Multiplex Real-Time PCR Assay for Six Major Carbapenemase Genes
Abstract
1. Introduction
2. Materials and Methods
2.1. Laboratory Examination
2.1.1. Bacterial Strains
2.1.2. Primers and Probes
2.1.3. PCR Conditions
2.1.4. Equipment
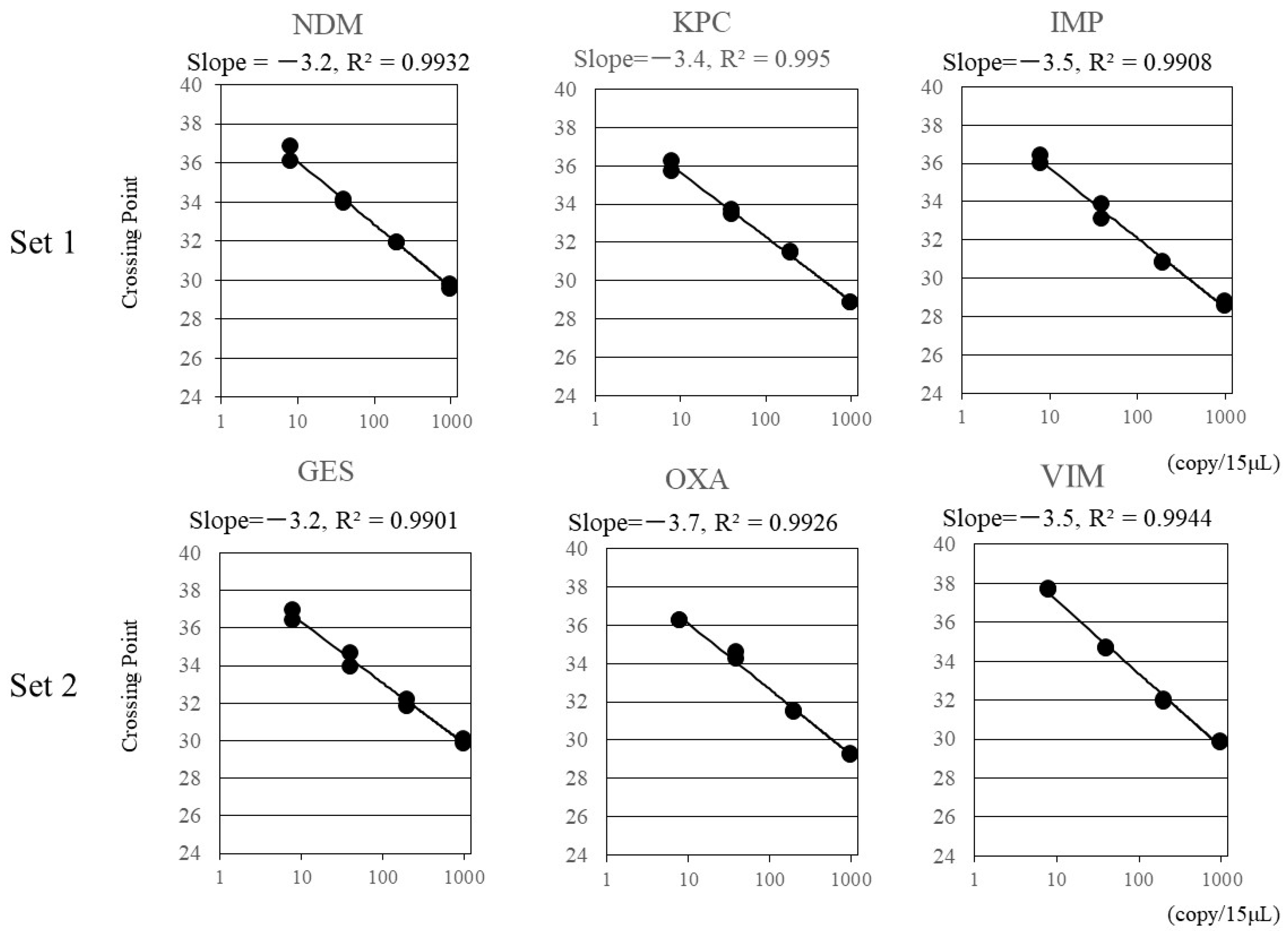
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Logan, L.K.; Weinstein, R.A. The epidemiology of Carbapenem-resistant enterobacteriaceae: The impact and evolution of a global menace. J. Infect. Dis. 2017, 215, S28–S36. [Google Scholar] [CrossRef]
- Köck, R.; Daniels-Haardt, I.; Becker, K.; Mellmann, A.; Friedrich, A.W.; Mevius, D.; Schwarz, S.; Jurke, A. Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing, and companion animals: A systematic review. Clin. Microbiol. Infect. 2018, 24, 1241–1250. [Google Scholar] [CrossRef]
- Yamamoto, N.; Asada, R.; Kawahara, R.; Hagiya, H.; Akeda, Y.; Shanmugakani, R.K.; Yoshidaab, H.; Yukawaa, S.; Yamamotoe, K.; Takayamac, Y.; et al. Prevalence of, and risk factors for, carriage of carbapenem-resistant Enterobacteriaceae among hospitalized patients in Japan. J. Hosp. Infect. 2017, 97, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Hagiya, H.; Yamamoto, N.; Kawahara, R.; Akeda, Y.; Shanmugakani, R.K.; Ueda, A.; Nishic, I.; Asadad, R.; Yoshidaa, H.; Tomonoa, K. Risk factors for fecal carriage of IMP-6-producing Enterobacteriaceae at a long-term care hospital in Japan: A follow-up report from the northern Osaka multicentre study group. J. Infect. Chemother. 2018, 24, 769–772. [Google Scholar] [CrossRef]
- Iovleva, A.; Doi, Y. Carbapenem-Resistant Enterobacteriaceae. Clin. Lab. Med. 2017, 37, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.; Humphries, R.M. Clinical laboratory detection of carbapenem-resistant and carbapenemase-producing Enterobacteriaceae. Expert Rev. Anti Infect. Ther. 2016, 14, 705–717. [Google Scholar] [CrossRef] [PubMed]
- Shanmugakani, R.K.; Akeda, Y.; Yamamoto, N.; Sakamoto, N.; Hagiya, H.; Yoshida, H.; Takeuchi, D.; Sugawara, Y.; Kodera, T.; Kawase, M.; et al. PCR-dipstick chromatography for differential detection of carbapenemase genes directly in stool specimens. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Greissl, C.; Saleh, A.; Hamprecht, A. Rapid detection of OXA-48-like, KPC, NDM, and VIM carbapenemases in Enterobacterales by a new multiplex immunochromatographic test. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 331–335. [Google Scholar] [CrossRef]
- Glupczynski, Y.; Jousset, A.; Evrard, S.; Bonnin, R.A.; Huang, T.D.; Dortet, L.; Bogaerts, P.; Naas, T. Prospective evaluation of the OKN K-SeT assay, a new multiplex immunochromatographic test for the rapid detection of OXA-48-like, KPC and NDM carbapenemases. J. Antimicrob. Chemother. 2017, 72, 1955–1960. [Google Scholar] [CrossRef]
- Wareham, D.W.; Momin, M.H.F.A. Rapid detection of carbapenemases in enterobacteriaceae: Evaluation of the resist-3 O.K.N. (OXA-48, KPC, NDM) lateral flow multiplexed assay. J. Clin. Microbiol. 2017, 55, 1223–1225. [Google Scholar] [CrossRef] [PubMed]
- Shanmugakani, R.K.; Akeda, Y.; Sugawara, Y.; Laolerd, W.; Chaihongsa, N.; Sirichot, S.; Yamamoto, N.; Hagiya, H.; Morii, D.; Fujiya, Y.; et al. PCR-dipstick-oriented surveillance and characterization of mcr-1- and carbapenemase-carrying Enterobacteriaceae in a Thai hospital. Front. Microbiol. 2019, 10, 149. [Google Scholar] [CrossRef]
- Longo, M.S.; Berninger, J.H. Use of Uracil DNA Glycosylase to Control Carry-Over Contamination in Polymerase Chain Reactions. Gene 1990, 93, 125–128. [Google Scholar] [CrossRef]
- Oviaño, M.; Torres, I.; González, M.; Bou, G. Evaluation of a novel procedure for rapid detection of carbapenemase-producing Enterobacteriaceae (CPE) using the LightMix® modular carbapenemase kits. J. Antimicrob. Chemother. 2016, 71, 3420–3423. [Google Scholar] [CrossRef][Green Version]
- Dortet, L.; Fusaro, M.; Naasa, T. Improvement of the Xpert carba-R kit for the detection of Carbapenemase-Producing Enterobacteriaceae. Antimicrob. Agents Chemother. 2016, 60, 3832–3837. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, J.; Widen, R.H.; Pignatari, A.C.; Kubasek, C.; Silbert, S. Rapid detection of carbapenemase genes by multiplex real-time PCR. J. Antimicrob. Chemother. 2012, 67, 906–909. [Google Scholar] [CrossRef] [PubMed]
- van der Zee, A.; Roorda, L.; Bosman, G.; Fluit, A.C.; Hermans, M.; Smits, P.H.; van der Zanden, A.G.M.; Witt, R.; van Coppenraet, L.E.S.B.; Stuart, J.C.; et al. Multi-centre evaluation of real-time multiplex PCR for detection of carbapenemase genes OXA-48, VIM, IMP, NDM and KPC. BMC Infect. Dis. 2014, 14, 27. [Google Scholar] [CrossRef]
- Smiljanic, M.; Kaase, M.; Ahmad-Nejad, P.; Ghebremedhin, B. Comparison of in-house and commercial real time-PCR based carbapenemase gene detection methods in Enterobacteriaceae and non-fermenting gram-negative bacterial isolates. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 48. [Google Scholar] [CrossRef] [PubMed]
- Teo, J.W.; La, M.V.; Lin, R.T. Development and evaluation of a multiplex real-time PCR for the detection of IMP, VIM, and OXA-23 carbapenemase gene families on the BD MAX open system. Diagn. Microbiol. Infect. Dis. 2016, 86, 358–361. [Google Scholar] [CrossRef]
- Oho, M.; Funashima, Y.; Nagasawa, Z.; Miyamoto, H.; Sueoka, E. Rapid detection method of carbapenemase-producing Enterobacteriaceae by MALDI-TOF MS with imipenem/cilastatin (KB) disc and zinc sulfate solution. J. Infect. Chemother. 2021, 27, 205–210. [Google Scholar] [CrossRef]
- Manageiro, V.; Romão, R.; Moura, I.B.; Sampaio, D.A.; Vieira, L.; Ferreira, E.; Network EuSCAPE-Portugal; Caniça, M. Molecular Epidemiology and Risk Factors of Carbapenemase-Producing Enterobacteriaceae Isolates in Portuguese Hospitals: Results from European Survey on Carbapenemase-Producing Enterobacteriaceae (EuSCAPE). Front. Microbiol. 2018, 9, 2834. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, G.R.; Jeong, J.; Kim, S.; Shin, J.H. Prevalence and Characteristics of Carbapenemase-Producing Enterobacteriaceae in Three Tertiary-Care Korean University Hospitals between 2017 and 2018. Jpn. J. Infect. Dis. 2020, 73, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Meunier, D.; Woodford, N.; Hopkins, K.L. Evaluation of the AusDiagnostics MT CRE EU assay for the detection of carbapenemase genes and transferable colistin resistance determinants mcr-1/-2 in MDR Gram-negative bacteria. J. Antimicrob. Chemother. 2018, 73, 3355–3358. [Google Scholar] [CrossRef] [PubMed]
| Gene | Primer Names | Probe | Sequences (5′-3′) | Wavelength (nm) |
|---|---|---|---|---|
| NDM | LightMix® Modular NDM Carbapenemase | FAM labeled hydrolysis probe | F: GGTTTGGCGATCTGGTTTTC R: CGGAATGGCTCATCACGATC | FAM (465–510) |
| KPC | LightMix® Modular KPC Carbapenemase | LC610 labeled hydrolysis probe | F: ATGTCACTGTATCGCCGTCT R: TTTTCAGAGCCTTACTGCCC | 610 (540–610) |
| IMP | LightMix® Modular IMP Carbapenemase | LC640 labeled probe | F: GAATAGRRTGGCTTAAYTCTC R: CCAAACYACTASGTTATC | 640 (610–645) |
| GES | LightMix® Modular GES Carbapenemase | LC640 labeled probe | F: GCTTCATTCACGCACTATT R: CGATGCTAGAAACCGCTC | 640 (610–645) |
| OXA | LightMix® Modular OXA-48 Carbapenemase | R6G labeled hydrolysis probe | F: TTGGTGGCATCGATTATCGG R: GAGCACTTCTTTTGTGATGGC | 580 (540–580) |
| VIM | LightMix® Modular VIM Carbapenemase | Cyan500 labeled hydrolysis probe | F: GTTTGGTCGCATATCGCAAC R: AATGCGCAGCACCAGGATAG | FAM (465–510) |
| PCR Set | Carbapenemase Type | Genotypes of the Tested Isolates | |||||
|---|---|---|---|---|---|---|---|
| blaNDM-5 | blaKPC-2 | blaIMP-6 | blaGES-24 | blaOXA-232 | blaVIM-1 | ||
| 1 | NDM | 19.52/19.36 | n.d. | n.d. | n.d. | n.d. | n.d. |
| KPC | n.d. | 18.33/18.46 | n.d. | n.d. | n.d. | n.d. | |
| IMP | n.d. | n.d. | 15.93/15.85 | n.d. | n.d. | n.d. | |
| 2 | GES | n.d. | n.d. | n.d. | 16.13/16.16 | n.d. | n.d. |
| OXA | n.d. | n.d. | n.d. | n.d. | 22.11/22/15 | n.d. | |
| VIM | n.d. | n.d. | n.d. | n.d. | n.d. | 17.67/17.62 | |
| PCR Set | Carbapenemase Type | Genotypes of the Tested Isolates | |||||
|---|---|---|---|---|---|---|---|
| Isolate 1 blaNDM-1 + blaOXA232 | Isolate 2 blaNDM-1 + blaOXA232 | Isolate 3 blaNDM-1 + blaOXA232 | Isolate 4 blaNDM-1 + blaOXA181 | Isolate 5 blaNDM-1 + blaOXA232 or 181 | Isolate 6 blaNDM-1 + blaOXA232 or 181 | ||
| 1 | NDM | 26.63/26.63 | 25.95/25.87 | 26.21/26.16 | 17.78/17.76 | 27.07/27.1 | 25.44/25.54 |
| KPC | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | |
| IMP | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | |
| 2 | GES | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| OXA | 27.63/27.85 | 26.95/26.94 | 27.34/27.31 | 25.95/25.72 | 29.55/29.56 | 30.42/30.2 | |
| VIM | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoshioka, N.; Hagiya, H.; Deguchi, M.; Hamaguchi, S.; Kagita, M.; Nishi, I.; Akeda, Y.; Tomono, K. Multiplex Real-Time PCR Assay for Six Major Carbapenemase Genes. Pathogens 2021, 10, 276. https://doi.org/10.3390/pathogens10030276
Yoshioka N, Hagiya H, Deguchi M, Hamaguchi S, Kagita M, Nishi I, Akeda Y, Tomono K. Multiplex Real-Time PCR Assay for Six Major Carbapenemase Genes. Pathogens. 2021; 10(3):276. https://doi.org/10.3390/pathogens10030276
Chicago/Turabian StyleYoshioka, Nori, Hideharu Hagiya, Matsuo Deguchi, Shigeto Hamaguchi, Masanori Kagita, Isao Nishi, Yukihiro Akeda, and Kazunori Tomono. 2021. "Multiplex Real-Time PCR Assay for Six Major Carbapenemase Genes" Pathogens 10, no. 3: 276. https://doi.org/10.3390/pathogens10030276
APA StyleYoshioka, N., Hagiya, H., Deguchi, M., Hamaguchi, S., Kagita, M., Nishi, I., Akeda, Y., & Tomono, K. (2021). Multiplex Real-Time PCR Assay for Six Major Carbapenemase Genes. Pathogens, 10(3), 276. https://doi.org/10.3390/pathogens10030276






