Phenotypic and Phylogenetic Characterization of Cu Homeostasis among Xylella fastidiosa Strains
Abstract
1. Introduction
2. Results and Discussion
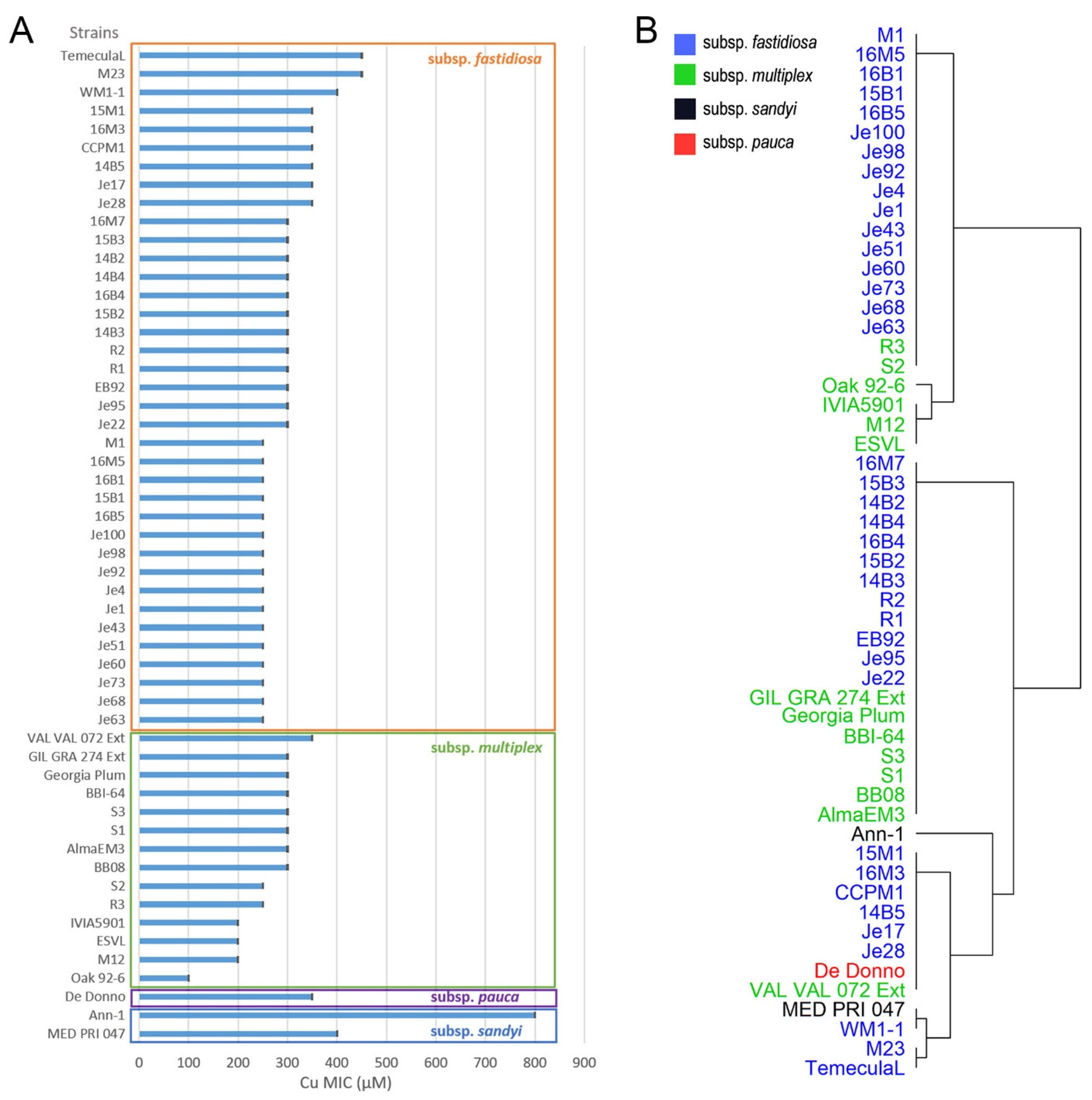
2.1. Cu Homeostasis Ability (Cu MIC) of X. fastidiosa Strains
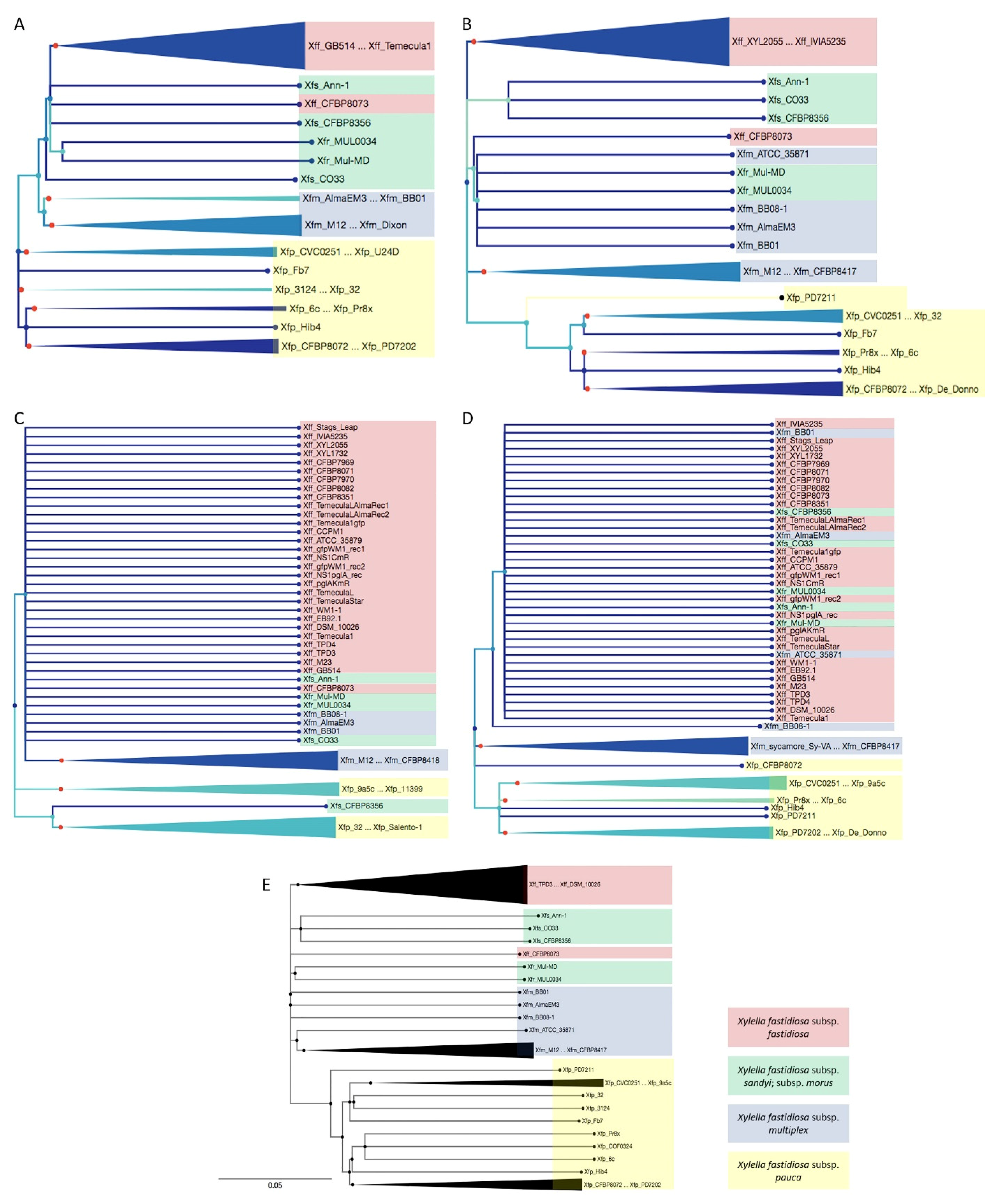
2.2. Phylogenetical Relationships of Cu-Related Genes copA, copB, copL, and cutC Follows X. fastidiosa Subspecies Classification and Was Not Related with Cu MIC
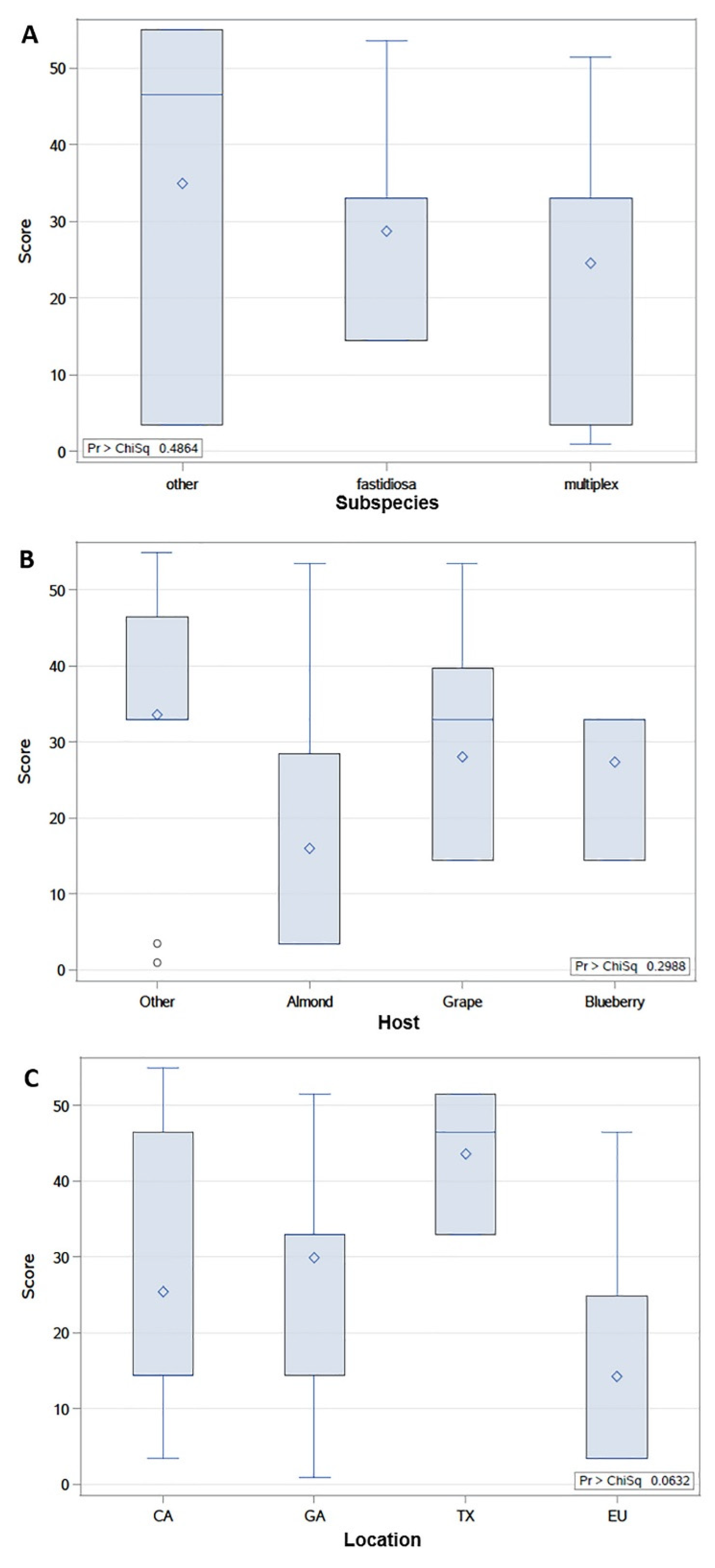
2.3. Cu MIC of X. fastidiosa Strains Is Not Significantly Associated with Subspecies, Host, or Location
2.4. X. fastidiosa Isolates from Georgia’s Vineyards Have Similar Responses of Cu Accumulation and Biofilm Formation under Cu Treatment
3. Materials and Methods
3.1. Cu MIC Measurements
3.2. Phylogenetic Analysis
3.3. Selective Pressure Analysis and Expression Prediction
3.4. Cu Accumulation in Cells
3.5. Biofilm/Total Cells Ratio
3.6. Association Tests
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- EFSA. Update of the Xylella spp. host plant database–systematic literature search up to 30 June 2019. EFSA J. 2020, 18, e06114. [Google Scholar]
- Hopkins, D.; Purcell, A. Xylella fastidiosa: Cause of Pierce’s disease of grapevine and other emergent diseases. Plant Dis. 2002, 86, 1056–1066. [Google Scholar] [CrossRef]
- Hopkins, D. Xylella fastidiosa: Xylem-limited bacterial pathogen of plants. Annu. Rev. Phytopathol. 1989, 27, 271–290. [Google Scholar] [CrossRef]
- Almeida, R.P.; Nascimento, F.E.; Chau, J.; Prado, S.S.; Tsai, C.-W.; Lopes, S.A.; Lopes, J.R. Genetic structure and biology of Xylella fastidiosa strains causing disease in citrus and coffee in Brazil. Appl. Environ. Microbiol. 2008, 74, 3690–3701. [Google Scholar] [CrossRef][Green Version]
- Cariddi, C.; Saponari, M.; Boscia, D.; De Stradis, A.; Loconsole, G.; Nigro, F.; Porcelli, F.; Potere, O.; Martelli, G. Isolation of a Xylella fastidiosa strain infecting olive and oleander in Apulia, Italy. J. Plant Pathol. 2014, 96, 425–429. [Google Scholar]
- Olmo, D.; Nieto, A.; Adrover, F.; Urbano, A.; Beidas, O.; Juan, A.; Marco-Noales, E.; López, M.M.; Navarro, I.; Monterde, A. First detection of Xylella fastidiosa infecting cherry (Prunus avium) and Polygala myrtifolia plants, in Mallorca Island, Spain. Plant Dis. 2017, 101, 1820. [Google Scholar] [CrossRef]
- Tumber, K.; Alston, J.; Fuller, K. Pierce’s disease costs California $104 million per year. Calif. Agric. 2014, 68, 20–29. [Google Scholar] [CrossRef]
- Schneider, K.; Van der Werf, W.; Cendoya, M.; Mourits, M.; Navas-Cortés, J.A.; Vicent, A.; Lansink, A.O. Impact of Xylella fastidiosa subspecies pauca in European olives. Proc. Natl. Acad. Sci. USA 2020, 117, 9250–9259. [Google Scholar] [CrossRef] [PubMed]
- Vanhove, M.; Sicard, A.; Ezennia, J.; Leviten, N.; Almeida, R.P. Population structure and adaptation of a bacterial pathogen in California grapevines. Environ. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Parat, C.; Chaussod, R.; Lévêque, J.; Dousset, S.; Andreux, F. The relationship between copper accumulated in vineyard calcareous soils and soil organic matter and iron. Eur. J. Soil Sci. 2002, 53, 663–670. [Google Scholar] [CrossRef]
- Romi, M.; Matijevi, L.; Baki, H.; Romi, D. Copper Accumulation in Vineyard Soils: Distribution, Fractionation and Bioavailability Assessment. In Environmental Risk Assessment of Soil Contamination; IntechOpen: London, UK, 2014. [Google Scholar] [CrossRef]
- Ge, Q.; Cobine, P.A.; De La Fuente, L. Copper supplementation in watering solution reaches the xylem but does not protect tobacco plants against Xylella fastidiosa infection. Plant Dis. 2020, 104, 724–730. [Google Scholar] [CrossRef]
- Behlau, F.; Hong, J.C.; Jones, J.B.; Graham, J.H. Evidence for acquisition of copper resistance genes from different sources in citrus-associated xanthomonads. Phytopathology 2013, 103, 409–418. [Google Scholar] [CrossRef]
- Graham, J.H.; Gottwald, T. Research perspectives on eradication of citrus bacterial diseases in Florida. Plant Dis. 1991, 75, 1193–1200. [Google Scholar] [CrossRef]
- Ge, Q.; Cobine, P.; De La Fuente, L. The influence of copper homeostasis genes copA and copB on Xylella fastidiosa virulence is affected by sap copper concentration. Phytopathology 2021. [Google Scholar] [CrossRef] [PubMed]
- Behlau, F. Risk Assessment of Copper and Streptomycin Resistance Developmentin Xanthomonas citri subsp. citri. Ph.D. Thesis, University of Florida, Gainesville, FL, USA, 2010. [Google Scholar]
- Robinson, O.; Dylus, D.; Dessimoz, C. Phylo.io: Interactive viewing and comparison of large phylogenetic trees on the web. Mol. Biol. Evol. 2016, 33, 2163–2166. [Google Scholar] [CrossRef]
- De Freitas, E.C.; Ucci, A.P.; Teixeira, E.C.; Pedroso, G.A.; Hilario, E.; Zocca, V.F.B.; De Paiva, G.B.; Ferreira, H.; Pedrolli, D.B.; Bertolini, M.C. The copper-inducible copAB operon in Xanthomonas citri subsp. citri is regulated at transcriptional and translational levels. Microbiology 2019, 165, 355–365. [Google Scholar]
- Poon, A.F.; Frost, S.D.; Pond, S.L.K. Detecting signatures of selection from DNA sequences using Datamonkey. In Bioinformatics for DNA Sequence Analysis; Springer: Berlin/Heidelberg, Germany, 2009; pp. 163–183. [Google Scholar]
- Delport, W.; Poon, A.F.; Frost, S.D.; Kosakovsky Pond, S.L. Datamonkey 2010: A suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 2010, 26, 2455–2457. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, R. Molecular signatures of natural selection. Annu. Rev. Genet. 2005, 39, 197–218. [Google Scholar] [CrossRef]
- Zhou, M.; Guo, J.; Cha, J.; Chae, M.; Chen, S.; Barral, J.M.; Sachs, M.S.; Liu, Y. Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature 2013, 495, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, Z.; Meng, D.; Liu, X.; Li, X.; Zhang, M.; Tao, J.; Gu, Y.; Zhong, S.; Yin, H. Comparative genomic analysis reveals the distribution, organization, and evolution of metal resistance genes in the genus Acidithiobacillus. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef]
- Puigbo, P.; Romeu, A.; Garcia-Vallve, S. HEG-DB: A database of predicted highly expressed genes in prokaryotic complete genomes under translational selection. Nucleic Acids Res. 2007, 36, D524–D527. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.W.; Wang, J.T.; Li, J.; Li, J.J.; Ma, Y.B.; Chen, D.; He, J.Z. Field-based evidence for copper contamination induced changes of antibiotic resistance in agricultural soils. Environ. Microbiol. 2016, 18, 3896–3909. [Google Scholar] [CrossRef]
- Kang, W.; Zhang, Y.-J.; Shi, X.; He, J.-Z.; Hu, H.-W. Short-term copper exposure as a selection pressure for antibiotic resistance and metal resistance in an agricultural soil. Environ. Sci. Pollut. Res. 2018, 25, 29314–29324. [Google Scholar] [CrossRef]
- Poole, K. At the nexus of antibiotics and metals: The impact of Cu and Zn on antibiotic activity and resistance. Trends Microbiol. 2017, 25, 820–832. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.K.; Havird, J.C.; De La Fuente, L. Differentiation of Xylella fastidiosa strains via multilocus sequence analysis of environmentally mediated genes (MLSA-E). Appl. Environ. Microbiol. 2012, 78, 1385–1396. [Google Scholar] [CrossRef] [PubMed]
- Ruyters, S.; Salaets, P.; Oorts, K.; Smolders, E. Copper toxicity in soils under established vineyards in Europe: A survey. Sci. Total Environ. 2013, 443, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Komárek, M.; Čadková, E.; Chrastný, V.; Bordas, F.; Bollinger, J.-C. Contamination of vineyard soils with fungicides: A review of environmental and toxicological aspects. Environ. Int. 2010, 36, 138–151. [Google Scholar] [CrossRef] [PubMed]
- Castillo, A.I.; Bojanini, I.; Chen, H.; Kandel, P.P.; De La Fuente, L.; Almeida, R.P.P. Allopatric Plant Pathogen Population Divergence following Disease Emergence. Appl. Environ. Microbiol. 2021, 87, e02095-20. [Google Scholar] [CrossRef] [PubMed]
- Marques, L.; Ceri, H.; Manfio, G.; Reid, D.; Olson, M. Characterization of biofilm formation by Xylella fastidiosa in vitro. Plant Dis. 2002, 86, 633–638. [Google Scholar] [CrossRef]
- Cobine, P.A.; Cruz, L.F.; Navarrete, F.; Duncan, D.; Tygart, M.; De La Fuente, L. Xylella fastidiosa differentially accumulates mineral elements in biofilm and planktonic cells. PLoS ONE 2013, 8, e54936. [Google Scholar] [CrossRef]
- Cruz, L.F.; Cobine, P.A.; De La Fuente, L. Calcium increases Xylella fastidiosa surface attachment, biofilm formation, and twitching motility. Appl. Environ. Microbiol. 2012, 78, 1321–1331. [Google Scholar] [CrossRef]
- Peterson, L.; Shanholtzer, C. Tests for bactericidal effects of antimicrobial agents: Technical performance and clinical relevance. Clin. Microbiol. Rev. 1992, 5, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Naranjo, E.; Merfa, M.V.; Santra, S.; Ozcan, A.; Johnson, E.; Cobine, P.A.; De La Fuente, L. Zinkicide is a ZnO-based nanoformulation with bactericidal activity against Liberibacter crescens in batch cultures and in microfluidic chambers simulating plant vascular systems. Appl. Environ. Microbiol. 2020, 86. [Google Scholar] [CrossRef] [PubMed]
- Puigbò, P.; Bravo, I.G.; Garcia-Vallve, S. CAIcal: A combined set of tools to assess codon usage adaptation. Biol. Direct. 2008, 3, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Carbone, A.; Zinovyev, A.; Képes, F. Codon adaptation index as a measure of dominating codon bias. Bioinformatics 2003, 19, 2005–2015. [Google Scholar] [CrossRef] [PubMed]


| Gene | # Positive Sites 1 | # Negative Sites 2 | # Total Sites 3 |
|---|---|---|---|
| copA | 1 | 14 | 611 |
| copB | 0 | 10 | 250 |
| copL | 0 | 2 | 126 |
| cutC | 0 | 3 | 246 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ge, Q.; Liu, R.; Cobine, P.A.; Potnis, N.; De La Fuente, L. Phenotypic and Phylogenetic Characterization of Cu Homeostasis among Xylella fastidiosa Strains. Pathogens 2021, 10, 495. https://doi.org/10.3390/pathogens10040495
Ge Q, Liu R, Cobine PA, Potnis N, De La Fuente L. Phenotypic and Phylogenetic Characterization of Cu Homeostasis among Xylella fastidiosa Strains. Pathogens. 2021; 10(4):495. https://doi.org/10.3390/pathogens10040495
Chicago/Turabian StyleGe, Qing, Ranlin Liu, Paul A. Cobine, Neha Potnis, and Leonardo De La Fuente. 2021. "Phenotypic and Phylogenetic Characterization of Cu Homeostasis among Xylella fastidiosa Strains" Pathogens 10, no. 4: 495. https://doi.org/10.3390/pathogens10040495
APA StyleGe, Q., Liu, R., Cobine, P. A., Potnis, N., & De La Fuente, L. (2021). Phenotypic and Phylogenetic Characterization of Cu Homeostasis among Xylella fastidiosa Strains. Pathogens, 10(4), 495. https://doi.org/10.3390/pathogens10040495






