China. bioRxiv 2020. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- He, Y.; Lu, H.; Siddiqui, P.; Zhou, Y.; Jiang, S. Receptor-binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation-dependent epitopes that induce highly potent neutralizing antibodies. J. Immunol. 2005, 174, 4908–4915. [Google Scholar] [CrossRef]
- Babcock, G.J.; Esshaki, D.J.; Thomas, W.D.; Ambrosino, D.M. Amino Acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J. Virol. 2004, 78, 4552–4560. [Google Scholar] [CrossRef] [PubMed]
- Saif, L.J. Coronavirus immunogens. Vet. Microbiol. 1993, 37, 285–297. [Google Scholar] [CrossRef]
- Buchholz, U.J.; Bukreyev, A.; Yang, L.; Lamirande, E.W.; Murphy, B.R.; Subbarao, K.; Collins, P.L. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc. Natl. Acad. Sci. USA 2004, 101, 9804–9809. [Google Scholar] [CrossRef] [PubMed]
- Purcell, A.W.; McCluskey, J.; Rossjohn, J. More than one reason to rethink the use of peptides in vaccine design. Nat. Rev. Drug Discov. 2007, 6, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Adam, K.-H.; Kaaden, O.; Strohmaier, K. Isolation of immunizing cyanogen bromide-peptides of foot-and-mouth disease virus. Biochem. Biophys. Res. Commun. 1978, 84, 677–683. [Google Scholar] [CrossRef]
- Lerner, R.A.; Green, N.; Alexander, H.; Liu, F.T.; Sutcliffe, J.G.; Shinnick, T.M. Chemically synthesized peptides predicted from the nucleotide sequence of the hepatitis B virus genome elicit antibodies reactive with the native envelope protein of Dane particles. Proc. Natl. Acad. Sci. USA 1981, 78, 3403–3407. [Google Scholar] [CrossRef]
- Fieser, T.M.; Tainer, J.; Geysen, H.M.; Houghten, R.A.; Lerner, R.A. Influence of protein flexibility and peptide conformation on reactivity of monoclonal anti-peptide antibodies with a protein alpha-helix. Proc. Natl. Acad. Sci. USA 1987, 84, 8568–8572. [Google Scholar] [CrossRef]
- Sanchez-Trincado, J.L.; Gomez-Perosanz, M.; Reche, P.A. Fundamentals and methods for T- and B-Cell epitope prediction. J. Immunol. Res. 2017, 2017, 1–14. [Google Scholar] [CrossRef]
- Black, M.; Trent, A.; Tirrell, M.; Olive, C. Advances in the design and delivery of peptide subunit vaccines with a focus on Toll-like receptor agonists. Expert Rev. Vaccines 2010, 9, 157–173. [Google Scholar] [CrossRef]
- Sesardic, D. Synthetic peptide vaccines. J. Med. Microbiol. 1993, 39, 241–242. [Google Scholar] [CrossRef]
- Skwarczynski, M.; Toth, I. Peptide-based synthetic vaccines. Chem. Sci. 2016, 7, 842–854. [Google Scholar] [CrossRef]
- Karim, S.S.A.; de Oliveira, T. New SARS-CoV-2 variants—Clinical, public health, and vaccine implications. N. Engl. J. Med. 2021, 384, 1866–1868. [Google Scholar] [CrossRef] [PubMed]
- Grifoni, A.; Sidney, J.; Zhang, Y.; Scheuermann, R.H.; Peters, B.; Sette, A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe 2020, 27, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Majumder, K.; Gutierrez, G.J.; Gupta, D.; Mittal, B. Immuno-informatics approach for multi-epitope vaccine designing against SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Dong, R.; Chu, Z.; Yu, F.; Zha, Y. Contriving Multi-Epitope Subunit of Vaccine for COVID-19: Immunoinformatics Approaches. Front. Immunol. 2020, 11, 1784. [Google Scholar] [CrossRef] [PubMed]
- Sanami, S.; Zandi, M.; Porhossein, B.; Mobini, G.-R.; Safaei, M.; Abed, A.; Arvejeh, P.M.; Chermahini, F.A.; Alizadeh, M. Design of a multi-epitope vaccine against SARS-CoV-2 using immunoinformatics approach. Int. J. Biol. Macromol. 2020, 164, 871–883. [Google Scholar] [CrossRef]
- Martin, W.R.; Cheng, F. A rational design of a multi-epitope vaccine against SARS-CoV-2 which accounts for the glycan shield of the spike glycoprotein. J. Biomol. Struct. Dyn. 2021, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Qamar, M.T.U.; Rehman, A.; Tusleem, K.; Ashfaq, U.A.; Qasim, M.; Zhu, X.; Fatima, I.; Shahid, F.; Chen, L.-L. Designing of a next generation multiepitope based vaccine (MEV) against SARS-COV-2: Immunoinformatics and in silico approaches. PLoS ONE 2020, 15, e0244176. [Google Scholar]
- Bhatnager, R.; Bhasin, M.; Arora, J.; Dang, A.S. Epitope based peptide vaccine against SARS-COV2: An immune-informatics approach. J. Biomol. Struct. Dyn. 2020, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Ojha, R.; Gupta, N.; Naik, B.; Singh, S.; Verma, V.K.; Prusty, D.; Prajapati, V.K. High throughput and comprehensive approach to develop multiepitope vaccine against minacious COVID-19. Eur. J. Pharm. Sci. 2020, 151, 105375. [Google Scholar] [CrossRef] [PubMed]
- Ranga, V.; Niemela, E.; Tamirat, M.Z.; Eriksson, J.E.; Airenne, T.T.; Johnson, M.S. Immunogenic SARS-CoV-2 Epitopes: In silico study towards better understanding of COVID-19 disease-paving the way for vaccine development. Vaccines 2020, 8, 408. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Mai, J.; Zhou, W.; Yu, W.; Zhan, Y.; Wang, N.; Epstein, N.; Yang, Y. Immunoinformatic analysis of T- and B-Cell Epitopes for SARS-CoV-2 vaccine design. Vaccines 2020, 8, 355. [Google Scholar] [CrossRef]
- Quadeer, A.A.; Ahmed, S.F.; McKay, M.R. Epitopes targeted by T cells in convalescent COVID-19 patients. bioRxiv 2020. [Google Scholar] [CrossRef]
- Schulien, I.; Kemming, J.; Oberhardt, V.; Wild, K.; Seidel, L.M.; Killmer, S.; Daul, F.; Lago, M.S.; Decker, A.; Luxenburger, H.; et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8+ T cells. Nat. Med. 2021, 27, 78–85. [Google Scholar] [CrossRef]
- Mo, F.; Chen, R.; Wu, J.; Yang, Q.; Fang, Y.; Dong, W.; Sun, Y.; Wang, K.; Chen, S. Abstract 4422: iNeo-PRED: A hybrid-model predictor for MHC class I neoantigens. Clin. Res. 2020, 80, 442. [Google Scholar]
- Fang, Y.; Mo, F.; Shou, J.; Wang, H.; Luo, K.; Zhang, S.; Han, N.; Li, H.; Ye, S.; Zhou, Z.; et al. A pan-cancer clinical study of personalized neoantigen vaccine monotherapy in treating patients with various types of advanced solid tumors. Clin. Cancer Res. 2020, 26, 4511. [Google Scholar] [CrossRef] [PubMed]
- Rauch, S.; Jasny, E.; Schmidt, K.E.; Petsch, B. New vaccine technologies to combat outbreak situations. Front. Immunol. 2018, 9, 1963. [Google Scholar] [CrossRef]
- Zhu, F.-C.; Li, Y.-H.; Guan, X.-H.; Hou, L.-H.; Wang, W.-J.; Li, J.-X.; Wu, S.-P.; Wang, B.-S.; Wang, Z.; Wang, L.; et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: A dose-escalation, open-label, non-randomised, first-in-human trial. Lancet 2020, 395, 1845–1854. [Google Scholar] [CrossRef]
- Jackson, N.A.C.; Kester, K.E.; Casimiro, D.; Gurunathan, S.; DeRosa, F. The promise of mRNA vaccines: A biotech and industrial perspective. NPJ Vaccines 2020, 5, 11. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Vita, R.; Overton, J.A.; Greenbaum, J.A.; Ponomarenko, J.; Clark, J.D.; Cantrell, J.R.; Wheeler, D.K.; Gabbard, J.L.; Hix, D.; Sette, A.; et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015, 43, D405–D412. [Google Scholar] [CrossRef]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, H.; Yang, J.; Chou, K.-C. Prediction of linear B-cell epitopes using amino acid pair antigenicity scale. Amino Acids 2007, 33, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Almofti, Y.A.; Abd-Elrahman, K.A.; Gassmallah, S.A.E.; Salih, M.A. Multi epitopes vaccine prediction against Severe Acute Respiratory Syndrome (SARS) coronavirus using immunoinformatics approaches. Am. J. Microbiol. Res. 2018, 6, 94–114. [Google Scholar] [CrossRef]
- Hoof, I.; Peters, B.; Sidney, J.; Pedersen, L.E.; Sette, A.; Lund, O.; Buus, S.; Nielsen, M. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 2008, 61, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Lamiable, A.; Thevenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tufféry, P. PEP-FOLD3: Fasterde novostructure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef]
- Xu, X.; Yan, C.; Zou, X. MDockPeP: An ab-initio protein-peptide docking server. J. Comput. Chem. 2018, 39, 2409–2413. [Google Scholar] [CrossRef] [PubMed]
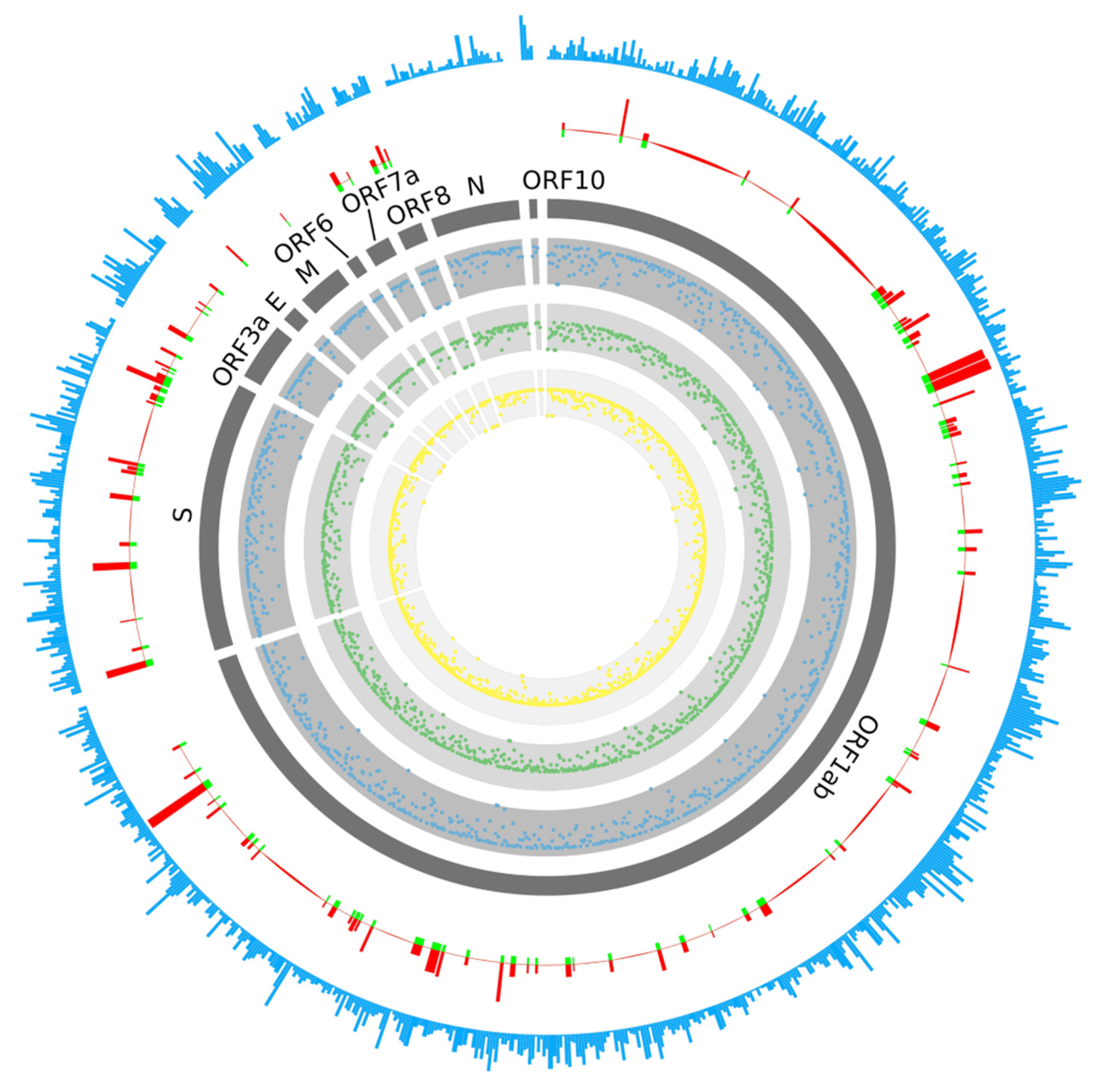



| Epitope | Protein | Start | End | Peptide | Emini | Kolaskar |
|---|---|---|---|---|---|---|
| B1 | Spike | 19 | 43 | TTRTQLPPAYTNSFTRGVYYPDKVF | 6.424 | 1.028 |
| B2 | Spike | 90 | 99 | VYFASTEKSN | 1.573 | 1.019 |
| B3 | Spike | 206 | 209 | KHTP | 2.463 | 1.002 |
| B4 | Spike | 405 | 430 | DEVRQIAPGQTGKIADYNYKLPDDFT | 5.81 | 1.001 |
| B5 | Spike | 494 | 507 | SYGFQPTNGVGYQP | 1.553 | 1.02 |
| B6 | Spike | 671 | 688 | CASYQTQTNSPRRARSVA | 3.531 | 1.027 |
| B7 | Spike | 771 | 782 | AVEQDKNTQEVF | 2.342 | 1.011 |
| B8 | Spike | 787 | 799 | QIYKTPPIKDFGG | 1.465 | 1.006 |
| B9 | Spike | 805 | 816 | ILPDPSKPSKRS | 4.69 | 1.019 |
| B10 | Spike | 1052 | 1058 | FPQSAPH | 1.381 | 1.059 |
| B11 | Spike | 1068 | 1091 | VPAQEKNFTTAPAICHDGKAHFPR | 1.063 | 1.03 |
| B12 | Spike | 1108 | 1123 | NFYEPQIITTDNTFVS | 1.039 | 1.007 |
| B13 | Spike | 1135 | 1151 | NTVYDPLQPELDSFKEE | 6.183 | 1.011 |
| B14 | Spike | 1153 | 1172 | DKYFKNHTSPDVDLGDISGI | 1.399 | 1.007 |
| B15 | Spike | 1190 | 1193 | AKNL | 1.087 | 1.005 |
| B16 | Spike | 1203 | 1209 | LGKYEQY | 2.512 | 1.035 |
| B17 | Spike | 1255 | 1265 | KFDEDDSEPVL | 2.654 | 1.003 |
| B18 | Spike | 63 | 70 | KNLNSSRV | 3.471 | 1.002 |
| B19 | Spike | 173 | 176 | SRTL | 1.504 | 1.011 |
| Protein | Count of T-Cell Epitopes | No. of Epitopes Per Residue | Epitope Overage | HLA Type Count |
|---|---|---|---|---|
| Spike | 378 | 0.297 | 93.01% | 33 |
| Membrane | 90 | 0.405 | 96.00% | 31 |
| Envelope | 31 | 0.413 | 94.14% | 32 |
| Peptide | Protein | Start | End | Vaccine Peptide | Count of T-Cell Epitopes | Count of B-Cell Epitopes | HLA Score |
|---|---|---|---|---|---|---|---|
| P1 | Spike | 19 | 46 | TTRTQLPPAYTNSFTRGVYYPDKVFRSS | 10 | 1 | 1.086 |
| P2 | Spike | 75 | 99 | GTKRFDNPVLPFNDGVYFASTEKSNK | 6 | 1 | 1.143 |
| P3 | Spike | 118 | 143 | LIVNNATNVVIKVCEFQFCNDPFLGVKK | 7 | 0 | 1.179 |
| P4 | Spike | 142 | 170 | GVYYHKNNKSWMESEFRVYSSANNCTFEY | 10 | 0 | 1.664 |
| P5 | Spike | 186 | 209 | FKNLREFVFKNIDGYFKIYSKHTP | 8 | 1 | 1.264 |
| P6 | Spike | 258 | 279 | WTAGAAAYYVGYLQPRTFLLKYKKKKK | 10 | 0 | 1.115 |
| P7 | Spike | 310 | 337 | KGIYQTSNFRVQPTESIVRFPNITNLCP | 10 | 0 | 1.012 |
| P8 * | Spike | 357 | 386 | RISNCVADYSVLYNSASFSTFKCYGVSPTK | 8 | 0 | 1.318 |
| P9 * | Spike | 405 | 433 | DEVRQIAPGQTGKIADYNYKLPDDFTGKKK | 7 | 1 | 0.928 |
| P10 * | Spike | 448 | 472 | NYNYLYRLFRKSNLKPFERDISTEI | 7 | 0 | 1.625 |
| P11 * | Spike | 478 | 505 | TPCNGVEGFNCYFPLQSYGFQPTNGVGYKK | 7 | 0 | 1.413 |
| P12 | Spike | 494 | 523 | SYGFQPTNGVGYQPYRVVVLSFELLHAPAT | 10 | 1 | 1.581 |
| P13 | Spike | 625 | 652 | HADQLTPTWRVYSTGSNVFQTRAGCLIG | 8 | 0 | 1.214 |
| P14 | Spike | 671 | 699 | CASYQTQTNSPRRARSVASQSIIAYTMSL | 8 | 1 | 1.234 |
| P15 | Spike | 771 | 799 | AVEQDKNTQEVFAQVKQIYKTPPIKDFGGK | 8 | 2 | 0.952 |
| P16 | Spike | 805 | 833 | ILPDPSKPSKRSFIEDLLFNKVTLADAGFK | 7 | 1 | 1.068 |
| P17 | Spike | 896 | 923 | IPFAMQMAYRFNGIGVTQNVLYENQKLI | 7 | 0 | 1.625 |
| P18 | Spike | 965 | 991 | QLSSNFGAISSVLNDILSRLDKVEAEVKKK | 9 | 0 | 1.012 |
| P19 | Spike | 1052 | 1073 | FPQSAPHGVVFLHVTYVPAQEK | 8 | 1 | 1.532 |
| P20 | Spike | 1068 | 1096 | VPAQEKNFTTAPAICHDGKAHFPREGVFV | 4 | 1 | 0.402 |
| P21 | Spike | 1095 | 1123 | FVSNGTHWFVTQRNFYEPQIITTDNTFVSK | 8 | 1 | 1.236 |
| P22 | Spike | 1135 | 1155 | NTVYDPLQPELDSFKEELDKYKKKKK | 2 | 1 | 0.254 |
| P23 | Spike | 1153 | 1181 | DKYFKNHTSPDVDLGDISGINASVVNIQKK | 5 | 1 | 0.322 |
| P24 | Spike | 1190 | 1217 | AKNLNESLIDLQELGKYEQYIKWPWYIWKK | 6 | 2 | 0.659 |
| P25 | Spike | 1216 | 1245 | IWLGFIAGLIAIVMVTIMLCKKKKKKKKKK | 5 | 0 | 1.394 |
| P26 | Spike | 1236 | 1265 | KKKKCCSCLKGCCSCGSCCKFDEDDSEPVL | 4 | 1 | 0.520 |
| P27 | Envelope | 4 | 33 | FVSEETGTLIVNSVLLFLAFVVFLKKKKKK | 11 | 0 | 1.133 |
| P28 | Envelope | 45 | 70 | NIVNVSLVKPSFYVYSRVKNLNSSRV | 9 | 1 | 1.455 |
| P29 | Membrane | 122 | 150 | VPLHGTILTRPLLESELVIGAVILRGHLRK | 9 | 0 | 1.508 |
| P30 | Membrane | 173 | 201 | SRTLSYYKLGASQRVAGDSGFAAYSRYRI | 6 | 1 | 0.902 |
| Panel | Protein | Start | Epitope | HLA Type | HLA Score | ITScorePeP | Contact Residues |
|---|---|---|---|---|---|---|---|
| a | Spike | 1220 | FIAGLIAIV | HLA-A*02:01 | 0.123 | −144.2 | PHE-1, GLY-4, LEU-5, ILE-6, ALA-7 |
| b | Spike | 1220 | FIAGLIAIV | HLA-B*46:01 | 0.102 | −138.2 | ILE-6, VAL-9 |
| c | Spike | 1220 | FIAGLIAIV | HLA-C*03:04 | 0.100 | −146.6 | PHE-1, ALA-3, ILE-8, VAL-9 |
| d | Envelope | 4 | FVSEETGTL | HLA-A*02:06 | 0.052 | −147.7 | PHE-1, VAL-2, SER-3, GLU-4, THR-6 |
| e | Envelope | 4 | FVSEETGTL | HLA-B*46:01 | 0.102 | −140.2 | PHE-1, SER-3, GLU-4, THR-6, THR-8 |
| f | Envelope | 4 | FVSEETGTL | HLA-C*07:02 | 0.152 | −136.7 | PHE-1, GLU-4, THR-8, LEU-9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, Y.; Jiang, H.; Qiu, M.; Liu, L.; Zou, S.; Li, Y.; Guo, Q.; Han, N.; Sun, Y.; Wang, K.; et al. Multi-Epitope Vaccine Design Using an Immunoinformatic Approach for SARS-CoV-2. Pathogens 2021, 10, 737. https://doi.org/10.3390/pathogens10060737
Feng Y, Jiang H, Qiu M, Liu L, Zou S, Li Y, Guo Q, Han N, Sun Y, Wang K, et al. Multi-Epitope Vaccine Design Using an Immunoinformatic Approach for SARS-CoV-2. Pathogens. 2021; 10(6):737. https://doi.org/10.3390/pathogens10060737
Chicago/Turabian StyleFeng, Ye, Haiping Jiang, Min Qiu, Liang Liu, Shengmei Zou, Yun Li, Qianpeng Guo, Ning Han, Yingqiang Sun, Kui Wang, and et al. 2021. "Multi-Epitope Vaccine Design Using an Immunoinformatic Approach for SARS-CoV-2" Pathogens 10, no. 6: 737. https://doi.org/10.3390/pathogens10060737
APA StyleFeng, Y., Jiang, H., Qiu, M., Liu, L., Zou, S., Li, Y., Guo, Q., Han, N., Sun, Y., Wang, K., Lu, L., Zhuang, X., Zhang, S., Chen, S., & Mo, F. (2021). Multi-Epitope Vaccine Design Using an Immunoinformatic Approach for SARS-CoV-2. Pathogens, 10(6), 737. https://doi.org/10.3390/pathogens10060737








