Abstract
Non-human primate populations act as potential reservoirs for human pathogens, including viruses, bacteria and parasites, which can lead to zoonotic infections. Furthermore, intestinal microorganisms may be pathogenic organisms to both non-human primates and humans. It is, therefore, essential to study the prevalence of these infectious agents in captive and wild non-human primates. This study aimed at showing the prevalence of the most frequently encountered human enteric protozoa in non-human primate populations based on qPCR detection. The three populations studied were common chimpanzees (Pan troglodytes) in Senegal and gorillas (Gorilla gorilla) in the Republic of the Congo and in the Beauval Zoo (France). Blastocystis spp. were mainly found, with an occurrence close to 100%, followed by Balantidium coli (23.7%), Giardia intestinalis (7.9%), Encephalitozoon intestinalis (1.3%) and Dientamoeba fragilis (0.2%). None of the following protozoa were detected: Entamoeba histolytica, Enterocytozoon bieneusi, Cryptosporidium parvum, C. hominis, Cyclospora cayetanensis or Cystoisospora belli. As chimpanzees and gorillas are genetically close to humans, it is important to monitor them frequently against different pathogens to protect these endangered species and to assess potential zoonotic transmissions to humans.
1. Introduction
Several enteric protozoa that can provoke zoonotic diseases are currently facilitated by and associated with the growing popularity of open farms and petting zoos, which are a possible source of outbreaks and transmission to humans [1,2,3,4]. The parasitic ecology of non-human primates varies depending on the wild or captive nature of the species studied [5]. Although several intestinal protozoan species have been involved in diseases affecting captive non-human primates [6], data on the pathogenicity of parasites in non-human primates are scarce. A few studies have reported that enteric protozoan pathogens, namely Giardia spp., Entamoeba histolytica and Cryptosporium spp., may cause gastrointestinal enteritis in non-human primates (gibbons, orangutans, marmosets, gorilla and chimpanzees) [7,8].
In recent years, molecular studies have identified common zoonotic parasites, including Giardia intestinalis, Entamoeba histolytica and Cryptosporidium sp. [5], in captive primates but not in the wild, suggesting possible transmission of these pathogens from humans. However, Cryptosporidium and Giardia have been isolated from wild primates living in disturbed forests, possibly as a result of their close contact with humans [9]. Moreover, most parasites carried by non-human primates pose a potential zoonotic risk to researchers and caretakers in breeding centers [10,11], acting as reservoirs for human infections [12]. It is, therefore, important to document and monitor protozoan parasites that are present in these primates, whether they are wild or captive. Regularly monitoring primates for pathogens, including protozoan parasites, is warranted and may prevent zoonotic transmission to humans and protect these endangered species. This study aimed at investigating the occurrence of the most common human enteric protozoa in non-human primate populations [13], namely Blastocystis spp., Cryptosporidium parvum, C. hominis, Cyclospora cayetanensis, Dientamoeba fragilis, Giardia intestinalis, Balantidium coli, Entamoeba histolytica and Cystoisospora belli, along with two microsporidia, namely Enterocytozoon bieneusi and Encephalitozoon intestinalis. These pathogens, which have been previously identified as the most frequent human parasites [13], lead to human diseases ranging from chronic intestinal colonization with abdominal discomfort and sporadic diarrhea to severe enteric disorders. In addition, many protozoa are overlooked, neglected or underestimated by traditional microscopic examination, depending on the microscopist’s experience and the sensitivity of the methods used. To overcome this, we employed molecular techniques to diagnose these parasites. Three primate populations were studied, namely common chimpanzees (Pan troglodytes) in Senegal and gorillas (Gorilla gorilla) in the Republic of the Congo and in the Beauval Zoo (France), both to improve the health and care of these great apes and to estimate the potential risk of zoonotic infection. Moreover, Blastocystis diversity (genotypes) was also studied in each primate population. This study was done to provide an update on the prevalence of the most frequently encountered human enteric protozoa in non-human primate populations based on qPCR detection.
2. Results
2.1. Parasitological Analysis
A total of 76 stool samples from great apes were analyzed using real-time PCR assays targeting ten protozoa. The parasitic infection rate (i.e., detection of at least one parasite per sample) was 97.4% (74/76 samples). The most prevalent protozoan parasite among all tested samples was Blastocystis, with a prevalence of 97.4%. Two other pathogenic protozoa also occurred with relatively high prevalence rates, namely Balantidium coli (at 23.7%) and Giardia intestinalis (at 7.9%). The remaining parasites were either detected at a low prevalence (<2%) or not detected at all.
2.2. Prevalence of Parasites by Primate Species and Site
2.2.1. Chimpanzees from Senegal
At least one parasitic infection was detected in 97.9% (47/48 samples) of the 13 individual chimpanzees tested from Senegal (Table 1). Blastocystis spp. was the most common protozoan with a prevalence rate of 97.9% (47/48), followed by Dientamoeba fragilis with a prevalence rate of 25% (12/48) and Giardia intestinalis with a prevalence rate of 2.1% (1/48) (Table 1).

Table 1.
Prevalence of ten protozoa in the studied non-human primates.
2.2.2. Gorillas from the Republic of the Congo
All gorilla samples collected from the Republic of the Congo were positive for at least one of the ten tested parasites (19/19, 100%) (Table 1). Blastocystis was reported in all tested samples, followed by Balantidium coli in 11 samples (57.9%), Giardia intestinalis in five samples (26.3%) and Encephalitozoon intestinalis in one sample (5.3%).
2.2.3. Gorillas from the Beauval Zoo
Of the nine stool samples collected from nine different gorillas in the Beauval Zoo, at least one of the ten tested parasites was detected in 8 samples (88.9%). In the same way, as the wild great apes from Senegal and the Republic of the Congo, Blastocystis spp. was the most common protozoan, with a prevalence of 88.9% (eight positive samples) (Table 1). The Balantidium coli infection rate was 77.8% (7/8 samples). No other parasite was detected.
2.3. Genotyping of Blastocystis spp.
Of the 74 samples positive for Blastocystis, 19 samples were from gorillas (the Republic of the Congo), 47 samples were from chimpanzees (Senegal) and eight samples were from gorillas (Beauval Zoo, France); three subtypes (STs) were detected (Table 2). ST1 was the most abundant (n = 60, 81.1%), followed by ST2 (n = 10, 13.5%) and ST5 (n = 4; 5.4%) (Table 2). A phylogenetic analysis was generated to illustrate the relationship between the different subtypes obtained in our study, along with other Blastocystis ST1, ST2, ST3 and ST5 sequences recovered from human, chimpanzee and gorilla species, which are available in the GenBank database (Figure 1). This figure shows that sequences from each ST lineage were clustered together as a monophyletic group and supported by bootstrap values higher than 50%. However, paraphyletic and polyphyletic clades resulting from certain nucleotide variations between sequences (bootstrap values below 50%) were formed within each ST branch (Figure 1).

Table 2.
Blastocystis subtypes in the studied non-human primates.
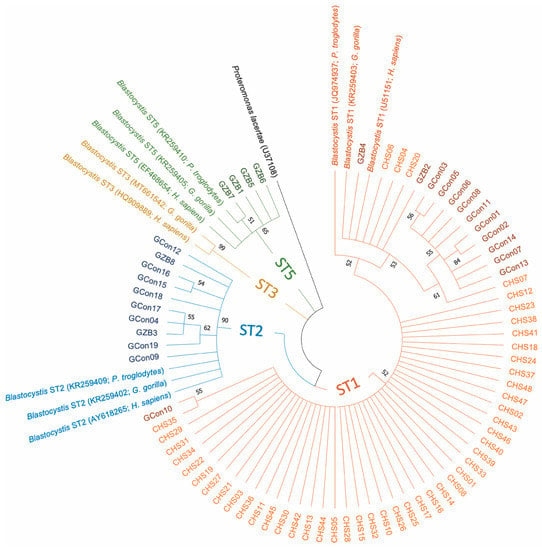
Figure 1.
Phylogenetic tree based on the small subunit ribosomal DNA (SSU rDNA) gene sequences, showing the relationships between the different subtypes found in our samples and reference sequences from GenBank. The molecular phylogenetic analysis was carried out using the Maximum Likelihood method based on the Tamura–Nei model in Molecular Evolutionary Genetics Analysis X (MEGA X) software. Bootstrap values (obtained from 1000 replicates) lower than 50% were removed. The branches are displayed with different colors according to the sequence subtype (ST): ST1, orange; ST2, blue; ST3, gold; ST5, green. The sequences obtained in this study from chimpanzees and gorillas are represented by lighter and darker colors than those of the corresponded STs. CHS, chimpanzees from Senegal; GCon, gorillas from the Republic of the Congo; GZB, gorillas from the Beauval Zoo. T5 13.m Beauval zoo, France (%).
3. Discussion
Our survey revealed a high rate of Balantidium coli infection in gorillas from the Republic of the Congo and captive gorillas from France, at 57.9% and 77.8%, respectively (Table 1). B. coli is a cosmopolitan ciliate that colonizes the intestines of many animals, including humans, with pigs serving as reservoir hosts [6,14]. In captive gorillas, infections with B. coli are usually without clinical signs. Although in some cases a mild or self-limiting disease may result upon first exposure to this protozoan, life-threatening B. coli infections have also occurred in gorilla populations [15,16].
Dientamoeba fragilis was found only in the chimpanzees from Senegal, with a prevalence of 25% (Table 1). This flagellate protozoan parasite has been reported in great apes [17]. Its pathogenicity has been demonstrated in Lowland gorillas from Cameroon [18].
We also found Giardia intestinalis in wild primates, with a prevalence of 2.1% in chimpanzees from Senegal and a much higher prevalence (26.3%) in gorillas from the Republic of the Congo (Table 1). This is in line with numerous previous studies that reported infection by this enteric protozoan in wild non-human primates and more frequently in captive ones [19,20,21,22,23]. It has previously been shown that the occurrence of Giardia in some wild primate populations is associated with a synanthropic lifestyle [9].
Encephalitozoon intestinalis was detected in one gorilla from the Republic of the Congo (Table 1). It is known that microsporidia can infect apes, as it was demonstrated through experimental infections [24], while E. intestinalis has previously been found in wild non-human primates [25,26].
The PCR-based diagnostic method used in our study highlighted a Blastocystis sp. prevalence of 97.4% in the great apes studied (Table 1). This ubiquitous microeukaryote is a commensal of the large intestine of more than one billion people in both developed and developing countries [27]. These data are consistent with previous studies showing that non-human primates are common hosts of this protozoon eukaryote [5], regardless of their lifestyle and geographical origin [28]. In the current study, Blastocystis ST1 occurred in 100% of chimpanzees from Senegal (Table 2; Figure 1). ST1 was also the major subtype found in gorillas, with a prevalence of 48.2% (13/27). Various epidemiological studies conducted in primates strongly suggest that the predominant subtypes, ST1, ST2 and ST3, are shared by captive and wild monkeys, and thus naturally occur in this host group [28,29,30]. While ST1 is the most frequent subtype in non-human primates, it is the second most common subtype in humans [28,29,30]. Finally, we found an additional subtype, ST5, in captive gorillas from France as compared to wild gorillas from the Republic of the Congo (Table 2; Figure 1). ST5 has previously been found in Western Lowland gorillas in French zoos [28]. One explanation might be that they are in close contact with other animals in captivity and also have direct and repeated contact with humans. In fact, the Beauval Zoo has the largest diversity of captive animals in France, with 600 species claimed. It is, therefore, not surprising that ST5 was only found in the gorillas from the zoo. Other subtypes not found in this study have been previously described in non-human primates, such as ST8 [30] and ST10-17 [31]. It is notable that ST8 has also been found in zookeepers working in close contact with non-human primates, highlighting the risk of zoonotic transmission [29]. Finally, our phylogenetic tree (Figure 1) shows the presence of some nucleotide variations between sequences from the same ST. This finding suggests the probable existence of “subgroups or subclades” within each Blastocystis ST, which require further investigation (e.g., by using longer small subunit ribosomal DNA gene sequences).
In summary, the present study illuminates our current understanding of parasitosis in ape populations and the occurrence of Blastocystis among these species. Furthermore, we think that an observational survey of pathogens is required at each opportunity and that similar studies should be conducted regularly, as the dynamics of microorganisms are not stable and each pathogen can emerge at any moment, leading to an epidemic situation and possible intra- or interspecies transmission.
4. Materials and Methods
4.1. Sample Collection and Information
Stool samples were collected from primates living in the Republic of the Congo, Senegal and at the Beauval Zoo (Saint-Aignan-sur-Cher, France). Nineteen gorilla samples were collected at the Lesio-Louna Lefini Gorilla Nature Reserve, located at 2°58′033.1″ S and 15°28′033.4″ E in the Republic of the Congo. Nine samples were collected at the Beauval Zoo, located at 47°14′51″ N and 1°21′12″ E, France, in 2015. Forty-eight chimpanzee stools were collected at the Dindefelo community natural reserve at 12°23′00″ N and 12°19′00″ W, Senegal, in 2016. Each collected sample was identified, placed in a 50-millilitre tube, stored in dry ice and then transported to the laboratory, where it was stored at −80 °C. All samples were collected around feeding sites or night nests, in line with standard practice.
4.2. Ethical Statement
No experimentation was conducted on the animals. All stool samples were collected from the soil. In the Republic of the Congo, the access permit to the Lesio-Louna Lefini Gorilla Nature Reserve was delivered by the Forestry Economy and Sustainable Development Ministry (0094/MEFDD/CAB/DGACFAP-DTS de la Direction Générale de l’Agence Congolaise de la Faune et Aires Protégées, on 24 August 2015). The permit to collect samples was delivered by the Environment and Sustainable Development Ministry of Senegal for chimpanzee stool samples (001914/DEF/DGF de la Direction des Eaux, Forêts, Chasses et de la Conservation des Sols, on 5 June 2016). No other permits were required, as no invasive procedure was used in this study and the collection of the samples did not disturb the wild fauna.
4.3. DNA Extraction from Stool Samples
DNA extraction was performed using the EZ1 (Qiagen GmbH, Hilden, Germany) automated protocol with pretreatment [14]. The extraction protocol was adapted for stool processing as follows: 200 mg of stool sample was added to 350 µL of G2 lysis buffer (Qiagen GmbH, Hilden, Germany) in a tube containing glass powder and then disrupted in a FastPrep-24 grinder (MP Biomedicals) at maximum power for 40 s. After 10 min of incubation at 100 °C to allow complete lysis, the tubes were centrifuged at 10,000 g for one minute. Then, 200 µL of supernatant was added to a tube containing 20 µL of Proteinase K, which was incubated overnight at 56 °C.
To control for DNA extraction quality and the absence of PCR inhibitors, universal eubacterial primers and probes were used to amplify 16S rRNA bacterial genes, and a qPCR technique named “all-bacteria” (TTB) was performed on each extracted DNA sample. Positive result indicated the absence of PCR inhibitors, while negative results indicated that the DNA extraction was repeated.
4.4. Singleplex Real-Time PCR Amplification and Detection
Ten different specific primers and TaqManTM hydrolysis probes targeting different sequence regions were used in multiparallel assays. All primers and probe sequences used in this study have previously been published and used [32,33] (Table 3). The real-time PCR reactions were conducted using 20 µL total volumes containing 10 µL of Master mix (Roche Diagnostics GmbH, Mannheim, Germany), 0.5 µL of each primer, 0.5 µL of probes, 3 µL of distilled water, 0.5 µL of UDG and 5 µL of DNA. Analyses were performed using a CFX96TM Real-Time PCR detection system (BIO-RAD, Life Science, Marnes-la-Coquette, France). Amplification reactions were performed as follows: two minutes of incubation at 50 °C, five minutes incubation at 95 °C, followed by 40 cycles of five seconds at 95 °C and 30 s at 60 °C. Real-time PCR results were considered negative when the Ct value was more than 38 or no amplification curve was obtained, as described in previous studies [32,34].

Table 3.
List of primers and probe sequences used in this study.
4.5. Amplification of the Small Subunit Ribosomal DNA (SSU rDNA) Gene and Blastocystis Typing
For each Blastocystis-positive sample, the extracted DNA was subjected to a standard polymerase chain reaction (PCR) assay using the Blastocystis-specific primers BL18SPPF1 (5′-AGTAGTCATACGCTCGTCTCAAA-3′) and BL18SR2PP (5′-TCTTCGTT ACCCGTTACTGC-3′) designed by Poirier et al. [44]. These primers target a 320 to 342 bp DNA fragment located in the Blastocystis sp. SSU rDNA gene. Amplification was performed in a total volume of 50 μL with AmpliTaq Gold® 360 (Thermo Fisher Scientific, Waltham, MA, USA). After denaturation at 95 °C for 15 min, 40 cycles of amplification were performed with a 2720 Thermal CyclerTM (Applied Biosystems, Courtaboeuf, France) as follows: 30 s at 95 °C, 30 s at 59 °C and 30 s at 72 °C. The amplification products were analyzed by electrophoresis using 1.5% weight-per-volume (w/v) agarose gel containing SYBRTM Safe DNA Gel Stain (ThermoFisher). The PCR products were purified using MultiScreen® PCR (Merck Millipore, Darmstadt, Germany) and sequencing reactions were carried out by using a DNA sequencing kit (BigDye Terminator Cycle Sequencing v1.1 Ready Reactions; PE Biosystems) according to the manufacturer’s instructions. Sequencing products were purified and electrophoresis was performed with a 3130 Genetic Analyzer (Applied Biosystems). The nucleotide sequences were analyzed with the CodonCode Aligner software (www.codoncode.com/aligner (accessed on 10 January 2021)) and compared with those available in the GenBank database by using the BLASTn programme (www.ncbi.nlm.nih.gov/BLAST (accessed on 10 January 2021)). Subtypes (STs) were then determined by selecting the best match (exact/closest similarity) with all Blastocystis sp. subtype sequences available in the GenBank database. Phylogenetic analyses were performed using the maximum likelihood method implemented in MEGA X software (https://www.megasoftware.net/ (accessed on 10 January 2021)). The maximum likelihood analysis was based on the Tamura–Nei model. Bootstrap values were calculated from 1000 replicates.
4.6. Accession Numbers
The nucleotide sequences for Blastocystis sp. were deposited in GenBank under the accession numbers MT676292 to MT676365.
5. Conclusions
Given the potential zoonotic risk of protozoan parasites and their pathogenicity, it is important to monitor their presence in captive and wild animals. Of the ten tested protozoa, five were detected in great apes. Therefore, non-human primates may present a potential reservoir for Balantidium coli, Blastocystis sp., Dientamoeba fragilis, Encephalitozoon intestinalis and Giardia intestinalis. This study highlights the high prevalence (close to 100%) of Blastocystis sp. colonization in both wild and captive primates, which is higher than previously reported. In line with previous studies, ST1 followed by ST2 were the predominant subtypes. Blastocystis ST5 was only detected in captive gorillas and might be acquired through close contact with human or other animal species.
Author Contributions
Conceptualization, S.R., D.R. and F.B.; methodology, F.B.; investigation, B.D., O.M., J.A., B.M., G.D., A.L. and D.R.; writing—original draft preparation, E.M.; writing—review and editing, F.B. and S.R.; supervision, F.B. and D.R. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the French Government under the Investissements d’avenir (Investments for the Future) program managed by the Agence Nationale de la Recherche (National Agency for Research—ANR), (Méditerranée Infection 10-IAHU-03). This work was supported by Région Provence-Alpes-Côte d’Azur and European funding (Fonds Européen de Développement Regional—FEDER; Plateformes de Recherche et d’Innovation Mutualisées Méditerranée Infection—PRIMMI).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chalmers, R.M.; Smith, R.P.; Hadfield, S.J.; Elwin, K.; Giles, M. Zoonotic Linkage and Variation in Cryptosporidium Parvum from Patients in the United Kingdom. Parasitol. Res. 2011, 108, 1321–1325. [Google Scholar] [CrossRef]
- Giangaspero, A.; Berrilli, F.; Brandonisio, O. Giardia and Cryptosporidium and Public Health: The Epidemiological Scenario from the Italian Perspective. Parasitol. Res. 2007, 101, 1169–1182. [Google Scholar] [CrossRef] [PubMed]
- Ng, J.; Yang, R.; Whiffin, V.; Cox, P.; Ryan, U. Identification of Zoonotic Cryptosporidium and Giardia Genotypes Infecting Animals in Sydney’s Water Catchments. Exp. Parasitol. 2011, 128, 138–144. [Google Scholar] [CrossRef]
- Rimšelienė, G.; Vold, L.; Robertson, L.; Nelke, C.; Søli, K.; Johansen, Ø.H.; Thrana, F.S.; Nygård, K. An Outbreak of Gastroenteritis among Schoolchildren Staying in a Wildlife Reserve: Thorough Investigation Reveals Norway’s Largest Cryptosporidiosis Outbreak. Scand. J. Public Health 2011, 39, 287–295. [Google Scholar] [CrossRef]
- Herrera, J.P.; Chakraborty, D.; Rushmore, J.; Altizer, S.; Nunn, C. The Changing Ecology of Primate Parasites: Insights from Wild-captive Comparisons. Am. J. Primatol. 2019, 81, e22991. [Google Scholar] [CrossRef]
- Zanzani, S.A.; Gazzonis, A.L.; Epis, S.; Manfredi, M.T. Study of the Gastrointestinal Parasitic Fauna of Captive Non-Human Primates (Macaca Fascicularis). Parasitol. Res. 2016, 115, 307–312. [Google Scholar] [CrossRef]
- Vlčková, K.; Kreisinger, J.; Pafčo, B.; Čížková, D.; Tagg, N.; Hehl, A.B.; Modrý, D. Diversity of Entamoeba Spp. in African Great Apes and Humans: An Insight from Illumina MiSeq High-Throughput Sequencing. Int. J. Parasitol. 2018, 48, 519–530. [Google Scholar] [CrossRef]
- Kalishman, J.; Paul-Murphy, J.; Scheffler, J.; Thomson, J.A. Survey of Cryptosporidium and Giardia Spp. in a Captive Population of Common Marmosets. Lab. Anim. Sci. 1996, 46, 116–119. [Google Scholar]
- Gillespie, T.R.; Chapman, C.A. Forest Fragmentation, the Decline of an Endangered Primate, and Changes in Host-Parasite Interactions Relative to an Unfragmented Forest. Am. J. Primatol. 2008, 70, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, H.; Wu, Z.; Pandey, K.; Pandey, B.D.; Sherchand, J.B.; Yanagi, T.; Kanbara, H. Molecular Characterization of Blastocystis Isolates from Children and Rhesus Monkeys in Kathmandu, Nepal. Vet. Parasitol. 2009, 160, 295–300. [Google Scholar] [CrossRef]
- Meloni, D.; Poirier, P.; Mantini, C.; Noël, C.; Gantois, N.; Wawrzyniak, I.; Delbac, F.; Chabé, M.; Delhaes, L.; Dei-Cas, E.; et al. Mixed Human Intra- and Inter-Subtype Infections with the Parasite Blastocystis Sp. Parasitol. Int. 2012, 61, 719–722. [Google Scholar] [CrossRef]
- Skotarczak, B. Genetic Diversity and Pathogenicity of Blastocystis. Ann. Agric. Environ. Med. 2018, 25, 411–416. [Google Scholar] [CrossRef]
- Fletcher, S.M.; Stark, D.; Harkness, J.; Ellis, J. Enteric Protozoa in the Developed World: A Public Health Perspective. Clin. Microbiol. Rev. 2012, 25, 420–449. [Google Scholar] [CrossRef]
- Menu, E.; Mary, C.; Toga, I.; Raoult, D.; Ranque, S.; Bittar, F. A Hospital QPCR-Based Survey of 10 Gastrointestinal Parasites in Routine Diagnostic Screening, Marseille, France. Epidemiol. Infect. 2019, 147. [Google Scholar] [CrossRef] [PubMed]
- Teare, J.A.; Loomis, M.R. Epizootic of Balantidiasis in Lowland Gorillas. J. Am. Vet. Med. Assoc. 1982, 181, 1345–1347. [Google Scholar] [PubMed]
- Lankester, F.; Mätz-Rensing, K.; Kiyang, J.; Jensen, S.A.; Weiss, S.; Leendertz, F.H. Fatal Ulcerative Colitis in a Western Lowland Gorilla (Gorilla Gorilla Gorilla). J. Med. Primatol. 2008, 37, 297–302. [Google Scholar] [CrossRef]
- Stark, D.; Phillips, O.; Peckett, D.; Munro, U.; Marriott, D.; Harkness, J.; Ellis, J. Gorillas Are a Host for Dientamoeba Fragilis: An Update on the Life Cycle and Host Distribution. Vet. Parasitol. 2008, 151, 21–26. [Google Scholar] [CrossRef]
- Lankester, F.; Kiyang, J.A.; Bailey, W.; Unwin, S. Dientamoeba Fragilis: Initial Evidence of Pathogenicity in the Western Lowland Gorilla (Gorilla Gorilla Gorilla). J. Zoo Wildl. Med. 2010, 41, 350–352. [Google Scholar] [CrossRef] [PubMed]
- Munene, E.; Otsyula, M.; Mbaabu, D.A.; Mutahi, W.T.; Muriuki, S.M.; Muchemi, G.M. Helminth and Protozoan Gastrointestinal Tract Parasites in Captive and Wild-Trapped African Non-Human Primates. Vet. Parasitol. 1998, 78, 195–201. [Google Scholar] [CrossRef]
- Berrilli, F.; Prisco, C.; Friedrich, K.G.; Di Cerbo, P.; Di Cave, D.; De Liberato, C. Giardia Duodenalis Assemblages and Entamoeba Species Infecting Non-Human Primates in an Italian Zoological Garden: Zoonotic Potential and Management Traits. Parasit. Vectors 2011, 4, 199. [Google Scholar] [CrossRef] [PubMed]
- Levecke, B.; Dorny, P.; Geurden, T.; Vercammen, F.; Vercruysse, J. Gastrointestinal Protozoa in Non-Human Primates of Four Zoological Gardens in Belgium. Vet. Parasitol. 2007, 148, 236–246. [Google Scholar] [CrossRef]
- Pérez Cordón, G.; Hitos Prados, A.; Romero, D.; Sánchez Moreno, M.; Pontes, A.; Osuna, A.; Rosales, M.J. Intestinal Parasitism in the Animals of the Zoological Garden “Peña Escrita” (Almuñecar, Spain). Vet. Parasitol. 2008, 156, 302–309. [Google Scholar] [CrossRef]
- Medkour, H.; Amona, I.; Laidoudi, Y.; Davoust, B.; Bitam, I.; Levasseur, A.; Akiana, J.; Diatta, G.; Pacheco, L.; Gorsane, S.; et al. Parasitic Infections in African Humans and Non-Human Primates. Pathogens 2020, 9, 561. [Google Scholar] [CrossRef]
- Wasson, K.; Peper, R.L. Mammalian Microsporidiosis. Vet. Pathol. 2000, 37, 113–128. [Google Scholar] [CrossRef]
- Graczyk, T.K.; Bosco-Nizeyi, J.; da Silva, A.J.; Moura, I.N.S.; Pieniazek, N.J.; Cranfield, M.R.; Lindquist, H.D.A. A Single Genotype of Encephalitozoon Intestinalis Infects Free-Ranging Gorillas and People Sharing Their Habitats in Uganda. Parasitol. Res. 2002, 88, 926–931. [Google Scholar] [CrossRef]
- Mynářová, A.; Foitová, I.; Kváč, M.; Květoňová, D.; Rost, M.; Morrogh-Bernard, H.; Nurcahyo, W.; Nguyen, C.; Supriyadi, S.; Sak, B. Prevalence of Cryptosporidium Spp., Enterocytozoon Bieneusi, Encephalitozoon Spp. and Giardia Intestinalis in Wild, Semi-Wild and Captive Orangutans (Pongo Abelii and Pongo Pygmaeus) on Sumatra and Borneo, Indonesia. PLoS ONE 2016, 11, e0152771. [Google Scholar] [CrossRef]
- Boorom, K.F.; Smith, H.; Nimri, L.; Viscogliosi, E.; Spanakos, G.; Parkar, U.; Li, L.-H.; Zhou, X.-N.; Ok, U.Z.; Leelayoova, S.; et al. Oh My Aching Gut: Irritable Bowel Syndrome, Blastocystis, and Asymptomatic Infection. Parasit. Vectors 2008, 1, 40. [Google Scholar] [CrossRef]
- Cian, A.; El Safadi, D.; Osman, M.; Moriniere, R.; Gantois, N.; Benamrouz-Vanneste, S.; Delgado-Viscogliosi, P.; Guyot, K.; Li, L.-L.; Monchy, S.; et al. Molecular Epidemiology of Blastocystis Sp. in Various Animal Groups from Two French Zoos and Evaluation of Potential Zoonotic Risk. PLoS ONE 2017, 12, e0169659. [Google Scholar] [CrossRef]
- Alfellani, M.A.; Jacob, A.S.; Perea, N.O.; Krecek, R.C.; Taner-Mulla, D.; Verweij, J.J.; Levecke, B.; Tannich, E.; Clark, C.G.; Stensvold, C.R. Diversity and Distribution of Blastocystis Sp. Subtypes in Non-Human Primates. Parasitology 2013, 140, 966–971. [Google Scholar] [CrossRef]
- Stensvold, C.R.; Clark, C.G. Current Status of Blastocystis: A Personal View. Parasitol. Int. 2016, 65, 763–771. [Google Scholar] [CrossRef]
- Li, J.; Karim, M.R.; Li, D.; Sumon, S.M.R.; Siddiki, S.F.; Rume, F.I.; Sun, R.; Jia, Y.; Zhang, L. Molecular Characterization of Blastocystis Sp. in Captive Wildlife in Bangladesh National Zoo: Non-Human Primates with High Prevalence and Zoonotic Significance. Int. J. Parasitol. Parasites Wildl. 2019, 10, 314–320. [Google Scholar] [CrossRef]
- Sow, D.; Parola, P.; Sylla, K.; Ndiaye, M.; Delaunay, P.; Halfon, P.; Camiade, S.; Dieng, T.; Tine, R.C.K.; Faye, B.; et al. Performance of Real-Time Polymerase Chain Reaction Assays for the Detection of 20 Gastrointestinal Parasites in Clinical Samples from Senegal. Am. J. Trop. Med. Hyg. 2017, 97, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Menu, E.; Mary, C.; Toga, I.; Raoult, D.; Ranque, S.; Bittar, F. Evaluation of Two DNA Extraction Methods for the PCR-Based Detection of Eukaryotic Enteric Pathogens in Fecal Samples. BMC Res. Notes 2018, 11, 206. [Google Scholar] [CrossRef]
- Mejia, R.; Vicuña, Y.; Broncano, N.; Sandoval, C.; Vaca, M.; Chico, M.; Cooper, P.J.; Nutman, T.B. A Novel, Multi-Parallel, Real-Time Polymerase Chain Reaction Approach for Eight Gastrointestinal Parasites Provides Improved Diagnostic Capabilities to Resource-Limited at-Risk Populations. Am. J. Trop. Med. Hyg. 2013, 88, 1041–1047. [Google Scholar] [CrossRef] [PubMed]
- Stensvold, C.R.; Ahmed, U.N.; Andersen, L.O.; Nielsen, H.V. Development and Evaluation of a Genus-Specific, Probe-Based, Internal-Process-Controlled Real-Time PCR Assay for Sensitive and Specific Detection of Blastocystis Spp. J. Clin. Microbiol. 2012, 50, 1847–1851. [Google Scholar] [CrossRef]
- Garcés-Sanchez, G.; Wilderer, P.A.; Munch, J.C.; Horn, H.; Lebuhn, M. Evaluation of Two Methods for Quantification of Hsp70 MRNA from the Waterborne Pathogen Cryptosporidium Parvum by Reverse Transcription Real-Time PCR in Environmental Samples. Water Res. 2009, 43, 2669–2678. [Google Scholar] [CrossRef]
- Verweij, J.J.; Laeijendecker, D.; Brienen, E.A.T.; van Lieshout, L.; Polderman, A.M. Detection of Cyclospora Cayetanensis in Travellers Returning from the Tropics and Subtropics Using Microscopy and Real-Time PCR. Int. J. Med. Microbiol. 2003, 293, 199–202. [Google Scholar] [CrossRef]
- Ten Hove, R.-J.; van Lieshout, L.; Brienen, E.A.T.; Perez, M.A.; Verweij, J.J. Real-Time Polymerase Chain Reaction for Detection of Isospora Belli in Stool Samples. Diagn. Microbiol. Infect. Dis. 2008, 61, 280–283. [Google Scholar] [CrossRef] [PubMed]
- Verweij, J.J.; Mulder, B.; Poell, B.; van Middelkoop, D.; Brienen, E.A.; van Lieshout, L. Real-Time PCR for the Detection of Dientamoeba Fragilis in Fecal Samples. Mol. Cell. Probes 2007, 21, 400–404. [Google Scholar] [CrossRef]
- Menotti, J.; Cassinat, B.; Sarfati, C.; Liguory, O.; Derouin, F.; Molina, J.-M. Development of a Real-Time PCR Assay for Quantitative Detection of Encephalitozoon Intestinalis DNA. J. Clin. Microbiol. 2003, 41, 1410–1413. [Google Scholar] [CrossRef]
- Roy, S.; Kabir, M.; Mondal, D.; Ali, I.K.M.; Petri, W.A.; Haque, R. Real-Time-PCR Assay for Diagnosis of Entamoeba Histolytica Infection. J. Clin. Microbiol. 2005, 43, 2168–2172. [Google Scholar] [CrossRef]
- Menotti, J.; Cassinat, B.; Porcher, R.; Sarfati, C.; Derouin, F.; Molina, J.-M. Development of a Real-Time Polymerase-Chain-Reaction Assay for Quantitative Detection of Enterocytozoon Bieneusi DNA in Stool Specimens from Immunocompromised Patients with Intestinal Microsporidiosis. J. Infect. Dis. 2003, 187, 1469–1474. [Google Scholar] [CrossRef]
- Verweij, J.J.; Blange, R.A.; Templeton, K.; Schinkel, J.; Brienen, E.A.; van Rooyen, M.A.; van Lieshout, L.; Polderman, A.M. Simultaneous Detection of Entamoeba Histolytica, Giardia Lamblia, and Cryptosporidium Parvum in Fecal Samples by Using Multiplex Real-Time PCR. J. Clin. Microbiol. 2004, 42, 1220–1223. [Google Scholar] [CrossRef]
- Poirier, P.; Wawrzyniak, I.; Albert, A.; El Alaoui, H.; Delbac, F.; Livrelli, V. Development and Evaluation of a Real-Time PCR Assay for Detection and Quantification of Blastocystis Parasites in Human Stool Samples: Prospective Study of Patients with Hematological Malignancies. J. Clin. Microbiol. 2011, 49, 975–983. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).