Genetic Listeria monocytogenes Types in the Pork Processing Plant Environment: From Occasional Introduction to Plausible Persistence in Harborage Sites
Abstract
1. Introduction
2. Results
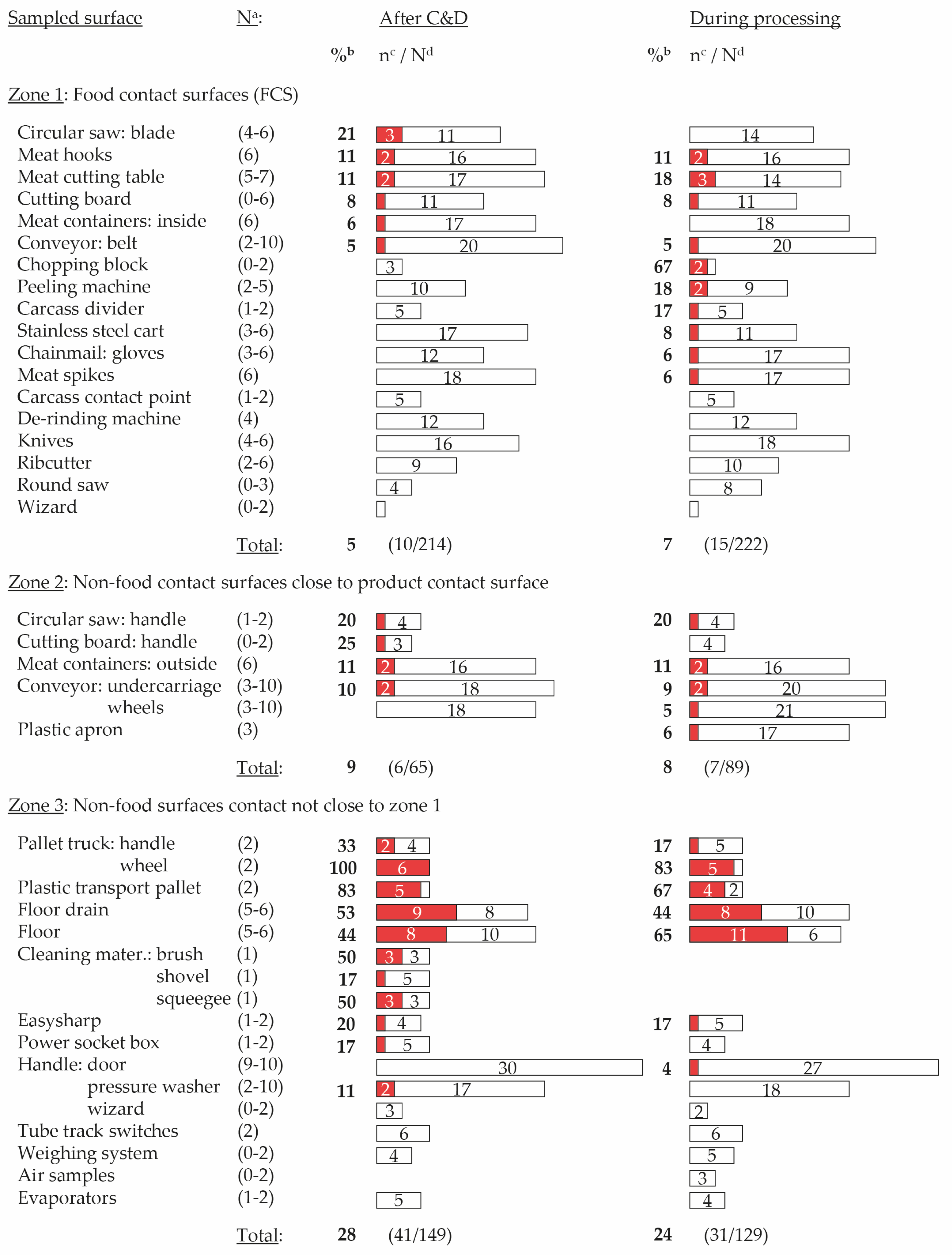
2.1. Contaminated Surfaces
2.1.1. General Results
2.1.2. Results at Plant Level
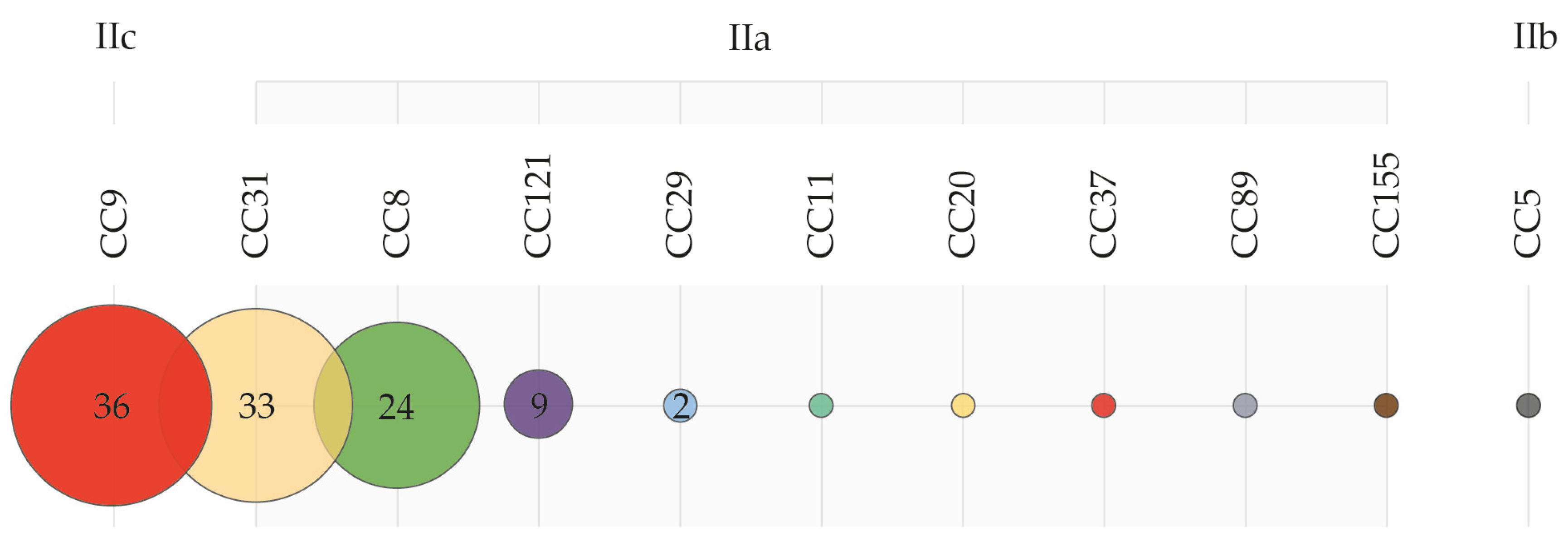
2.2. Molecular Typing
2.2.1. Genetic Diversity
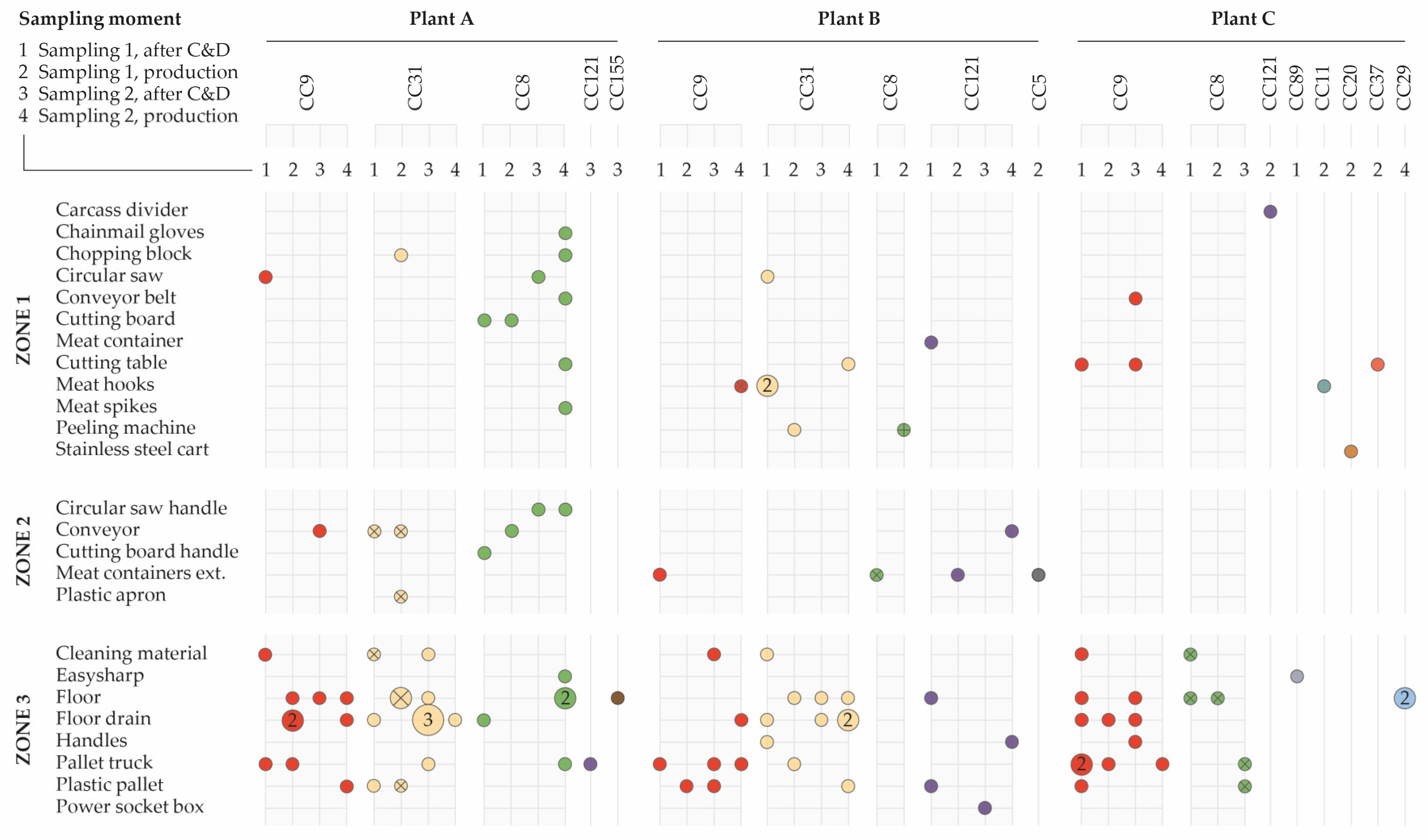
2.2.2. Distribution of Clonal Complexes in the Environment
3. Discussion
3.1. Meat Processing Facilities Are Expected to Be Colonized by L. monocytogenes
3.2. Food Contact Surfaces Are Contaminated, Even after C&D
3.3. Most Frequently Contaminated Zone 3 Surfaces Are Typical Harborage Sites in Processing Environments
3.4. Low Genetic Heterogeneity and the Widespread Prevalence of Meat-Adapted Clones
3.5. Continual Reintroduction (i) or Actual Persistence (ii)
3.6. Evaluation of Persistence Demonstrated by PFGE
4. Materials and Methods
4.1. Meat Processing Plant Selection and Description
4.2. Environmental Sampling
4.3. Microbiological Analyses
4.4. Molecular Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- EFSA; ECDC. The European Union one health 2018 zoonoses report. EFSA J. 2019, 17, e05926. [Google Scholar] [CrossRef]
- EFSA BIOHAZ Panel, (EFSA Panel on Biological Hazards); Riccci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Girones, R.; Herman, L.; Salvador, P.; Koutsoumanis, K.; et al. Listeria monocytogenes contamination of ready-to-eat foods and the risk for human health in the EU. EFSA J. 2018, 16, 5134. [Google Scholar] [CrossRef]
- Muhterem-uyar, M.; Dalmasso, M.; Sorin, A.; Manios, S.G.; Hernandez, M.; Kapetanakou, A.E.; Melero, B.; Minarovi, J.; Stessl, B.; Skandamis, P.N.; et al. Environmental sampling for Listeria monocytogenes control in food processing facilities reveals three contamination scenarios. Food Control 2015, 51, 94–107. [Google Scholar] [CrossRef]
- Malley, T.J.V.; Butts, J.; Wiedmann, M. Seek and Destroy Process: Listeria monocytogenes Process Controls in the Ready-to-Eat Meat and Poultry Industry. J. Food Prot. 2015, 78, 436–445. [Google Scholar] [CrossRef]
- Hellström, S.; Laukkanen, R.; Siekkinen, K.; Ranta, J.; Maijala, R.; Korkeala, H. Listeria monocytogenes contamination in pork can originate from farms. J. Food Prot. 2010, 73, 641–648. [Google Scholar] [CrossRef]
- Demaître, N.; Van Damme, I.; De Zutter, L.; Geeraerd, A.H.; Rasschaert, G.; De Reu, K. Occurrence, distribution and diversity of Listeria monocytogenes contamination on beef and pig carcasses after slaughter. Meat Sci. 2020, 169, 108177. [Google Scholar] [CrossRef]
- Kathariou, S. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 2002, 65, 1811–1829. [Google Scholar] [CrossRef] [PubMed]
- Annunziata, V.; Rizzi, V.; Acciari, V.; Iannetti, L.; Giovannini, A.; Serraino, A.; Calderone, D.; Rossi, A.; Morelli, D.; Marino, L.; et al. Listeria monocytogenes prevalence, contamination levels and strains characterization throughout the Parma ham processing chain. Food Control 2012, 25, 150–158. [Google Scholar] [CrossRef]
- Thévenot, D.; Dernburg, A.; Vernozy-Rozand, C. An updated review of Listeria monocytogenes in the pork meat industry and its products. J. Appl. Microbiol. 2006, 101, 7–17. [Google Scholar] [CrossRef]
- Larivière-Gauthier, G.; Letellier, A.; Kérouanton, A.; Bekal, S.; Quessy, S.; Fournaise, S.; Fravalo, P. Analysis of Listeria monocytogenes Strain Distribution in a Pork Slaughter and Cutting Plant in the Province of Quebec. J. Food Prot. 2014, 77, 2121–2128. [Google Scholar] [CrossRef]
- Arrigo, M.D.; Mateo-vivaracho, L.; Guillam, E.; Bravo, D.; Peirot, A.; Fernanda, M.; Medina, M.; García-lafuente, A. Characterization of persistent Listeria monocytogenes strains from ten dry-cured ham processing facilities. Food Microbiol. 2020, 92, 103581. [Google Scholar] [CrossRef]
- Thévenot, D.; Delignette, M.L.; Christieans, S.; Vernozy-Rozand, C. Prevalence of Listeria monocytogenes in 13 dried sausage processing plants and their products. International 2005, 102, 85–94. [Google Scholar] [CrossRef]
- Bolocan, A.S.; Nicolai, A.I.; Alvarez-ordóñez, A.; Borda, D.; Oniciuc, E.A.; Stessl, B.; Gurgu, L.; Wagner, M.; Jordan, K. Dynamics of Listeria monocytogenes colonisation in a newly-opened meat processing facility. Meat Sci. 2016, 113, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Lunden, J.M.; Autio, T.J.; Sjo, A.; Korkeala, H.J. Persistent and Nonpersistent Listeria monocytogenes Contamination in Meat and Poultry Processing Plants. J. Food Prot. 2003, 66, 2062–2069. [Google Scholar] [CrossRef] [PubMed]
- Spanu, C.; Jordan, K. Listeria monocytogenes environmental sampling program in ready-to-eat processing facilities: A practical approach. Compr. Rev. Food Sci. Food Saf. 2020, 19, 2843–2861. [Google Scholar] [CrossRef]
- Carpentier, B.; Barre, L. Guidelines on Sampling the Food Processing Area and Equipment for the Detection of Listeria monocytogenes. Available online: https://ec.europa.eu/food/sites/food/files/safety/docs/biosafety_fh_mc_guidelines_on_sampling.pdf (accessed on 7 June 2021).
- Burnett, J.; Wu, S.T.; den Bakker, H.C.; Cook, P.W.; Veenhuizen, D.R.; Hammons, S.R.; Singh, M.; Oliver, H.F. Listeria monocytogenes is prevalent in retail produce environments but Salmonella enterica is rare. Food Control 2020, 113, 107173. [Google Scholar] [CrossRef]
- Dzieciol, M.; Schornsteiner, E.; Muhterem-Uyar, M.; Stessl, B.; Wagner, M.; Schmitz-Esser, S. Bacterial diversity of floor drain biofilms and drain waters in a Listeria monocytogenes contaminated food processing environment. Int. J. Food Microbiol. 2016, 223, 33–40. [Google Scholar] [CrossRef]
- Meloni, D.; Piras, F.; Mureddu, A.; Mazza, R.; Nucera, D.; Mazzette, R.; Vienna, V.; Animale, P.; Veterinaria, M.; Torino, U. Contamination in traditional fermented sausage processing plants in Italy. Italy J. Food Sci. 2012, 24, 214–223. [Google Scholar]
- Barros, M.; Nero, L.; Silva, L.; D’Ovidio, L.; Monteiro, F.; Tamanini, R.; Fagnani, R.; Hofer, E.; Beloti, V. Listeria monocytogenes: Occurrence in beef and identification of the main contamination points in processing plants. Meat Sci. 2007, 76, 591–596. [Google Scholar] [CrossRef]
- Tompkin, R.B.; Scott, V.N.; Bernard, D.T.; Sveum, W.S.; Gombas, K.S. Guidelines to prevent post-processing contamination from Listeria monocytogenes. Dairy Food Environ. Sanit. 1999, 19, 551–562. [Google Scholar]
- Meloni, D.; Consolati, S.G.; Mazza, R.; Mureddu, A.; Fois, F.; Piras, F.; Mazzette, R. Presence and molecular characterization of the major serovars of Listeria monocytogenes in ten Sardinian fermented sausage processing plants. Meat Sci. 2014, 97, 443–450. [Google Scholar] [CrossRef]
- Meloni, D.; Piras, F.; Mureddu, A.; Fois, F.; Consolati, S.G.; Lamon, S.; Mazzette, R. Listeria monocytogenes in five Sardinian swine slaughterhouses: Prevalence, serotype, and genotype characterization. J. Food Prot. 2013, 76, 1863–1867. [Google Scholar] [CrossRef]
- Félix, B.; Feurer, C.; Maillet, A.; Guillier, L.; Boscher, E.; Kerouanton, A.; Denis, M.; Roussel, S. Population genetic structure of Listeria monocytogenes strains isolated from the pig and pork production chain in France. Front. Microbiol. 2018, 9, 684. [Google Scholar] [CrossRef]
- Verghese, B.; Lok, M.; Alessandria, V.; Chen, Y.; Kathariou, S.; Knabel, S. comK Prphage Junction Fragments as Markers for Listeria monocytogenes Genotypes Unque to Individual Meat and Poultry Processing Plants and a Model for Rapid Niche-Specific Adaptation, Biofilm Formation, and Persistence. Appl. Environ. Microbiol. 2011, 77, 3279–3292. [Google Scholar] [CrossRef]
- Knabel, S.J.; Reimer, A.; Verghese, B.; Lok, M.; Ziegler, J.; Farber, J.; Pagotto, F.; Graham, M.; Nadon, C.A.; Gilmour, M.W. Sequence typing confirms that a predominant Listeria monocytogenes clone caused human listeriosis cases and outbreaks in Canada from 1988 to 2010. J. Clin. Microbiol. 2012, 50, 1748–1751. [Google Scholar] [CrossRef]
- Fagerlund, A.; Langsrud, S.; Møretrø, T. In depth longitudinal study of Listeria monocytogenes ST9 isolates from meat processing industry: Resolving diversity and transmission patterns using WGS. Appl. Environ. Microbiol. 2020. [Google Scholar] [CrossRef]
- Maury, M.; Bracq-dieye, H.; Huang, L.; Vales, G.; Lavina, M.; Thouvenot, P.; Disson, O.; Leclercq, A.; Brisse, S.; Lecuit, M. Hypervirulent Listeria monocytogenes clones’ adaption to mammalian gut accounts for their association with dairy products. Nat. Commun. 2019, 10, 2488. [Google Scholar] [CrossRef] [PubMed]
- Stoller, A.; Stevens, M.; Stephan, R.; Guldimann, C. Characteristics of Listeria monocytogenes strains persisting in a meat processing facility over a 4-year period. Pathogens 2019, 8, 32. [Google Scholar] [CrossRef]
- Guidi, F.; Orsini, M.; Chiaverini, A.; Torresi, M.; Centorame, P.; Acciari, V.A.; Salini, R.; Palombo, B.; Brandi, G.; Amagliani, G.; et al. Hypo- and Hyper-Virulent Listeria monocytogenes Clones Persisting in Two Different Food Processing Plants of Central Italy. Microorganisms 2021, 9, 376. [Google Scholar] [CrossRef] [PubMed]
- Autio, T.; Lundén, J.; Fredriksson-Ahomaa, M.; Björkroth, J.; Sjöberg, A.M.; Korkeala, H. Similar Listeria monocytogenes pulsotypes detected in several foods originating from different sources. Int. J. Food Microbiol. 2002, 77, 83–90. [Google Scholar] [CrossRef]
- Nastasijevic, I.; Milanov, D.; Velebit, B.; Djordjevic, V.; Swift, C. Tracking of Listeria monocytogenes in meat establishment using Whole Genome Sequencing as a food safety management tool: A proof of concept. Int. J. Food Microbiol. 2017, 257, 157–164. [Google Scholar] [CrossRef]
- Carpentier, B.; Cerf, O. Review—Persistence of Listeria monocytogenes in food industry equipment and premises. Int. J. Food Microbiol. 2011, 145, 1–8. [Google Scholar] [CrossRef]
- Stasiewicz, M.J.; Oliver, H.F.; Wiedmann, M.; den Bakker, H.C. Whole-genome sequencing allows for improved identification of persistent Listeria monocytogenes in food-associated environments. Appl. Environ. Microbiol. 2015, 81, 6024–6037. [Google Scholar] [CrossRef] [PubMed]
- Halbedel, S.; Prager, R.; Fuchs, S.; Trost, E.; Werner, G.; Flieger, A. Whole-Genome Sequencing of Recent Listeria monocytogenes Isolates from Germany Reveals Population Structure and Disease Clusters. J. Clin. Microbiol. 2018, 56, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Luning, P.A.; Jacxsens, L.; Rovira, J.; Osés, S.M.; Uyttendaele, M.; Marcelis, W.J. A concurrent diagnosis of microbiological food safety output and food safety management system performance: Cases from meat processing industries. Food Control 2011, 22, 555–565. [Google Scholar] [CrossRef]
- Herman, L.; De Ridder, H.; Vlaemynck, G. A multiplex PCR method for the identification of Listeria spp. and Listeria monocytogenes in dairy samples. J. Food Prot. 1995, 58, 867–872. [Google Scholar] [CrossRef]
- Borucki, M.; Call, D. Listeria monocytogenes serotype identification by PCR. J. Clin. Microbiol. 2003, 41, 5537–5540. [Google Scholar] [CrossRef]
- Graves, L.M.; Swaminathan, B. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 2001, 65, 55–62. [Google Scholar] [CrossRef]
- Maury, M.; Tsai, Y.; Charlier, C.; Touchon, M.; Chenal-francisque, V.; Leclercq, A.; Criscuolo, A.; Gaultier, C.; Roussel, S.; Brisabois, A.; et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 2016, 48, 308–313. [Google Scholar] [CrossRef]
- Henri, C.; Félix, B.; Guillier, L.; Leekitcharoenphon, P.; Michelon, D.; Aarestrup, F.M.; Mistou, M.; Roussel, S. Population Genetic Structure of Listeria monocytogenes Strains as Determined by Pulsed-Field Gel Electrophoresis and Multilocus. Appl. Environ. Microbiol. 2016, 82, 5720–5728. [Google Scholar] [CrossRef]
| Plant A | Plant B | Plant C | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Zone | Day | After C&D | During Production | After C&D | During Production | After C&D | During Production | Totals | |||||||
| 1 | 1 | 5% | (2/38) | 6% | (2/35) | 11% | (4/36) | 5% | (2/41) | 3% | (1/36) | 11% | (4/37) | 7% | (15/223) |
| 2 | 3% | (1/38) | 14% | (5/35) | 0% | (0/30) | 5% | (2/37) | 6% | (2/36) | 0% | (0/37) | 5% | (10/213) | |
| 2 | 1 | 17% | (2/12) | 19% | (3/16) | 33% | (2/6) | 17% | (2/12) | 0% | (0/14) | 0% | (0/17) | 12% | (9/77) |
| 2 | 17% | (2/12) | 6% | (1/16) | 0% | (0/7) | 8% | (1/12) | 0% | (0/14) | 0% | (0/16) | 5% | (4/77) | |
| 3 | 1 | 25% | (6/24) | 35% | (7/20) | 22% | (6/27) | 11% | (3/27) | 36% | (9/25) | 14% | (3/21) | 24% | (34/144) |
| 2 | 38% | (9/24) | 38% | (8/21) | 27% | (6/22) | 35% | (7/20) | 19% | (5/27) | 15% | (3/20) | 28% | (38/134) | |
| Totals | 15% | (22/148) | 18% | (26/143) | 14% | (18/128) | 11% | (17/149) | 11% | (17/152) | 7% | (10/148) | 13% | (110/868) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Demaître, N.; Rasschaert, G.; De Zutter, L.; Geeraerd, A.; De Reu, K. Genetic Listeria monocytogenes Types in the Pork Processing Plant Environment: From Occasional Introduction to Plausible Persistence in Harborage Sites. Pathogens 2021, 10, 717. https://doi.org/10.3390/pathogens10060717
Demaître N, Rasschaert G, De Zutter L, Geeraerd A, De Reu K. Genetic Listeria monocytogenes Types in the Pork Processing Plant Environment: From Occasional Introduction to Plausible Persistence in Harborage Sites. Pathogens. 2021; 10(6):717. https://doi.org/10.3390/pathogens10060717
Chicago/Turabian StyleDemaître, Niels, Geertrui Rasschaert, Lieven De Zutter, Annemie Geeraerd, and Koen De Reu. 2021. "Genetic Listeria monocytogenes Types in the Pork Processing Plant Environment: From Occasional Introduction to Plausible Persistence in Harborage Sites" Pathogens 10, no. 6: 717. https://doi.org/10.3390/pathogens10060717
APA StyleDemaître, N., Rasschaert, G., De Zutter, L., Geeraerd, A., & De Reu, K. (2021). Genetic Listeria monocytogenes Types in the Pork Processing Plant Environment: From Occasional Introduction to Plausible Persistence in Harborage Sites. Pathogens, 10(6), 717. https://doi.org/10.3390/pathogens10060717






