Strong Inhibitory Activity and Action Modes of Synthetic Maslinic Acid Derivative on Highly Pathogenic Coronaviruses: COVID-19 Drug Candidate
Abstract
1. Introduction
2. Results and Discussion
2.1. Chemistry
2.1.1. Green Chemistry Extraction of Compounds 1 and 2
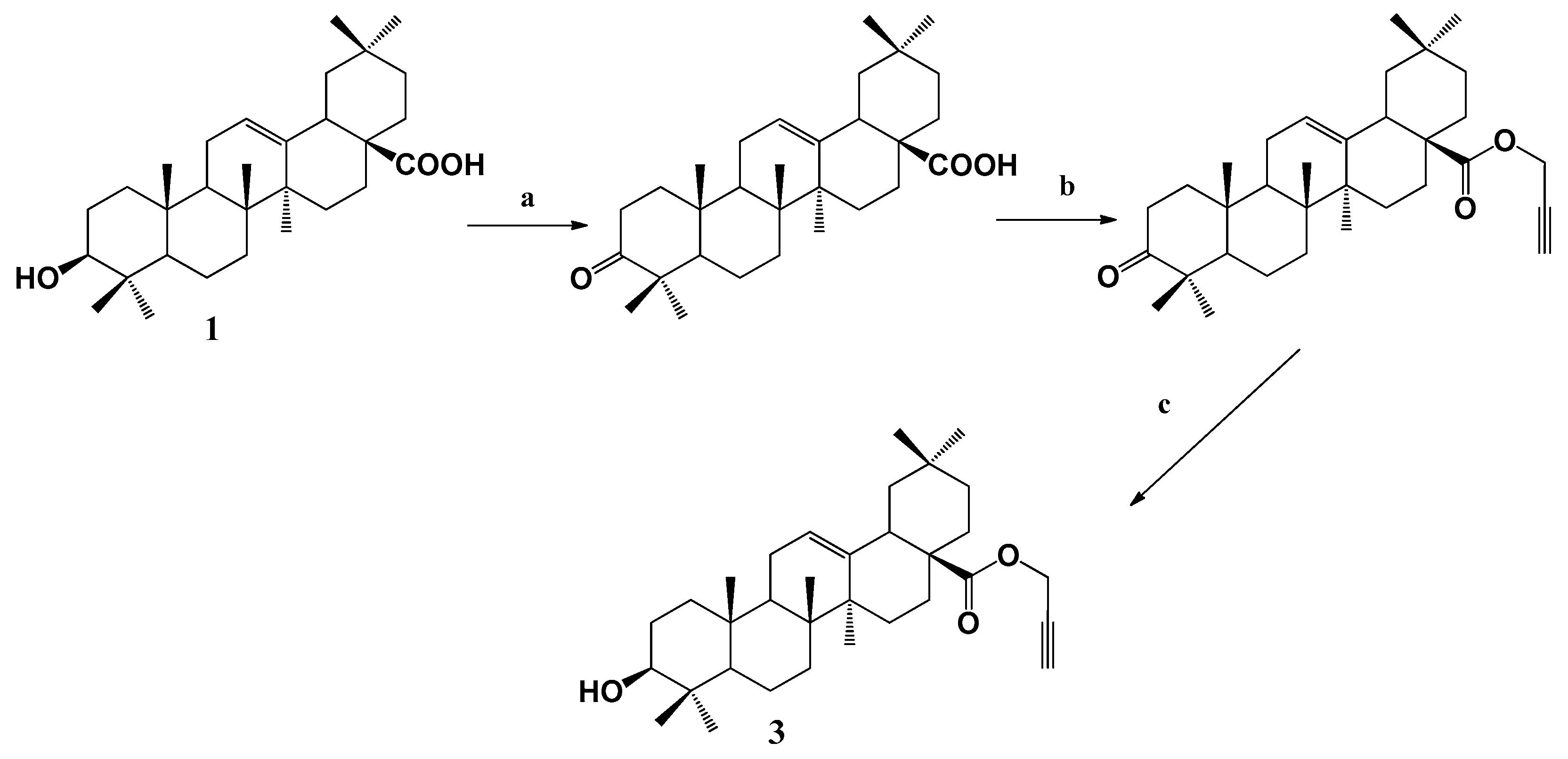
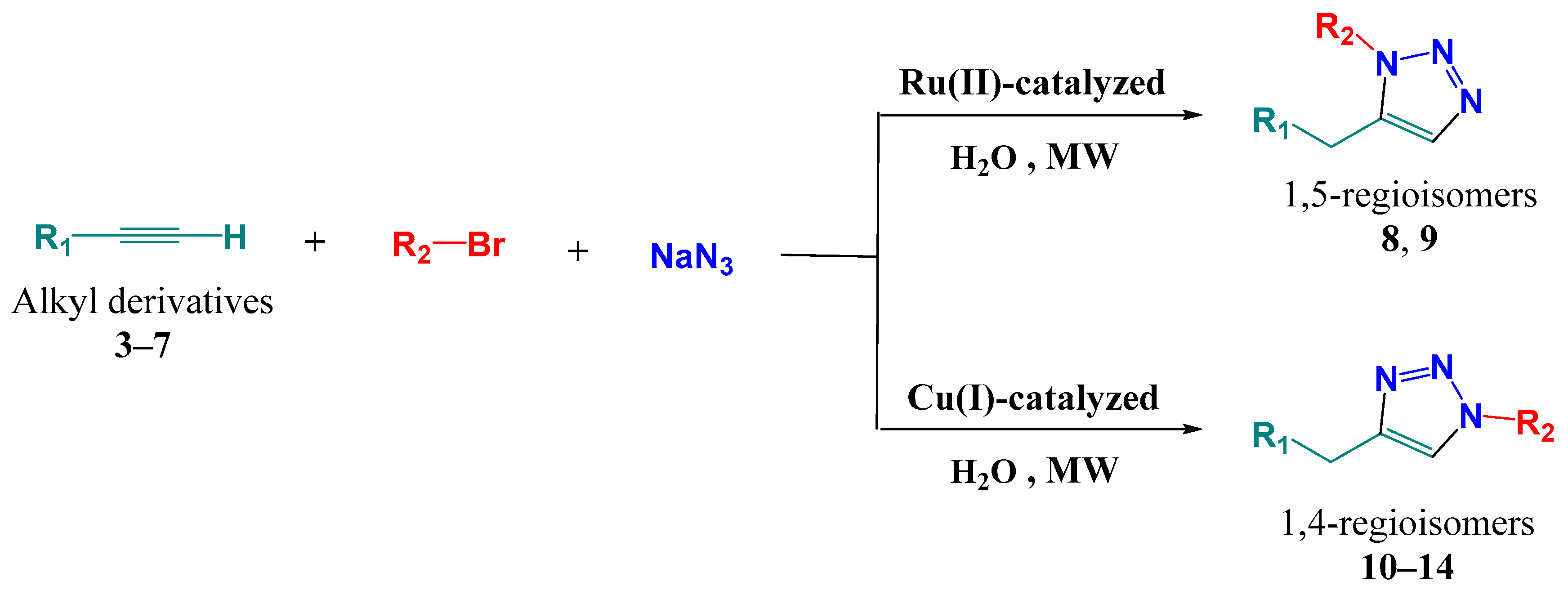
2.1.2. Synthesis
2.2. Anti-SARS-CoV-2 Activity
2.2.1. Molecular Docking Studies
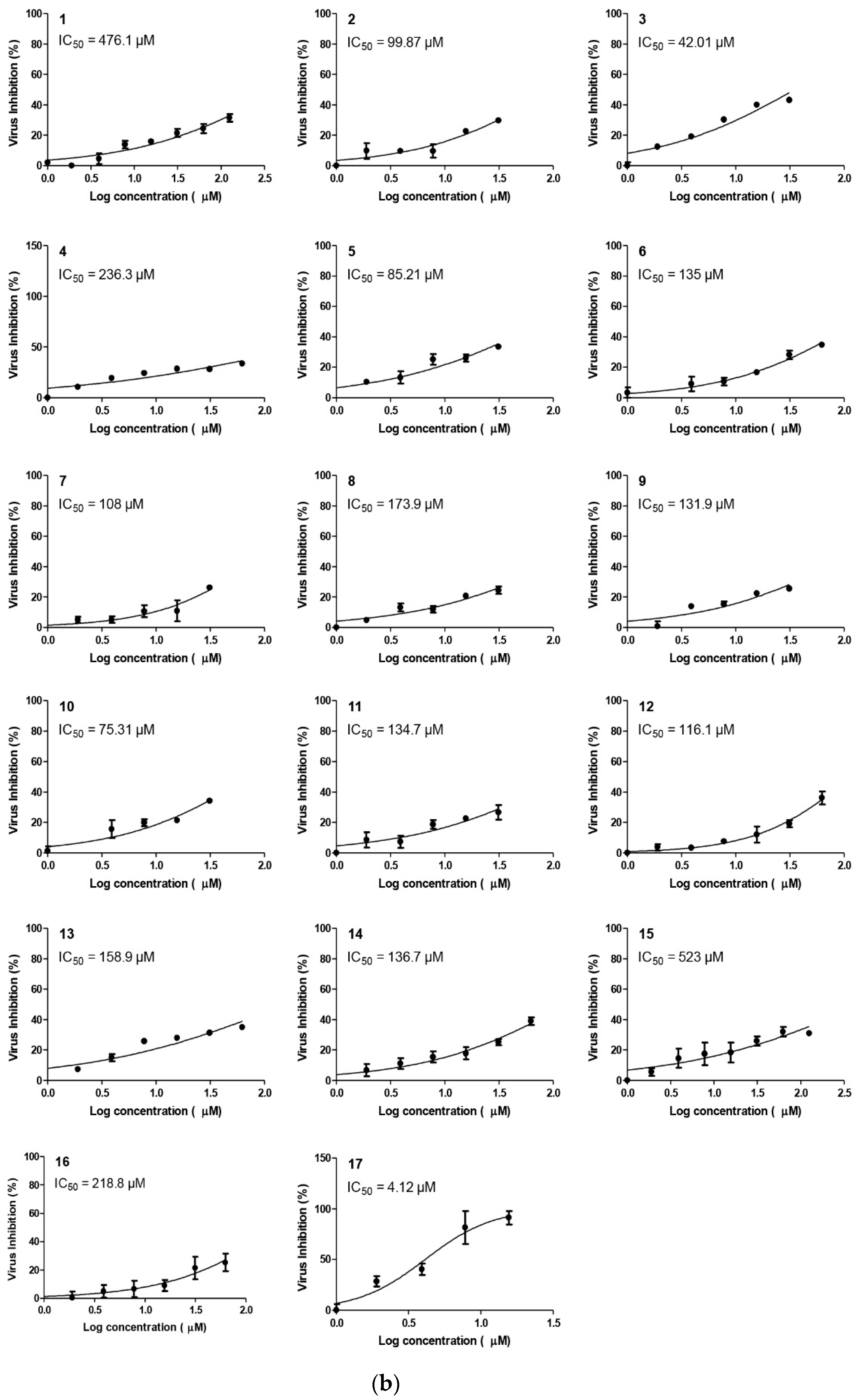
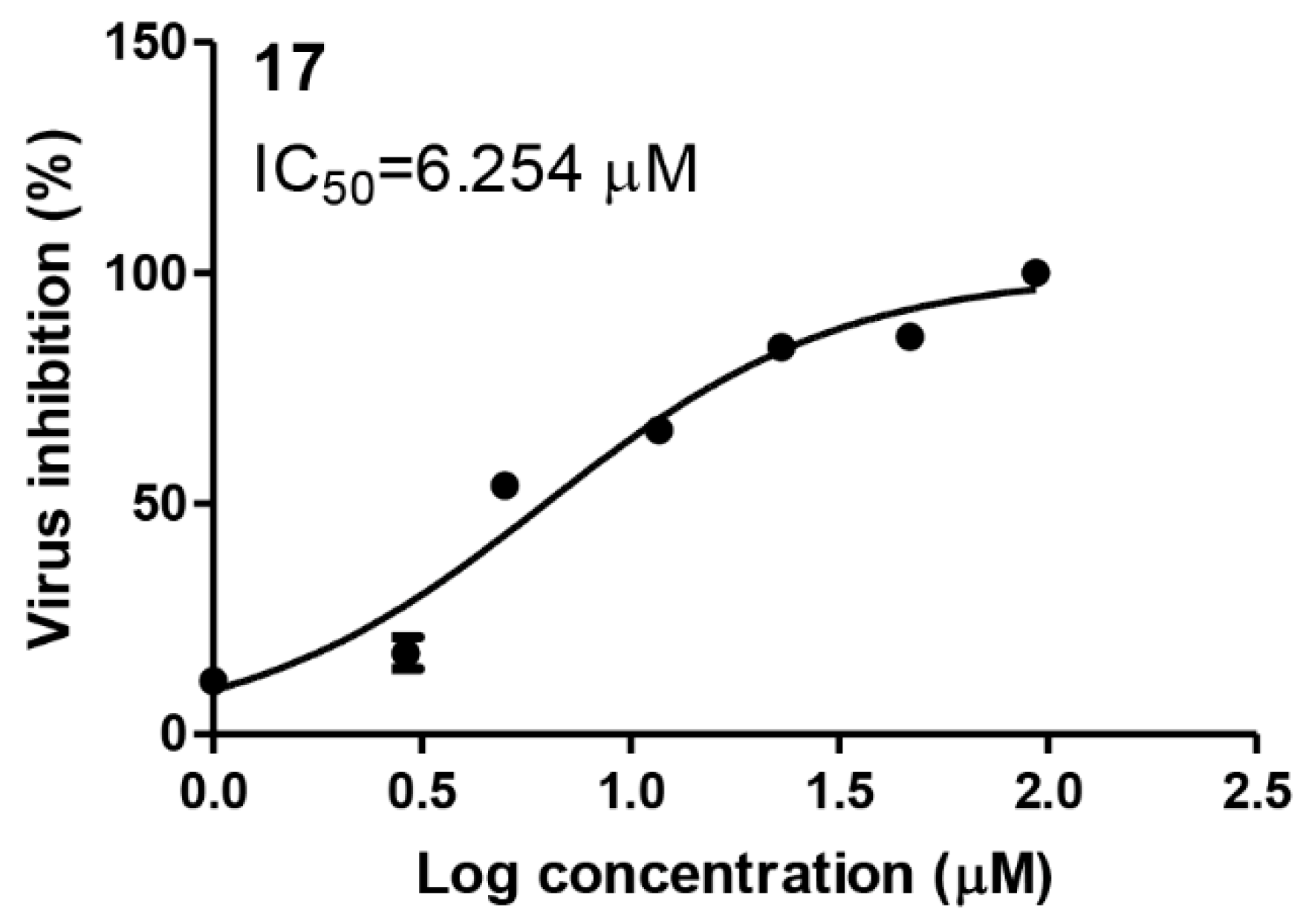
2.2.2. Compound 17 Showed Promising Anti-SARS-CoV-2 Activity at Non-Toxic Concentrations
2.2.3. Compound 17 Showed Promising Anti-MERS-CoV Activity and Non-Toxic Concentrations
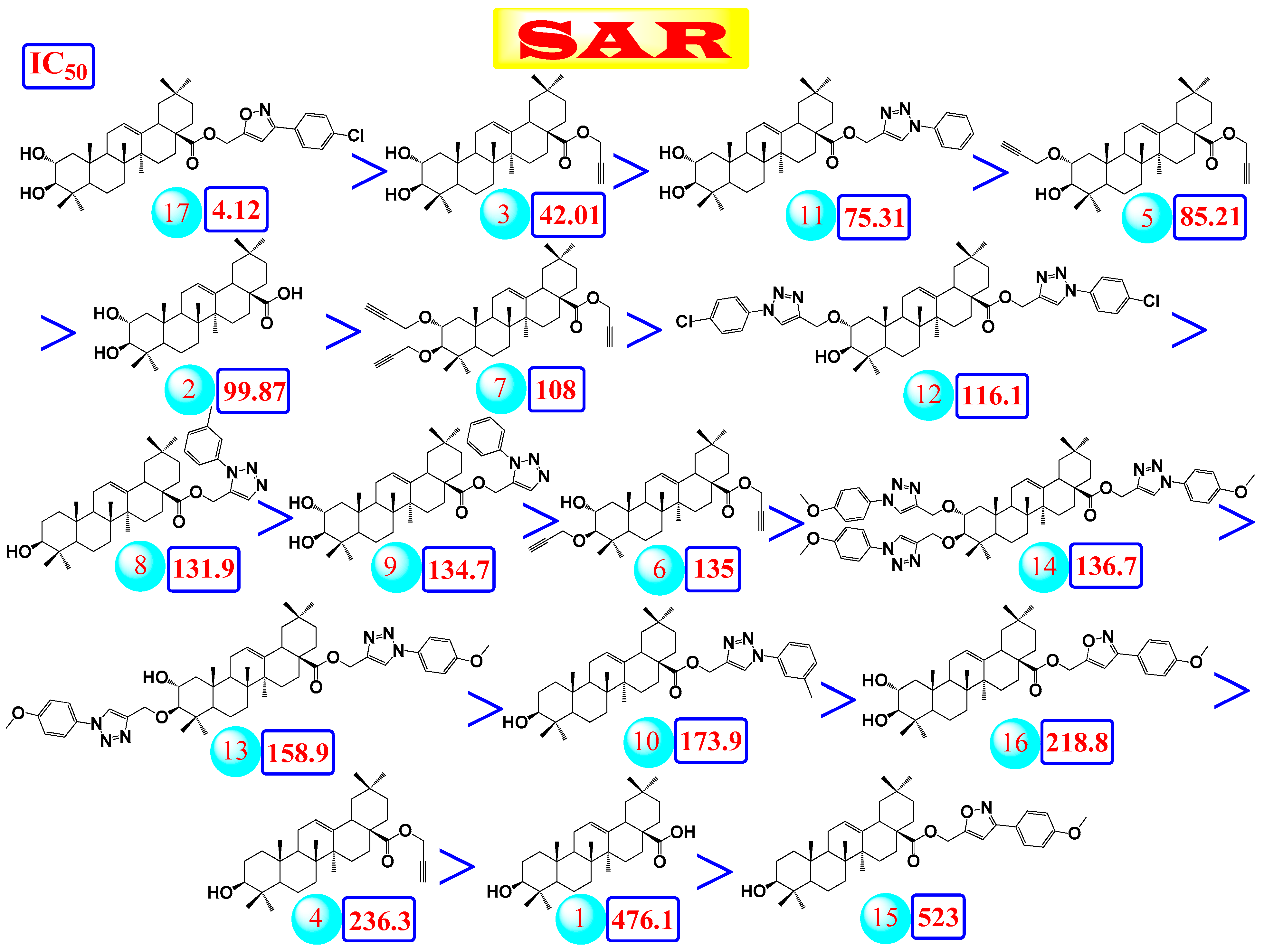
2.2.4. Structure–Activity Relationship (SAR) Study
- (a)
- The common shared steroidal nucleus of the selected tested compounds (1–17) appeared to be very promising for their antiviral activities. Molecular docking revealed the great stabilization of the tested compounds inside the branched large pocket of SARS-CoV-2 main protease as a proposed mechanism of action.
- (b)
- The presence of OH groups at positions 2 and 3 of the steroidal nucleus seems to be very crucial for the antiviral activity. Most compounds (17, 3, and 10) containing both 2- and 3-OH groups achieved high to moderate activities (4.12–99.87 µM). Docking studies referred to the involvement of both OH groups in hydrogen bond formations with Thr24 and/or Thr26 amino acids of the binding pocket.
- (c)
- Compound (17) containing the isoxazole side chain with a p-chlorophenyl substitution (an electron withdrawing lipophilic group) achieved a very high cytotoxic activity (IC50 = 4.12 µM). However, both compounds (16 and 15) containing the isoxazole side chain with a p-methoxyphenyl substitution (an electron donating hydrophilic group) achieved low cytotoxic activities (IC50 = 218.8 and 523 µM, respectively). Therefore, the lipophilic p-chloro group is very important for the antiviral activity. Again, compound (16) containing both 2- and 3-OH groups showed a higher cytotoxic activity compared to compound (15) with only 3-OH group as mentioned before.
- (d)
- The presence of the acetyl methylene group at positions 2 and/or 3 in compounds (5, 7, and 6) gave better cytotoxic activities compared to the heteroaromatic ring substitutions at the same positions in compounds (12, 14, and 13).
- (e)
- The cytotoxic activity of compound (17) containing the isoxazole side chain with a p-chlorophenyl substitution (IC50 = 4.12 µM) was clearly higher than the respective same compound (10) with a phenyl triazole side chain instead (IC50 = 75.31 µM).
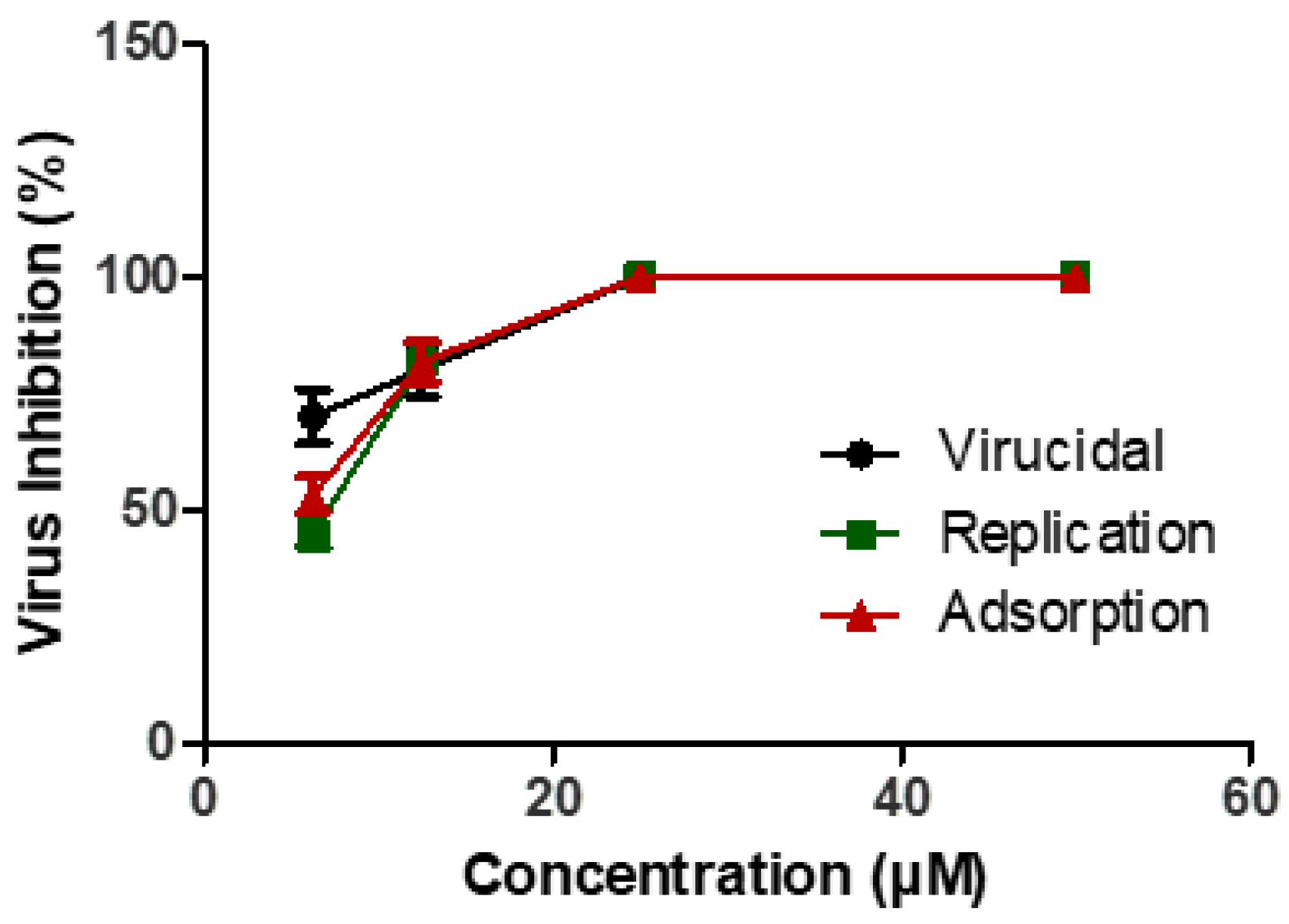
2.2.5. Mechanism of Actions
3. Materials and Methods
3.1. Chemistry
3.1.1. Green Chemistry Condition for Extraction and Purification of OA (1) and MA (2) from the Pomace Olive of Olea europaea L.
3.1.2. Synthesis
Multicomponent Synthesis of 1,5-Disubstituted Triazoles in Water Catalyzed by Cp*RuCl (PPh3)2
Cu(I))-Cuatalyzed Multicomponent Huisgen 1,3-Dipolar Cycloaddition Reaction
Multicomponent Synthesis of 3,5-Disubstituted Isoxazoles in Water Catalyzed by CuI
3.2. Biological
3.2.1. Molecular Docking Studies
3.2.2. Preparation of the Selected Tested Compounds
3.2.3. Preparation of the Active Site of the Applied SARS-CoV-2 Mpro Receptor
3.2.4. Validation of the Applied MOE Program
3.2.5. Docking of the Prepared Database to the Binding Pocket of SARS-CoV-2 Mpro Receptor
3.2.6. In Vitro Antiviral Bioassay
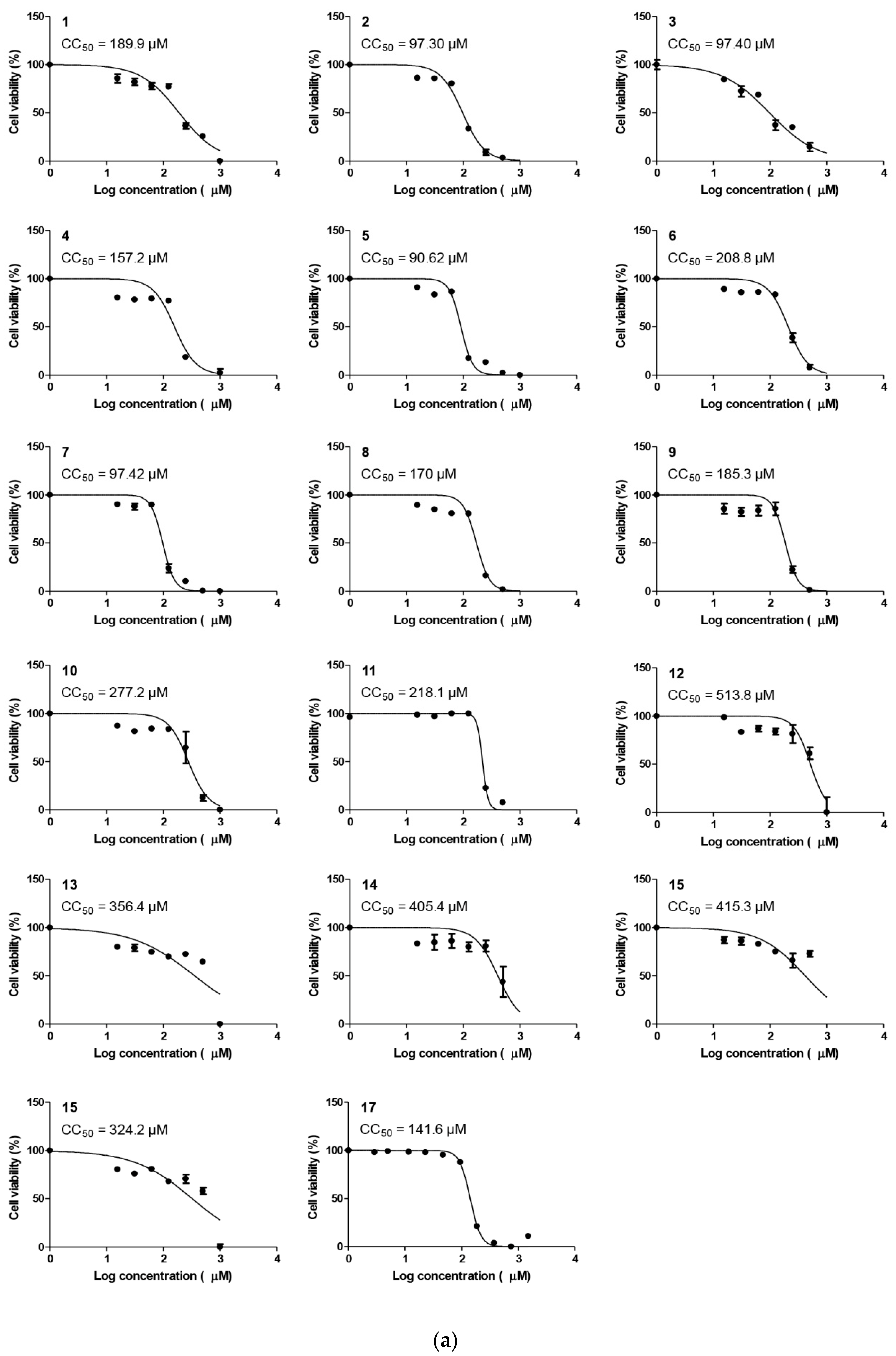
Cytotoxicity (CC50) Determination
Inhibitory Concentration 50 (IC50) Determination
Mechanism of Action(s) for Compound 17
- (a)
- Viral adsorption mechanism
- (b)
- Viral replication mechanism
- (c)
- Virucidal mechanism
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, S.; Singh, R.; Kumari, N.; Karmakar, S.; Behera, M.; Siddiqui, A.J.; Rajput, V.D.; Minkina, T.; Bauddh, K.; Kumar, N. Current understanding of the influence of environmental factors on sars-cov-2 transmission, persistence, and infectivity. Environ. Sci. Pollut. Res. Int. 2021, 28, 6267–6288. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, A.; Kandeil, A.; Elshaier, Y.A.M.M.; Kutkat, O.; Moatasim, Y.; Rashad, A.A.; Shehata, M.; Gomaa, M.R.; Mahrous, N.; Mahmoud, S.H.; et al. Fda-approved drugs with potent in vitro antiviral activity against severe acute respiratory syndrome coronavirus 2. Pharmaceuticals 2020, 13, 443. [Google Scholar] [CrossRef] [PubMed]
- Alnajjar, R.; Mostafa, A.; Kandeil, A.; Al-Karmalawy, A.A. Molecular docking, molecular dynamics, and in vitro studies reveal the potential of angiotensin ii receptor blockers to inhibit the covid-19 main protease. Heliyon 2020, 6, e05641. [Google Scholar] [CrossRef]
- Mahmoud, D.B.; Shitu, Z.; Mostafa, A. Drug repurposing of nitazoxanide: Can it be an effective therapy for covid-19? J. Genet. Eng. Biotechnol. 2020, 18, 35. [Google Scholar] [CrossRef]
- Surti, M.; Patel, M.; Adnan, M.; Moin, A.; Ashraf, S.A.; Siddiqui, A.J.; Snoussi, M.; Deshpande, S.; Reddy, M.N. Ilimaquinone (marine sponge metabolite) as a novel inhibitor of sars-cov-2 key target proteins in comparison with suggested covid-19 drugs: Designing, docking and molecular dynamics simulation study. RSC Adv. 2020, 10, 37707–37720. [Google Scholar] [CrossRef]
- Alzahrani, F.A.; Saadeldin, I.M.; Ahmad, A.; Kumar, D.; Azhar, E.I.; Siddiqui, A.J.; Kurdi, B.; Sajini, A.; Alrefaei, A.F.; Jahan, S. The potential use of mesenchymal stem cells and their derived exosomes as immunomodulatory agents for covid-19 patients. Stem Cells Int. 2020, 2020, 8835986. [Google Scholar] [CrossRef] [PubMed]
- Fayed, M.A.A.; El-Behairy, M.F.; Abdallah, I.A.; Abdel-Bar, H.M.; Elimam, H.; Mostafa, A.; Moatasim, Y.; Abouzid, K.A.M.; Elshaier, Y.A.M.M. Structure- and ligand-based in silico studies towards the repurposing of marine bioactive compounds to target sars-cov-2. Arab. J. Chem. 2021, 14, 103092. [Google Scholar] [CrossRef]
- Xu, R.; Fazio, G.C.; Matsuda, S.P.T. On the origins of triterpenoid skeletal diversity. Phytochemistry 2004, 65, 261–291. [Google Scholar] [CrossRef] [PubMed]
- Bruneton, J. Pharmacognosie, Phytochimie, Plantes Médicinales; Technique et Documentation Lavoisier: Paris, France, 1999. [Google Scholar]
- Muffler, K.; Leipold, D.; Scheller, M.-C.; Haas, C.; Steingroewer, J.; Bley, T.; Neuhaus, H.E.; Mirata, M.A.; Schrader, J.; Ulber, R. Biotransformation of triterpenes. Process Biochem. 2011, 46, 1–15. [Google Scholar] [CrossRef]
- Hisham Shady, N.; Youssif, K.A.; Sayed, A.M.; Belbahri, L.; Oszako, T.; Hassan, H.M.; Abdelmohsen, U.R. Sterols and triterpenes: Antiviral potential supported by in-silico analysis. Plants 2021, 10, 41. [Google Scholar] [CrossRef]
- Dzubak, P.; Hajduch, M.; Vydra, D.; Hustova, A.; Kvasnica, M.; Biedermann, D.; Markova, L.; Urban, M.; Sarek, J. Pharmacological activities of natural triterpenoids and their therapeutic implications. Nat. Prod. Rep. 2006, 23, 394–411. [Google Scholar] [CrossRef] [PubMed]
- Sparg, S.G.; Light, M.E.; van Staden, J. Biological activities and distribution of plant saponins. J. Ethnopharmacol. 2004, 94, 219–243. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.; Liu, J.; Liu, X.; Li, H.; Sun, H.; Xie, J. Synthesis of glucoconjugates of oleanolic acid as inhibitors of glycogen phosphorylase. Carbohydr. Res. 2009, 344, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Chouaïb, K.; Hichri, F.; Nguir, A.; Daami-Remadi, M.; Elie, N.; Touboul, D.; Ben Jannet, H.; Hamza, M.A. Semi-synthesis of new antimicrobial esters from the natural oleanolic and maslinic acids. Food Chem. 2015, 183, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Chouaïb, K.; Delemasure, S.; Dutartre, P.; Jannet, H.B. Microwave-assisted synthesis, anti-inflammatory and anti-proliferative activities of new maslinic acid derivatives bearing 1,5- and 1,4-disubstituted triazoles. J. Enzym. Inhib. Med. Chem. 2016, 31, 130–147. [Google Scholar] [CrossRef] [PubMed]
- Chouaïb, K.; Romdhane, A.; Delemasure, S.; Dutartre, P.; Elie, N.; Touboul, D.; Ben jannet, H.; Ali Hamza, M.h. Regiospecific synthesis, anti-inflammatory and anticancer evaluation of novel 3,5-disubstituted isoxazoles from the natural maslinic and oleanolic acids. Ind. Crop. Prod. 2016, 85, 287–299. [Google Scholar] [CrossRef]
- Chouaïb, K.; Romdhane, A.; Delemasure, S.; Dutartre, P.; Elie, N.; Touboul, D.; Ben Jannet, H. Regiospecific synthesis by copper- and ruthenium-catalyzed azide–alkyne 1,3-dipolar cycloaddition, anticancer and anti-inflammatory activities of oleanolic acid triazole derivatives. Arab. J. Chem. 2019, 12, 3732–3742. [Google Scholar] [CrossRef]
- Chen, P.; Zeng, H.; Wang, Y.; Fan, X.; Xu, C.; Deng, R.; Zhou, X.; Bi, H.; Huang, M. Low dose of oleanolic acid protects against lithocholic acid-induced cholestasis in mice: Potential involvement of nuclear factor-e2-related factor 2-mediated upregulation of multidrug resistance-associated proteins. Drug Metab. Dispos. Biol. Fate Chem. 2014, 42, 844–852. [Google Scholar] [CrossRef]
- Sheng, H.; Sun, H. Synthesis, biology and clinical significance of pentacyclic triterpenes: A multi-target approach to prevention and treatment of metabolic and vascular diseases. Nat. Prod. Rep. 2011, 28, 543–593. [Google Scholar] [CrossRef]
- Chen, D.-F.; Zhang, S.-X.; Wang, H.-K.; Zhang, S.-Y.; Sun, Q.-Z.; Cosentino, L.M.; Lee, K.-H. Novel anti-hiv lancilactone c and related triterpenes from kadsura lancilimba. J. Nat. Prod. 1999, 62, 94–97. [Google Scholar] [CrossRef]
- Kashiwada, Y.; Wang, H.K.; Nagao, T.; Kitanaka, S.; Yasuda, I.; Fujioka, T.; Yamagishi, T.; Cosentino, L.M.; Kozuka, M.; Okabe, H.; et al. Anti-aids agents. 30. Anti-hiv activity of oleanolic acid, pomolic acid, and structurally related triterpenoids. J. Nat. Prod. 1998, 61, 1090–1095. [Google Scholar] [CrossRef]
- Zhu, Y.M.; Shen, J.K.; Wang, H.K.; Cosentino, L.M.; Lee, K.H. Synthesis and anti-hiv activity of oleanolic acid derivatives. Bioorg. Med. Chem. Lett. 2001, 11, 3115–3118. [Google Scholar] [CrossRef]
- Shanmugam, M.K.; Dai, X.; Kumar, A.P.; Tan, B.K.; Sethi, G.; Bishayee, A. Oleanolic acid and its synthetic derivatives for the prevention and therapy of cancer: Preclinical and clinical evidence. Cancer Lett. 2014, 346, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Hichri, F.; Jannet, H.B.; Cheriaa, J.; Jegham, S.; Mighri, Z. Antibacterial activities of a few prepared derivatives of oleanolic acid and of other natural triterpenic compounds. Comptes Rendus Chim. 2003, 6, 473–483. [Google Scholar] [CrossRef]
- Tsuji, M.; Sriwilaijaroen, N.; Inoue, H.; Miki, K.; Kinoshita, K.; Koyama, K.; Furuhata, K.; Suzuki, Y.; Takahashi, K. Synthesis and anti-influenza virus evaluation of triterpene-sialic acid conjugates. Bioorg. Med. Chem. 2018, 26, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Cabra, N.; Vega-Granados, K.; Moya-Andérico, L.; Vukomanovic, M.; Parra, A.; Álvarez de Cienfuegos, L.; Torrents, E. Novel oleanolic and maslinic acid derivatives as a promising treatment against bacterial biofilm in nosocomial infections: An in vitro and in vivo study. ACS Infect. Dis. 2019, 5, 1581–1589. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Zhang, P.; Liu, J.; Zhang, L.; Wu, X.; Ni, P.; Sun, H. Pentacyclic triterpenes. Part 2: Synthesis and biological evaluation of maslinic acid derivatives as glycogen phosphorylase inhibitors. Bioorg. Med. Chem. Lett. 2006, 16, 722–726. [Google Scholar] [CrossRef]
- Qiu, W.-W.; Shen, Q.; Yang, F.; Wang, B.; Zou, H.; Li, J.-Y.; Li, J.; Tang, J. Synthesis and biological evaluation of heterocyclic ring-substituted maslinic acid derivatives as novel inhibitors of protein tyrosine phosphatase 1b. Bioorg. Med. Chem. Lett. 2009, 19, 6618–6622. [Google Scholar] [CrossRef]
- Taniguchi, S.; Imayoshi, Y.; Kobayashi, E.; Takamatsu, Y.; Ito, H.; Hatano, T.; Sakagami, H.; Tokuda, H.; Nishino, H.; Sugita, D.; et al. Production of bioactive triterpenes by eriobotrya japonica calli. Phytochemistry 2002, 59, 315–323. [Google Scholar] [CrossRef]
- Parra, A.; Rivas, F.; Lopez, P.E.; Garcia-Granados, A.; Martinez, A.; Albericio, F.; Marquez, N.; Muñoz, E. Solution- and solid-phase synthesis and anti-hiv activity of maslinic acid derivatives containing amino acids and peptides. Bioorg. Med. Chem. 2009, 17, 1139–1145. [Google Scholar] [CrossRef]
- Agrawal, N.; Mishra, P. The synthetic and therapeutic expedition of isoxazole and its analogs. Med. Chem. Res. 2018, 27, 1309–1344. [Google Scholar] [CrossRef] [PubMed]
- Kónya, B.; Docsa, T.; Gergely, P.; Somsák, L. Synthesis of heterocyclic n-(β-d-glucopyranosyl)carboxamides for inhibition of glycogen phosphorylase. Carbohydr. Res. 2012, 351, 56–63. [Google Scholar] [CrossRef][Green Version]
- Koufaki, M.; Tsatsaroni, A.; Alexi, X.; Guerrand, H.; Zerva, S.; Alexis, M.N. Isoxazole substituted chromans against oxidative stress-induced neuronal damage. Bioorg. Med. Chem. 2011, 19, 4841–4850. [Google Scholar] [CrossRef] [PubMed]
- Filali, I.; Romdhane, A.; Znati, M.; Jannet, H.B.; Bouajila, J. Synthesis of new harmine isoxazoles and evaluation of their potential anti-alzheimer, anti-inflammatory, and anticancer activities. Med. Chem. 2016, 12, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Agalave, S.G.; Maujan, S.R.; Pore, V.S. Click chemistry: 1,2,3-triazoles as pharmacophores. Chem. Asian J. 2011, 6, 2696–2718. [Google Scholar] [CrossRef] [PubMed]
- Gonzaga, D.T.G.; Souza, T.M.L.; Andrade, V.M.M.; Ferreira, V.F.; de C da Silva, F. Identification of 1-aryl-1h-1,2,3-triazoles as potential new antiretroviral agents. Med. Chem. 2018, 14, 242–248. [Google Scholar] [CrossRef]
- El-Sebaey, S.A. Recent advances in 1,2,4-triazole scaffolds as antiviral agents. ChemistrySelect 2020, 5, 11654–11680. [Google Scholar] [CrossRef]
- Wang, X.L.; Wan, K.; Zhou, C.H. Synthesis of novel sulfanilamide-derived 1,2,3-triazoles and their evaluation for antibacterial and antifungal activities. Eur. J. Med. Chem. 2010, 45, 4631–4639. [Google Scholar] [CrossRef] [PubMed]
- Kandeil, A.; Mostafa, A.; El-Shesheny, R.; Shehata, M.; Roshdy, W.H.; Ahmed, S.S.; Gomaa, M.; Taweel, A.E.; Kayed, A.E.; Mahmoud, S.H.; et al. Coding-complete genome sequences of two sars-cov-2 isolates from egypt. Microbiol. Resour. Announc. 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Guinda, A.; Rada, M.; Delgado, T.; Gutiérrez-Adánez, P.; Castellano, J.M. Pentacyclic triterpenoids from olive fruit and leaf. J. Agric. Food Chem. 2010, 58, 9685–9691. [Google Scholar] [CrossRef] [PubMed]
- Gil, M.; Haïdour, A.; Ramos, J.L. Identification of two triterpenoids in solid wastes from olive cake. J. Agric. Food Chem. 1997, 45, 4490–4494. [Google Scholar] [CrossRef]
- Romero, C.; García, A.; Medina, E.; Ruíz-Méndez, M.V.; Castro, A.d.; Brenes, M. Triterpenic acids in table olives. Food Chem. 2010, 118, 670–674. [Google Scholar] [CrossRef]
- Goulas, V.; Manganaris, G.A. Towards an efficient protocol for the determination of triterpenic acids in olive fruit: A comparative study of drying and extraction methods. Phytochem. Anal. 2012, 23, 444–449. [Google Scholar] [CrossRef]
- Chan, R.W.Y.; Hemida, M.G.; Kayali, G.; Chu, D.K.W.; Poon, L.L.M.; Alnaeem, A.; Ali, M.A.; Tao, K.P.; Ng, H.Y.; Chan, M.C.W.; et al. Tropism and replication of middle east respiratory syndrome coronavirus from dromedary camels in the human respiratory tract: An in-vitro and ex-vivo study. Lancet Respir. Med. 2014, 2, 813–822. [Google Scholar] [CrossRef]
- Oakdale, J.S.; Fokin, V.V.; Umezaki, S.; Fukuyama, T. Preparation of 1,5-disubstituted 1,2,3-triazoles via ruthenium-catalyzed azide alkyne cycloaddition. Org. Synth 2013, 90, 96–104. [Google Scholar] [PubMed]
- Takasu, K.; Azuma, T.; Takemoto, Y. Synthesis of trifunctional thioureas bearing 1,5-disubstituted triazole tether by ru-catalyzed huisgen cycloaddition. Tetrahedron Lett. 2010, 51, 2737–2740. [Google Scholar] [CrossRef]
- Nador, F.; Volpe, M.A.; Alonso, F.; Feldhoff, A.; Kirschning, A.; Radivoy, G. Copper nanoparticles supported on silica coated maghemite as versatile, magnetically recoverable and reusable catalyst for alkyne coupling and cycloaddition reactions. Appl. Catal. A Gen. 2013, 455, 39–45. [Google Scholar] [CrossRef]
- Hansen, T.V.; Wu, P.; Fokin, V.V. One-pot copper(i)-catalyzed synthesis of 3,5-disubstituted isoxazoles. J. Org. Chem. 2005, 70, 7761–7764. [Google Scholar] [CrossRef]
- Kuo, Y.C.; Lin, L.C.; Tsai, W.J.; Chou, C.J.; Kung, S.H.; Ho, Y.H. Samarangenin b from limonium sinense suppresses herpes simplex virus type 1 replication in vero cells by regulation of viral macromolecular synthesis. Antimicrob. Agents Chemother. 2002, 46, 2854–2864. [Google Scholar] [CrossRef]
- Vilar, S.; Cozza, G.; Moro, S. Medicinal chemistry and the molecular operating environment (moe): Application of qsar and molecular docking to drug discovery. Curr. Top. Med. Chem. 2008, 8, 1555–1572. [Google Scholar] [CrossRef]
- Al-Karmalawy, A.A.; Khattab, M. Molecular modelling of mebendazole polymorphs as a potential colchicine binding site inhibitor. New J. Chem. 2020, 44, 13990–13996. [Google Scholar] [CrossRef]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of mpro from sars-cov-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef]
- Ghanem, A.; Emara, H.A.; Muawia, S.; Abd El Maksoud, A.I.; Al-Karmalawy, A.A.; Elshal, M.F. Tanshinone iia synergistically enhances the antitumor activity of doxorubicin by interfering with the pi3k/akt/mtor pathway and inhibition of topoisomerase ii: In vitro and molecular docking studies. New J. Chem. 2020, 44, 17374–17381. [Google Scholar] [CrossRef]
- Khattab, M.; Al-Karmalawy, A. Revisiting activity of some nocodazole analogues as a potential anticancer drugs using molecular docking and dft calculations. Front. Chem. 2021, 9, 92. [Google Scholar] [CrossRef] [PubMed]
- McConkey, B.J.; Sobolev, V.; Edelman, M. The performance of current methods in ligand–protein docking. Curr. Sci. 2002, 83, 845–856. [Google Scholar]
- Eliaa, S.G.; Al-Karmalawy, A.A.; Saleh, R.M.; Elshal, M.F. Empagliflozin and doxorubicin synergistically inhibit the survival of triple-negative breast cancer cells via interfering with the mtor pathway and inhibition of calmodulin: In vitro and molecular docking studies. ACS Pharmacol. Transl. Sci. 2020, 3, 1330–1338. [Google Scholar] [CrossRef]
- Al-Rabia, M.W.; Alhakamy, N.A.; Ahmed, O.A.A.; Eljaaly, K.; Aloafi, A.L.; Mostafa, A.; Asfour, H.Z.; Aldarmahi, A.A.; Darwish, K.M.; Ibrahim, T.S.; et al. Repurposing of sitagliptin- melittin optimized nanoformula against sars-cov-2; antiviral screening and molecular docking studies. Pharmaceutics 2021, 13, 307. [Google Scholar] [CrossRef]
- Feoktistova, M.; Geserick, P.; Leverkus, M. Crystal violet assay for determining viability of cultured cells. Cold Spring Harb. Protoc. 2016, 2016. [Google Scholar] [CrossRef]
- Zhang, J.; Zhan, B.; Yao, X.; Gao, Y.; Shong, J. Antiviral activity of tannin from the pericarp of punica granatum l. Against genital herpes virus in vitro. China J. Chin. Mater. Med. 1995, 20, 556–558, 576, inside backcover. [Google Scholar]
- Schuhmacher, A.; Reichling, J.; Schnitzler, P. Virucidal effect of peppermint oil on the enveloped viruses herpes simplex virus type 1 and type 2 in vitro. Phytomed. Int. J. Phytother. Phytopharm. 2003, 10, 504–510. [Google Scholar] [CrossRef]

| Cultivar | Extraction Methods | Yield of Extraction (mg/g DW) | References | |
|---|---|---|---|---|
| Oleanolic Acid 1 | Maslinic Acid 2 | |||
| Picual | Solid–liquid extraction (maceration) | 0.500 | 1.200 | [41] |
| Hojiblanca | 0.500 | 1.300 | ||
| Arbequina | 0.400 | 1.500 | ||
| Non-indicated | 0.015 | 0.034 | [42] | |
| Manzanilla | Solid–liquid extraction (centrifugation) | 0.274 | 0.824 | [43] |
| Hojiblanca | 0.565 | 0.904 | ||
| Cacereña | 0.185 | 0.295 | ||
| Kalamata | 0.841 | 1.318 | ||
| Picual | Ultrasonic-assisted extraction | 1.003 | 2.440 | [44] |
| Kalamon | 0.838 | 2.100 | ||
| Chemlali | Solid–liquid, then ultrasonic-assisted extractions | 3.400 | 8.500 | [15] |
| Chemlali | Ultrasonic-assisted extractions, then centrifugation isolation | 3.6 | 9.2 | This work |
| Code | Nomenclature | Chemical Structures | Yield (%) |
|---|---|---|---|
| 1 | Oleanolic acid | 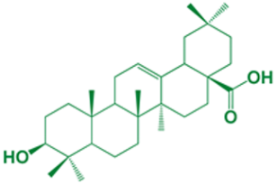 | - |
| 2 | Maslinic acid | 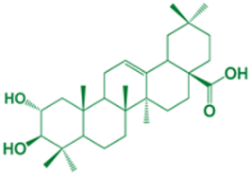 | - |
| 3 | Propargyl-(3β)-3-hydroxyolean-12-en-28-oate |  | 99 |
| 4 | Propargyl-(2α,3β)-2,3-dihydroxyolean-12-en-28-oate |  | 25 |
| 5 | Propargyl-(2α)-2-(propargyloxy)-(3β)-3-hydroxy-olean-12-en-28-oate | 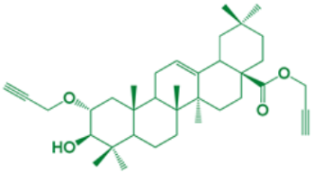 | 23 |
| 6 | Propargyl-(3α)-3-(propargyloxy)-(2β)-2-hydroxy-olean-12-en-28-oate | 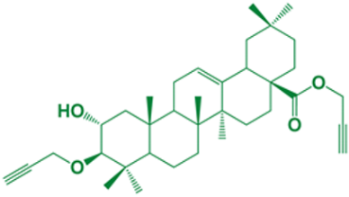 | 20 |
| 7 | Propargyl-(2α,3β)-2,3-bis(propargyloxy)-olean-12-en-28-oate |  | 31 |
| 8 | (1-(3-methylphenyl)-1H-1,2,3-triazol-5-yl)methyl-(3β)-3-hydroxyolean-12-en-28-oate | 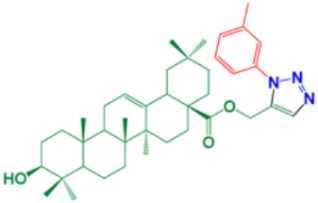 | 90 |
| 9 | (1-(3-methylphenyl)-1H-1,2,3-triazol-5-yl)methyl-(2α,3β)-2,3-dihydroxyolean-12-en-28-oate | 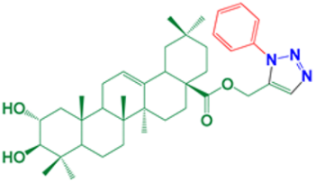 | 92 |
| 10 | (1-(3-methylphenyl)-1H-1,2,3-triazol-4-yl)methyl-(3β)-3-hydroxyolean-12-en-28-oate | 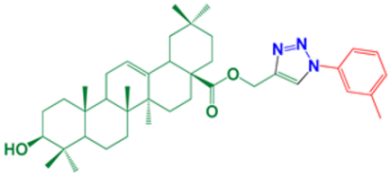 | 98 |
| 11 | (1-Phenyl-1H-1,2,3-triazol-4-yl)methyl-(2α,3β)-2,3-dihydroxyolean-12-en-28-oate | 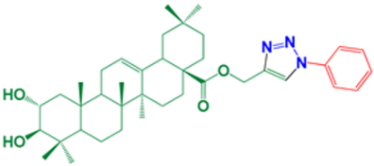 | 96 |
| 12 | (1-(4-Chlorophenyl)-1H-1,2,3-triazol-4-yl)methyl-(2α,3β)-2-((1-(4-chlorophenyl)-1H-1,2,3-triazol-4-yl)methoxy)-3-hydroxyolean-12-en-28-oate |  | 94 |
| 13 | (1-(4-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)methyl-(2α,3β)-2-hydroxy-3-((1-(4-methoxyphenyl)-1H-1,2,3-triazol-4-yl)methoxy)-olean-12-en-28-oate | 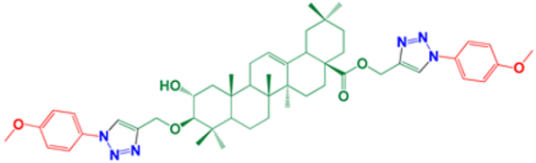 | 96 |
| 14 | (1-(4-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)methyl-(2α,3β)-2,3-bis((1-(4-methoxyphenyl)-1H-1,2,3-triazol-4-yl)methoxy)-olean-12-en-28-oate |  | 92 |
| 15 | (3-(4-methoxyphenyl) isoxazol-5-yl) methyl-(3β)-3-hydroxyolean-12-en-28-oate |  | 98 |
| 16 | (3-(4-methoxyphenyl) isoxazol-5-yl) methyl-(2α,3β)-2,3-dihydroxyolean-12-en-28-oate | 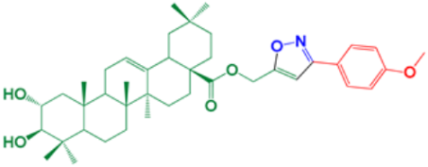 | 87 |
| 17 | (3-(4-chlorophenyl) isoxazol-5-yl) methyl-(2α,3β)-2,3-dihydroxyolean-12-en-28-oate | 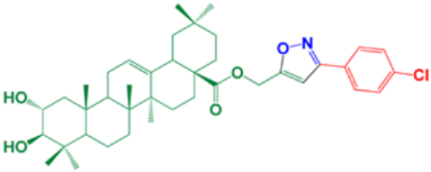 | 96 |
| 18 | N3 (co-crystallized native inhibitor of SARS-CoV-2) |  | - |
| Code | 3D Binding Interactions | 3D Pocket Positioning |
|---|---|---|
| 17 | 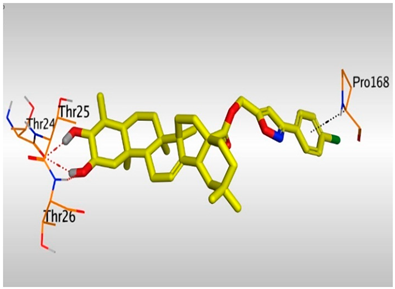 | 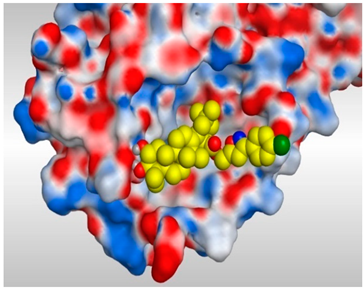 |
| 3 |  |  |
| N3 (18) | 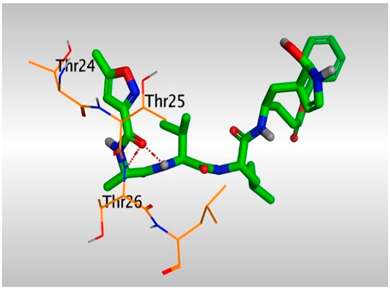 |  |
| Compound | CC50 | IC50 | Selectivity Index (CC50/IC50) |
|---|---|---|---|
| 1 | 189.9 | 476.1 | 0.39 |
| 2 | 97.30 | 99.87 | 0.97 |
| 3 | 97.40 | 42.01 | 2.32 |
| 4 | 157.2 | 236.3 | 0.67 |
| 5 | 90.62 | 85.21 | 1.06 |
| 6 | 208.8 | 135 | 1.55 |
| 7 | 97.42 | 108 | 0.90 |
| 8 | 170 | 173.9 | 0.98 |
| 9 | 185.3 | 131.9 | 1.40 |
| 10 | 277.2 | 75.31 | 3.68 |
| 11 | 218.1 | 134.7 | 1.62 |
| 12 | 513.8 | 116.1 | 4.42 |
| 13 | 356.4 | 158.9 | 2.24 |
| 14 | 405.4 | 136.7 | 2.96 |
| 15 | 415.3 | 523 | 0.79 |
| 16 | 324.2 | 218.6 | 1.48 |
| 17 | 141.6 | 4.12 | 34.36 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soltane, R.; Chrouda, A.; Mostafa, A.; Al-Karmalawy, A.A.; Chouaïb, K.; dhahri, A.; Pashameah, R.A.; Alasiri, A.; Kutkat, O.; Shehata, M.; et al. Strong Inhibitory Activity and Action Modes of Synthetic Maslinic Acid Derivative on Highly Pathogenic Coronaviruses: COVID-19 Drug Candidate. Pathogens 2021, 10, 623. https://doi.org/10.3390/pathogens10050623
Soltane R, Chrouda A, Mostafa A, Al-Karmalawy AA, Chouaïb K, dhahri A, Pashameah RA, Alasiri A, Kutkat O, Shehata M, et al. Strong Inhibitory Activity and Action Modes of Synthetic Maslinic Acid Derivative on Highly Pathogenic Coronaviruses: COVID-19 Drug Candidate. Pathogens. 2021; 10(5):623. https://doi.org/10.3390/pathogens10050623
Chicago/Turabian StyleSoltane, Raya, Amani Chrouda, Ahmed Mostafa, Ahmed A. Al-Karmalawy, Karim Chouaïb, Abdelwaheb dhahri, Rami Adel Pashameah, Ahlam Alasiri, Omnia Kutkat, Mahmoud Shehata, and et al. 2021. "Strong Inhibitory Activity and Action Modes of Synthetic Maslinic Acid Derivative on Highly Pathogenic Coronaviruses: COVID-19 Drug Candidate" Pathogens 10, no. 5: 623. https://doi.org/10.3390/pathogens10050623
APA StyleSoltane, R., Chrouda, A., Mostafa, A., Al-Karmalawy, A. A., Chouaïb, K., dhahri, A., Pashameah, R. A., Alasiri, A., Kutkat, O., Shehata, M., Jannet, H. B., Gharbi, J., & Ali, M. A. (2021). Strong Inhibitory Activity and Action Modes of Synthetic Maslinic Acid Derivative on Highly Pathogenic Coronaviruses: COVID-19 Drug Candidate. Pathogens, 10(5), 623. https://doi.org/10.3390/pathogens10050623








