Molecular Characterization of 4/91 Infectious Bronchitis Virus Leading to Studies of Pathogenesis and Host Responses in Laying Hens
Abstract
1. Introduction
2. Results
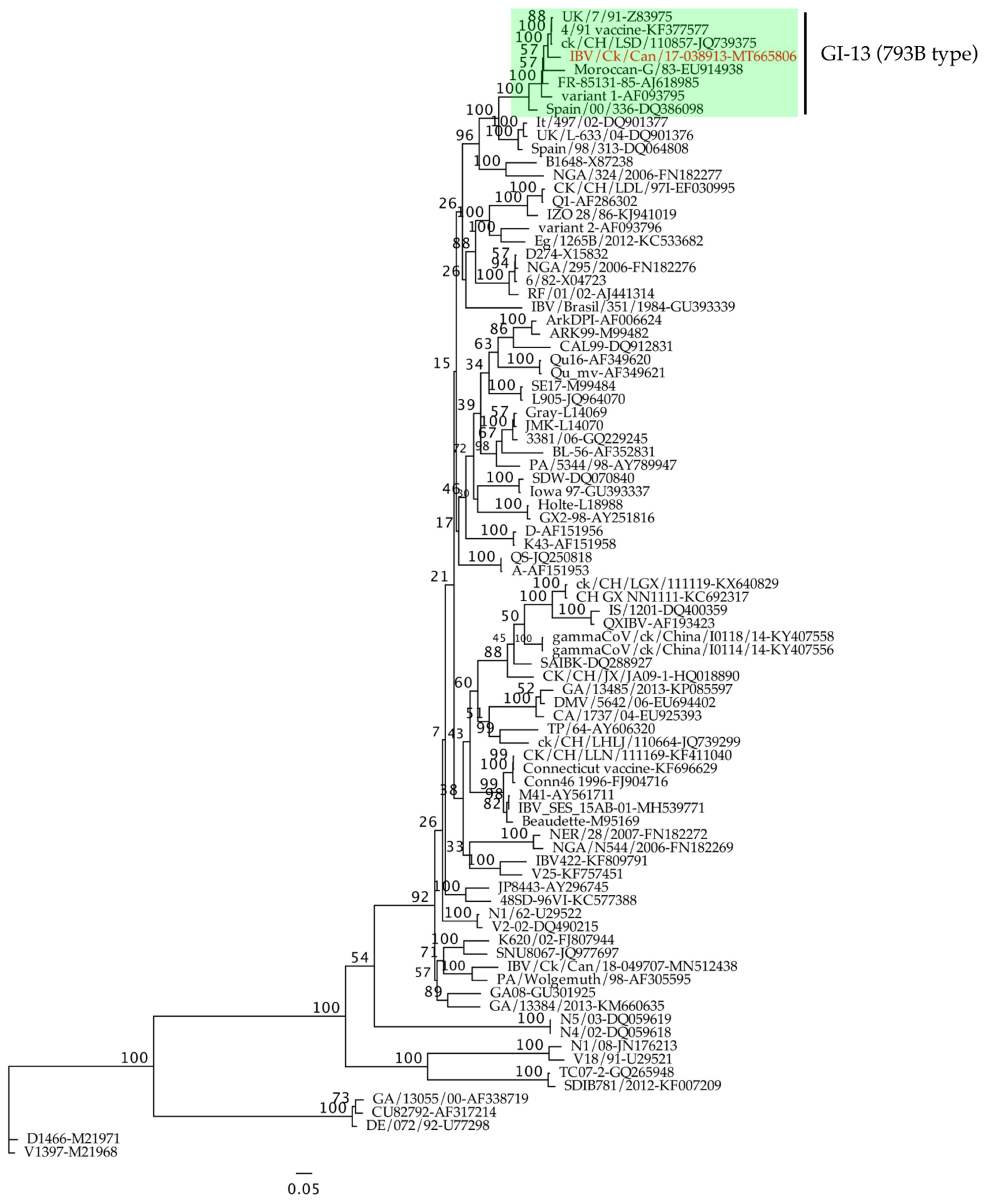
2.1. Phylogenetic Analysis and Full Genome Characterization
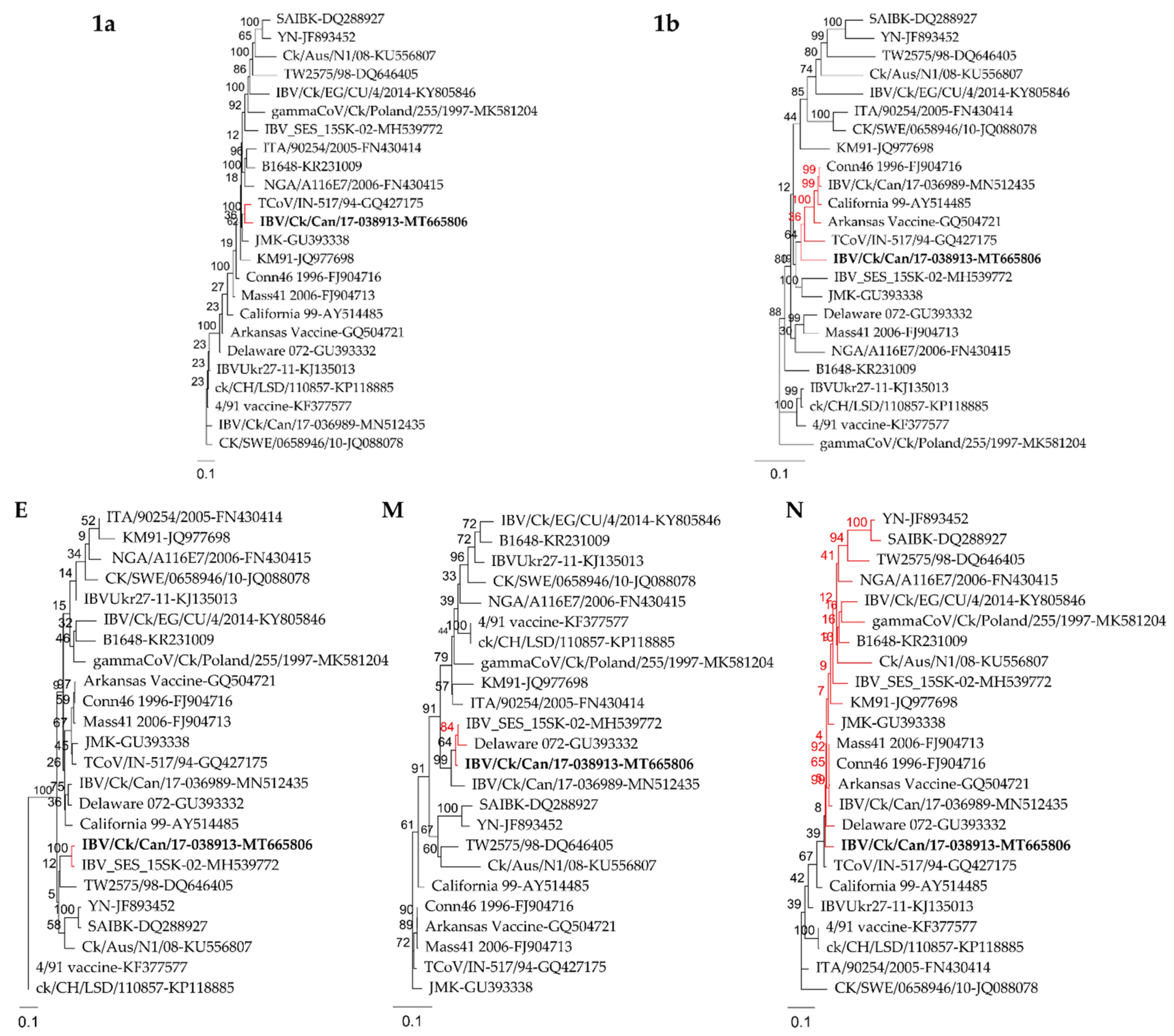
2.2. Recombination Analysis
2.3. Embryo Lesions
2.4. Clinical and Pathological Manifestations in Laying Hens
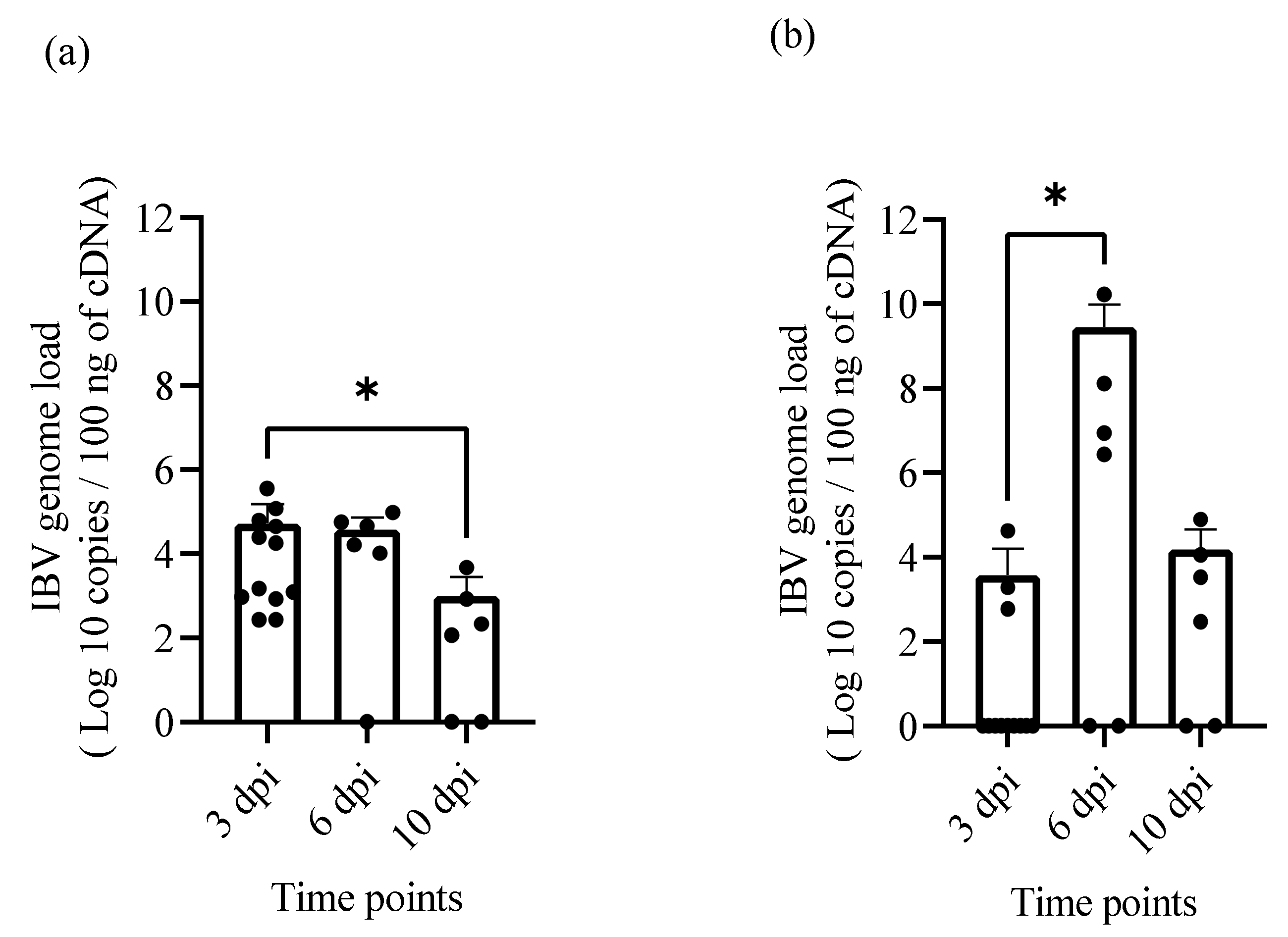
2.5. IBV Genome Loads in Oropharyngeal, Cloacal Swabs and Tissue Samples
2.6. Host Responses
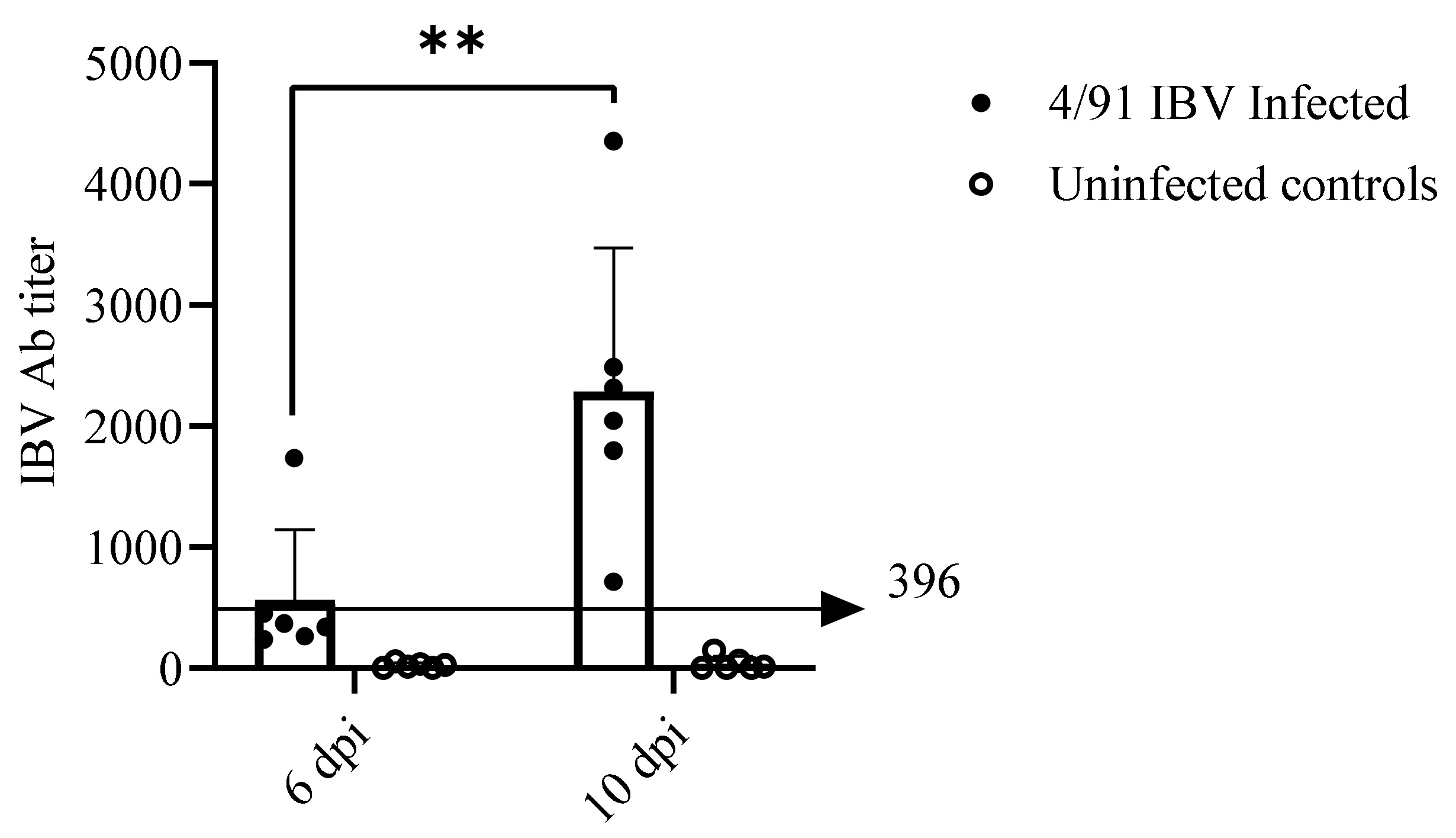
2.6.1. Serum Anti-IBV Antibody (Ab) Response
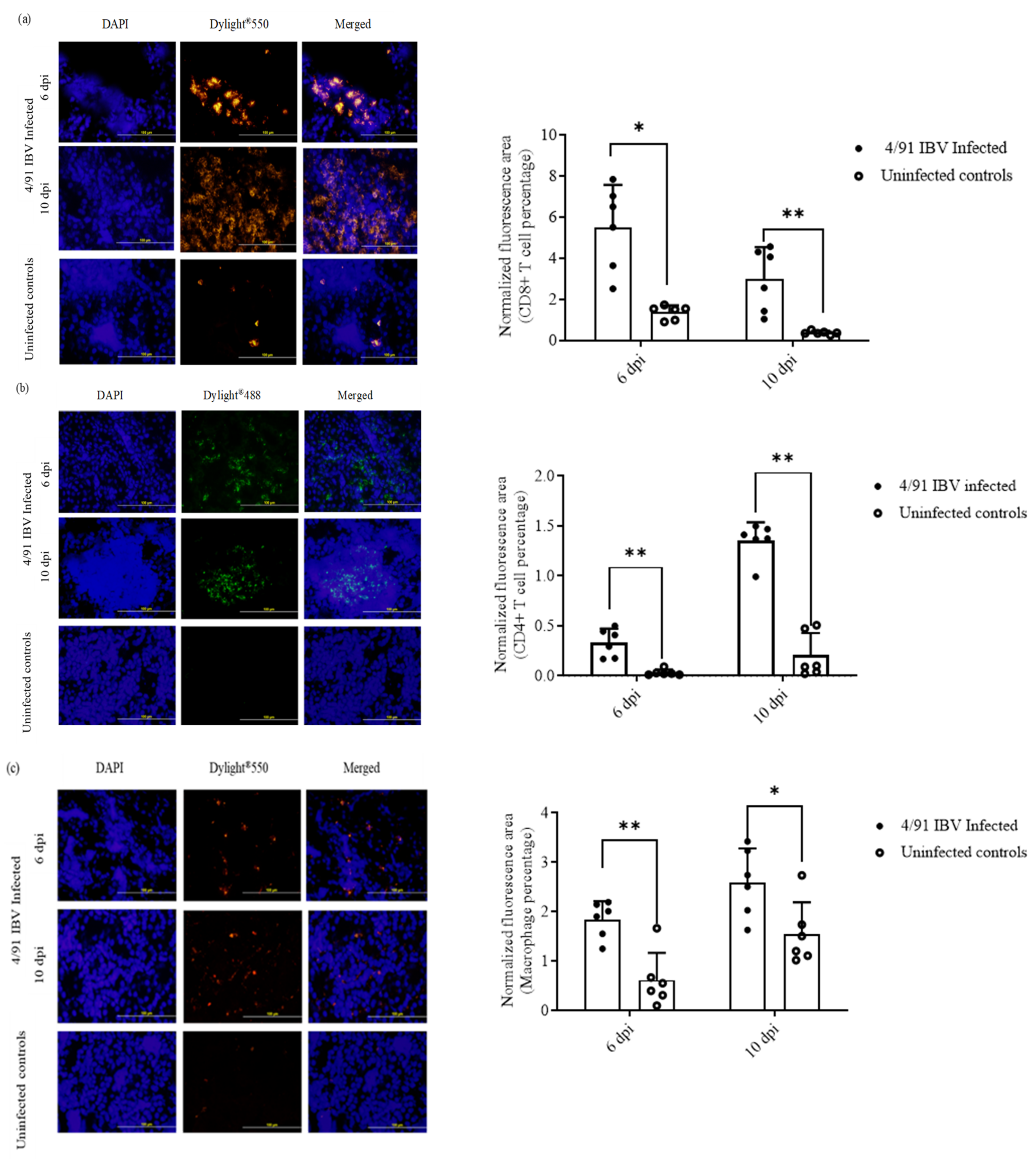
2.6.2. Recruitment of CD4+ T Cells, CD8+ T Cells and Macrophages
2.6.3. Histopathological Findings in the Kidney
3. Discussion
4. Materials and Methods
4.1. Animals, Ethics and Virus
4.2. Ribonucleic Acid (RNA) Extraction and Sequencing
4.3. Genotyping and Sequence Analysis
4.4. Recombination Analysis
4.5. Experimental Infection of Laying Hens
4.6. Techniques
4.6.1. RNA Extraction and cDNA Synthesis
4.6.2. IBV Genome Load Quantification by qRT-PCR
4.6.3. Histology
4.6.4. Enzyme-Linked Immunosorbent Assay (ELISA)
4.6.5. Immunofluorescent Assay
4.7. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Milek, J.; Blicharz-Domanska, K. Coronaviruses in Avian Species-Review with Focus on Epidemiology and Diagnosis in Wild Birds. J. Vet. Res. 2018, 62, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Schalk, A.F. An Apparently New Respiratory Disease of Baby Chicks. J. Am. Vet. Med. Assoc. 1931, 78, 413–423. [Google Scholar]
- Lin, S.-Y.; Chen, H.-W. Infectious Bronchitis Virus Variants: Molecular Analysis and Pathogenicity Investigation. Int. J. Mol. Sci. 2017, 18, 2030. [Google Scholar] [CrossRef] [PubMed]
- Martin, E.A.K.; Brash, M.L.; Hoyland, S.K.; Coventry, J.M.; Sandrock, C.; Guerin, M.T.; Ojkic, D. Genotyping of Infectious Bronchitis Viruses Identified in Canada between 2000 and 2013. Avian Pathol. 2014, 43, 264–268. [Google Scholar] [CrossRef]
- Bande, F.; Arshad, S.S.; Omar, A.R.; Hair-Bejo, M.; Mahmuda, A.; Nair, V. Global Distributions and Strain Diversity of Avian Infectious Bronchitis Virus: A Review. Anim. Health Res. Rev. 2017, 18, 70–83. [Google Scholar] [CrossRef]
- Haji-Abdolvahab, H.; Ghalyanchilangeroudi, A.; Bahonar, A.; Ghafouri, S.A.; Vasfi Marandi, M.; Mehrabadi, M.H.F.; Tehrani, F. Prevalence of Avian Influenza, Newcastle Disease, and Infectious Bronchitis Viruses in Broiler Flocks Infected with Multifactorial Respiratory Diseases in Iran, 2015–2016. Trop. Anim. Health Prod. 2019, 51, 689–695. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Wang, F.; Xue, Y.; Zhou, Q.; Chen, F.; Bi, Y.; Xie, Q. Epidemiology and Characterization of Avian Infectious Bronchitis Virus Strains Circulating in Southern China during the Period from 2013–2015. Sci. Rep. 2017, 7, 6576. [Google Scholar] [CrossRef]
- Khataby, K.; Souiri, A.; Kasmi, Y.; Loutfi, C.; Ennaji, M.M. Current Situation, Genetic Relationship and Control Measures of Infectious Bronchitis Virus Variants Circulating in African Regions. J. Basic Appl. Zool. 2016, 76, 20–30. [Google Scholar] [CrossRef]
- Lai, M.M.C.; Cavanagh, D. The Molecular Biology of Coronaviruses. In Advances in Virus Research; Maramorosch, K., Murphy, F.A., Shatkin, A.J., Eds.; Academic Press: Cambridge, MA, USA, 1997; Volume 48, pp. 1–100. [Google Scholar]
- Armesto, M.; Cavanagh, D.; Britton, P. The Replicase Gene of Avian Coronavirus Infectious Bronchitis Virus Is a Determinant of Pathogenicity. PLoS ONE 2009, 4, e7384. [Google Scholar] [CrossRef]
- Cavanagh, D. Coronavirus Avian Infectious Bronchitis Virus. Vet. Res. 2007, 38, 281–297. [Google Scholar] [CrossRef]
- Casais, R.; Davies, M.; Cavanagh, D.; Britton, P. Gene 5 of the Avian Coronavirus Infectious Bronchitis Virus is not Essential for Replication. J. Virol. 2005, 79, 8065–8078. [Google Scholar] [CrossRef] [PubMed]
- Sawicki, S.G.; Sawicki, D.L.; Siddell, S.G. A Contemporary View of Coronavirus Transcription. J. Virol. 2007, 81, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Wickramasinghe, I.N.A.; de Vries, R.P.; Gröne, A.; de Haan, C.A.M.; Verheije, M.H. Binding of Avian Coronavirus Spike Proteins to Host Factors Reflects Virus Tropism and Pathogenicity. J. Virol. 2011, 85, 8903–8912. [Google Scholar] [CrossRef]
- Arshad, S.; Al-Salihi, K.; Noordin, M.M. Ultrastructural Pathology of Trachea in Chicken Experimentally Infected with Infectious Bronchitis Virus-MH-5365/95. Ann. Microsc. 2002, 3, 43–47. [Google Scholar]
- Ambali, A.G.; Jones, R.C. Early Pathogenesis in Chicks of Infection with an Enterotropic Strain of Infectious Bronchitis Virus. Avian Dis. 1990, 34, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.R.A.P.; Trus, I.; Desmarets, L.M.B.; Li, Y.; Theuns, S.; Nauwynck, H.J. Productive Replication of Nephropathogenic Infectious Bronchitis Virus in Peripheral Blood Monocytic Cells, a Strategy for Viral Dissemination and Kidney Infection in Chickens. Vet. Res. 2016, 47, 70. [Google Scholar] [CrossRef]
- Chong, K.T.; Apostolov, K. The Pathogenesis of Nephritis in Chickens Induced by Infectious Bronchitis Virus. J. Comp. Pathol. 1982, 92, 199–211. [Google Scholar] [CrossRef]
- Crinion, R.A.P.; Ball, R.A.; Hofstad, M.S. Pathogenesis of Oviduct Lesions in Immature Chickens Following Exposure to Infectious Bronchitis Virus at One Day Old. Avian Dis. 1971, 15, 32–41. [Google Scholar] [CrossRef]
- Gough, R.E.; Randall, C.J.; Dagless, M.; Alexander, D.J.; Cox, W.J.; Pearson, D. A ‘New’ Strain of Infectious Bronchitis Virus Infecting Domestic Fowl in Great Britain. Vet. Rec. 1992, 130, 493–494. [Google Scholar] [CrossRef] [PubMed]
- Raj, G.D.; Jones, R.C. Immunopathogenesis of Infection in SPF Chicks and Commercial Broiler Chickens of a Variant Infectious Bronchitis Virus of Economic Importance. Avian Pathol. 1996, 25, 481–501. [Google Scholar] [CrossRef]
- Van Roekel, H.; Clarke, M.K.; Bullis, K.L.; Olesiuk, O.M.; Sperling, F.G. Infectious Bronchitis. Am. J. Vet. Res. 1951, 12, 140–146. [Google Scholar] [PubMed]
- Winterfield, R.W.; Albassam, M.A. Nephropathogenicity of Infectious Bronchitis Virus. Poult. Sci. 1984, 63, 2358–2363. [Google Scholar] [CrossRef]
- Crinion, R.A.P.; Hofstad, M.S. Pathogenicity of Four Serotypes of Avian Infectious Bronchitis Virus for the Oviduct of Young Chickens of Various Ages. Avian Dis. 1972, 16, 351–363. [Google Scholar] [CrossRef]
- Otsuki, K.; Huggins, M.B.; Cook, J.K.A. Comparison of the Susceptibility to Avian Infectious Bronchitis Virus Infection of Two Inbred Lines of White Leghorn Chickens. Avian Pathol. 1990, 19, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Albassam, M.A.; Winterfield, R.W.; Thacker, H.L. Comparison of the Nephropathogenicity of Four Strains of Infectious Bronchitis Virus. Avian Dis. 1986, 30, 468–476. [Google Scholar] [CrossRef]
- Benyeda, Z.; Szeredi, L.; Mató, T.; Süveges, T.; Balka, G.; Abonyi-Tóth, Z.; Rusvai, M.; Palya, V. Comparative Histopathology and Immunohistochemistry of QX-like, Massachusetts and 793/B Serotypes of Infectious Bronchitis Virus Infection in Chickens. J. Comp. Pathol. 2010, 143, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Chousalkar, K.K.; Roberts, J.R.; Reece, R. Comparative Histopathology of Two Serotypes of Infectious Bronchitis Virus (T and n1/88) in Laying Hens and Cockerels. Poult. Sci. 2007, 86, 50–58. [Google Scholar] [CrossRef]
- Butcher, G.D.; Winterfield, R.W.; Shapiro, D.P. Pathogenesis of H13 Nephropathogenic Infectious Bronchitis Virus. Avian Dis. 1990, 34, 916–921. [Google Scholar] [CrossRef]
- Mork, A.-K.; Hesse, M.; Abd El Rahman, S.; Rautenschlein, S.; Herrler, G.; Winter, C. Differences in the Tissue Tropism to Chicken Oviduct Epithelial Cells between Avian Coronavirus IBV Strains QX and B1648 are not Related to the Sialic Acid Binding Properties of Their Spike Proteins. Vet. Res. 2014, 45, 67. [Google Scholar] [CrossRef] [PubMed]
- Cook, J.K.A.; Huggins, M.B. Newly Isolated Serotypes of Infectious Bronchitis Virus: Their Role in Disease. Avian Pathol. 1986, 15, 129–138. [Google Scholar] [CrossRef]
- Broadfoot, D.I.; Pomeroy, B.S.; Smith, W.M., Jr. Effects of Infectious Bronchitis in Baby Chicks. Poult. Sci. 1956, 35, 757–762. [Google Scholar] [CrossRef]
- Jones, R.C.; Jordan, F.T.W. Persistence of Virus in the Tissues and Development of the Oviduct in the Fowl Following Infection at Day Old with Infectious Bronchitis Virus. Res. Vet. Sci. 1972, 13, 52–60. [Google Scholar] [CrossRef]
- Capua, I.; Minta, Z.; Karpinska, E.; Mawditt, K.; Britton, P.; Cavanagh, D.; Gough, R.E. Co-circulation of Four Types of Infectious Bronchitis Virus (793/B, 624/I, B1648 and Massachusetts). Avian Pathol. 1999, 28, 587–592. [Google Scholar] [CrossRef]
- Jones, R. Europe: History, Current Situation and Control Measures for Infectious Bronchitis. Braz. J. Poult. Sci. 2010, 12, 125–128. [Google Scholar] [CrossRef]
- Lounas, A.; Oumouna-Benachour, K.; Medkour, H.; Oumouna, M. The First Evidence of a New Genotype of Nephropathogenic Infectious Bronchitis Virus Circulating in Vaccinated and Unvaccinated Broiler Flocks in Algeria. Vet. World 2018, 11, 1630–1636. [Google Scholar] [CrossRef]
- Martin, E.A.K.; Brash, M.L.; Stalker, M.; Ojkic, D. Using Phylogenetic Analysis to Examine the Changing Strains of Infectious Bronchitis Virus Infections in Ontario Over Time. In Proceedings of the 16th Annual Meeting of the Canadian Animal Health Laboratorians Network (CAHLN), Guelph, ON, Canada, 4–7 June 2017. [Google Scholar]
- Amarasinghe, A.; de Silva Senapathi, U.; Abdul-Cader, M.S.; Popowich, S.; Marshall, F.; Cork, S.C.; van der Meer, F.; Gomis, S.; Abdul-Careem, M.F. Comparative Features of Infections of Two Massachusetts (Mass) Infectious Bronchitis Virus (IBV) Variants Isolated from Western Canadian Layer Flocks. BMC Vet. Res. 2018, 14, 391. [Google Scholar] [CrossRef]
- Hassan, M.S.H.; Ojkic, D.; Coffin, C.S.; Cork, S.C.; van der Meer, F.; Abdul-Careem, M.F. Delmarva (DMV/1639) Infectious Bronchitis Virus (IBV) Variants Isolated in Eastern Canada Show Evidence of Recombination. Viruses 2019, 11, 1054. [Google Scholar] [CrossRef]
- Jackwood, M.W.; Boynton, T.O.; Hilt, D.A.; McKinley, E.T.; Kissinger, J.C.; Paterson, A.H.; Robertson, J.; Lemke, C.; McCall, A.W.; Williams, S.M.; et al. Emergence of a Group 3 Coronavirus through Recombination. Virology 2010, 398, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Han, Z.; Xu, Q.; Wang, Q.; Gao, M.; Wu, W.; Shao, Y.; Li, H.; Kong, X.; Liu, S. Serotype Shift of a 793/B Genotype Infectious Bronchitis Coronavirus by Natural Recombination. Infect. Genet. Evol. 2015, 32, 377–387. [Google Scholar] [CrossRef]
- Pohjola, L.K.; Ek-Kommonen, S.C.; Tammiranta, N.E.; Kaukonen, E.S.; Rossow, L.M.; Huovilainen, T.A. Emergence of Avian Infectious Bronchitis in a Non-vaccinating Country. Avian Pathol. 2014, 43, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Kalokhoran, A.Y.; Ghalyanchilangeroudi, A.; Hosseini, H.; Madadgar, O.; Karimi, V.; Hashemzadeh, M.; Hesari, P.; Zabihi Petroudi, M.T.; Najafi, H. Co-circulation of Three Clusters of 793/B-like Avian Infectious Bronchitis Virus Genotypes in Iranian Chicken Flocks. Arch. Virol. 2017, 162, 3183–3189. [Google Scholar] [CrossRef]
- Benyeda, Z.; Mató, T.; Süveges, T.; Szabó, É.; Kardi, V.; Abonyi-Tóth, Z.; Rusvai, M.; Palya, V. Comparison of the Pathogenicity of QX-like, M41 and 793/B Infectious Bronchitis Strains from Different Pathological Conditions. Avian Pathol. 2009, 38, 449–456. [Google Scholar] [CrossRef]
- Boroomand, Z.; Asasi, K.; Mohammadi, A. Pathogenesis and Tissue Distribution of Avian Infectious Bronchitis Virus Isolate IRFIBV32 (793/B serotype) in Experimentally Infected Broiler Chickens. Sci. World J. 2012, 2012, 402537. [Google Scholar] [CrossRef] [PubMed]
- Pourbakhsh, S.A.; Tavasoly, A.; Mahdavi, S.; Momayez, R. Experimental Histopathologic Study of the Lesions Induced by Serotype 793/B (4/91) Infectious Bronchitis Virus. Arch. Razi Inst. 2007, 62, 15–22. [Google Scholar] [CrossRef]
- Yan, S.-h.; Chen, Y.; Zhao, J.; Xu, G.; Zhao, Y.; Zhang, G.-z. Pathogenicity of a TW-Like Strain of Infectious Bronchitis Virus and Evaluation of the Protection Induced against It by a QX-Like Strain. Front. Microbiol. 2016, 7, 1653. [Google Scholar] [CrossRef]
- Alexander, D.J.; Gough, R.E. Isolation of Avian Infectious Bronchitis Virus from Experimentally Infected Chickens. Res. Vet. Sci. 1977, 23, 344–347. [Google Scholar] [CrossRef]
- Alexander, D.J.; Gough, R.E.; Pattison, M. A Long-term Study of the Pathogenesis of Infection of Fowls with Three Strains of Avian Infectious Bronchitis Virus. Res. Vet. Sci. 1978, 24, 228–233. [Google Scholar] [CrossRef]
- Crinion, R.A. Egg Quality and Production Following Infectious Bronchitis Virus Exposure at One Day Old. Poult. Sci. 1972, 51, 582–585. [Google Scholar] [CrossRef] [PubMed]
- Bisgaard, M. The Influence of Infectious Bronchitis Virus on Egg Production, Fertility, Hatchability and Mortality Rate in Chickens (Author’s Transl.). Nord. Vet. 1976, 28, 368–376. [Google Scholar]
- Sevoian, M.; Levine, P.P. Effects of Infectious Bronchitis on the Reproductive Tracts, Egg Production, and Egg Quality of Laying Chickens. Avian Dis. 1957, 1, 136–164. [Google Scholar] [CrossRef]
- Valastro, V.; Holmes, E.C.; Britton, P.; Fusaro, A.; Jackwood, M.W.; Cattoli, G.; Monne, I. S1 Gene-based Phylogeny of Infectious Bronchitis Virus: An Attempt to Harmonize Virus Classification. Infect. Genet. Evol. 2016, 39, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, Scalable Generation of High-quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and Analysis of Recombination Patterns in Virus Genomes. Virus Evol. 2015, 1. [Google Scholar] [CrossRef]
- Amarasinghe, A.; Popowich, S.; de Silva Senapathi, U.; Abdul-Cader, M.S.; Marshall, F.; van der Meer, F.; Cork, S.C.; Gomis, S.; Abdul-Careem, M.F. Shell-Less Egg Syndrome (SES) Widespread in Western Canadian Layer Operations Is Linked to a Massachusetts (Mass) Type Infectious Bronchitis Virus (IBV) Isolate. Viruses 2018, 10, 437. [Google Scholar] [CrossRef]
- Chandra, M. Comparative Nephropathogenicity of Different Strains of Infectious Bronchitis Virus in Chickens. Poult. Sci. 1987, 66, 954–959. [Google Scholar] [CrossRef]
- Kameka, A.M.; Haddadi, S.; Kim, D.S.; Cork, S.C.; Abdul-Careem, M.F. Induction of Innate Immune Response Following Infectious Bronchitis Corona Virus Infection in the Respiratory Tract of Chickens. Virology 2014, 450, 114–121. [Google Scholar] [CrossRef] [PubMed]



| UTR or ORF | Nucleotide Position | Nucleotide Length | Amino Acid Length |
|---|---|---|---|
| 5′UTR | 1–528 | 528 | - |
| 1a | 529–12,384 | 11,856 | 3951 |
| 1b | 12,459–20,417 | 7959 | 2652 |
| Spike | 20,368–23,862 | 3495 | 1164 |
| 3a | 23,862–24,035 | 174 | 57 |
| 3b | 24,035–24,229 | 195 | 64 |
| E (3c) | 24,210–24,539 | 330 | 109 |
| M | 24,511–25,188 | 678 | 225 |
| 4b | 25,189–25,473 | 285 | 94 |
| 4c | 25,394–25,555 | 162 | 53 |
| 5a | 25,539–25,736 | 198 | 65 |
| 5b | 25,733–25,981 | 249 | 82 |
| N | 25,924–27,153 | 1230 | 409 |
| 3′UTR | 27,154–27,472 | 319 | - |
| IBV Strain-Database Accession No. | Genome | 1a | 1b | Spike | 3a | 3b | E (3c) | M | 4b | 4c | 5a | 5b | N |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4/91 vaccine-KF377577 | 90.7 | 88.5 | 93.3 | 96.6 | 93.7 | 89.7 | 83.0 | 92.4 | 88.1 | 83.0 | 84.3 | 96.4 | 90.9 |
| Arkansas Vaccine-GQ504721 | 90.5 | 90.3 | 94.1 | 83.5 | 93.7 | 90.3 | 86.7 | 90.6 | 91.9 | 85.4 | 88.4 | 97.6 | 96.4 |
| B1648-KR231009 | 90.4 | 91.4 | 92.4 | 85.3 | 93.7 | 88.2 | 72.4 | 91.6 | 91.6 | 83.6 | 86.9 | 95.2 | 93.3 |
| California 99-AY514485 | 90.8 | 91.0 | 94.4 | 82.9 | 94.8 | 89.2 | 90.6 | 92.9 | 91.9 | 84.8 | 88.4 | 98.0 | 94.7 |
| Ck/Aus/N1/08-KU556807 | 85.5 | 86.5 | 90.6 | 70.9 | 87.4 | 86.7 | 83.8 | 87.0 | 86.0 | 80.1 | 83.3 | 91.6 | 88.5 |
| ck/CH/LSD/110857-KP118885 | 91.1 | 89.1 | 93.3 | 96.7 | 93.7 | 89.7 | 83.0 | 92.4 | 88.1 | 83.0 | 84.3 | 96.4 | 90.8 |
| CK/SWE/0658946/10-JQ088078 | 88.3 | 89.7 | 91.0 | 81.0 | 84.5 | 77.4 | 84.7 | 86.1 | 91.9 | 88.3 | 86.9 | 94.4 | 88.9 |
| Conn46 1996-FJ904716 | 91.7 | 92.5 | 94.5 | 83.7 | 94.8 | 89.2 | 86.4 | 90.6 | 91.9 | 85.4 | 88.4 | 98.4 | 96.7 |
| Delaware 072-GU393332 | 87.6 | 89.3 | 92.6 | 66.8 | 91.4 | 90.3 | 90.6 | 97.2 | 85.3 | 78.9 | 87.4 | 98.4 | 95.1 |
| γCoV/Ck/Poland/255/1997-MK581204 | 89.9 | 88.3 | 92.6 | 93.0 | 93.1 | 89.2 | 73.3 | 91.5 | 90.5 | 82.5 | 84.8 | 96.4 | 89.5 |
| IBV_SES_15SK-02-MH539772 | 90.8 | 90.0 | 92.5 | 83.6 | 86.8 | 93.3 | 97.9 | 99.0 | 97.9 | 98.1 | 97.5 | 98.4 | 93.2 |
| IBV/Ck/Can/17-036989-MN512435 | 90.3 | 90.0 | 94.6 | 81.0 | 94.8 | 89.2 | 91.5 | 96.2 | 92.6 | 85.4 | 88.4 | 98.4 | 95.7 |
| IBV/Ck/EG/CU/4/2014-KY805846 | 87.3 | 86.7 | 90.4 | 82.4 | 90.8 | 88.7 | 73.3 | 90.9 | 88.4 | 78.4 | 84.3 | 95.6 | 91.6 |
| IBVUkr27-11-KJ135013 | 90.9 | 89.1 | 93.3 | 95.1 | 93.1 | 88.7 | 76.7 | 90.9 | 90.9 | 83.6 | 85.9 | 97.2 | 92.7 |
| ITA/90254/2005-FN430414 | 89.2 | 90.8 | 90.8 | 81.5 | 85.6 | 77.9 | 88.6 | 92.5 | 90.5 | 83.0 | 85.9 | 96.8 | 92.1 |
| JMK-GU393338 | 92.0 | 93.0 | 93.2 | 83.3 | 93.7 | 90.3 | 87.6 | 91.7 | 91.9 | 84.8 | 88.4 | 97.2 | 95.5 |
| KM91-JQ977698 | 90.0 | 91.5 | 92.6 | 82.1 | 79.6 | 82.6 | 87.3 | 90.7 | 93.0 | 86.0 | 87.4 | 97.2 | 93.7 |
| Mass41 2006-FJ904713 | 91.3 | 92.1 | 93.8 | 84.1 | 94.8 | 89.2 | 85.5 | 90.5 | 91.9 | 85.4 | 88.4 | 98.4 | 96.7 |
| NGA/A116E7/2006-FN430415 | 90.3 | 91.4 | 92.5 | 85.6 | 85.6 | 85.1 | 73.9 | 91.2 | 89.4 | 76.6 | 85.9 | 95.6 | 92.1 |
| SAIBK-DQ288927 | 87.5 | 87.4 | 90.1 | 81.9 | 84.5 | 83.3 | 88.7 | 88.9 | 85.3 | 83.6 | 82.3 | 96.0 | 86.8 |
| TCoV/IN-517/94-GQ427175 | 87.2 | 93.5 | 94.3 | 48.3 | 93.7 | 88.7 | 90.0 | 90.8 | 91.6 | 83.6 | 87.4 | 97.6 | 96.6 |
| TW2575/98-DQ646405 | 86.1 | 85.1 | 89.5 | 81.7 | 87.9 | 89.2 | 88.8 | 89.1 | 84.6 | 81.3 | 81.3 | 96.4 | 89.9 |
| YN-JF893452 | 86.8 | 87.3 | 89.4 | 81.9 | 83.9 | 82.1 | 89.3 | 89.2 | 86.3 | 83.6 | 80.8 | 96.0 | 87.5 |
| Histopathological Finding | 4/91 IBV Infected | Control |
|---|---|---|
| Dilation of collecting duct | 6/6 | 1/6 |
| Increased urate spherules | 6/6 | 0/6 |
| Mononuclear cell infiltration | 2/6 | 0/6 |
| Sloughed epithelial cells in the lumen of collecting duct | 4/6 | 0/6 |
| Hyper eosinophilic necrotic cellular debris | 2/6 | 0/6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Najimudeen, S.M.; Hassan, M.S.H.; Goldsmith, D.; Ojkic, D.; Cork, S.C.; Boulianne, M.; Abdul-Careem, M.F. Molecular Characterization of 4/91 Infectious Bronchitis Virus Leading to Studies of Pathogenesis and Host Responses in Laying Hens. Pathogens 2021, 10, 624. https://doi.org/10.3390/pathogens10050624
Najimudeen SM, Hassan MSH, Goldsmith D, Ojkic D, Cork SC, Boulianne M, Abdul-Careem MF. Molecular Characterization of 4/91 Infectious Bronchitis Virus Leading to Studies of Pathogenesis and Host Responses in Laying Hens. Pathogens. 2021; 10(5):624. https://doi.org/10.3390/pathogens10050624
Chicago/Turabian StyleNajimudeen, Shahnas M., Mohamed S. H. Hassan, Dayna Goldsmith, Davor Ojkic, Susan C. Cork, Martine Boulianne, and Mohamed Faizal Abdul-Careem. 2021. "Molecular Characterization of 4/91 Infectious Bronchitis Virus Leading to Studies of Pathogenesis and Host Responses in Laying Hens" Pathogens 10, no. 5: 624. https://doi.org/10.3390/pathogens10050624
APA StyleNajimudeen, S. M., Hassan, M. S. H., Goldsmith, D., Ojkic, D., Cork, S. C., Boulianne, M., & Abdul-Careem, M. F. (2021). Molecular Characterization of 4/91 Infectious Bronchitis Virus Leading to Studies of Pathogenesis and Host Responses in Laying Hens. Pathogens, 10(5), 624. https://doi.org/10.3390/pathogens10050624







