Comparative Analysis of Hepatitis C Virus NS5A Dynamics and Localization in Assembly-Deficient Mutants
Abstract
1. Introduction
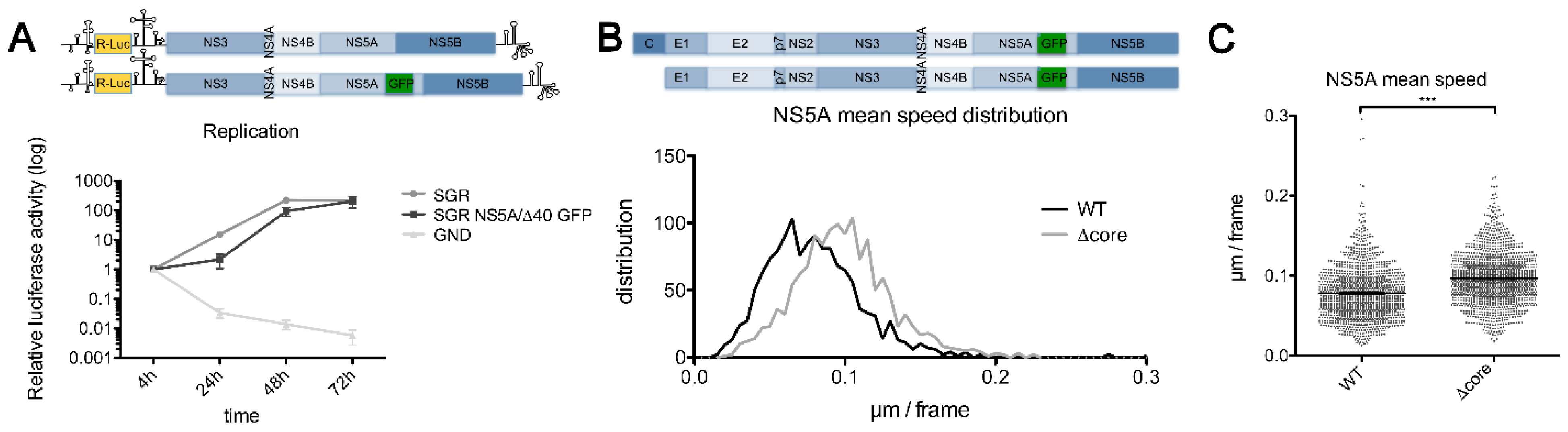
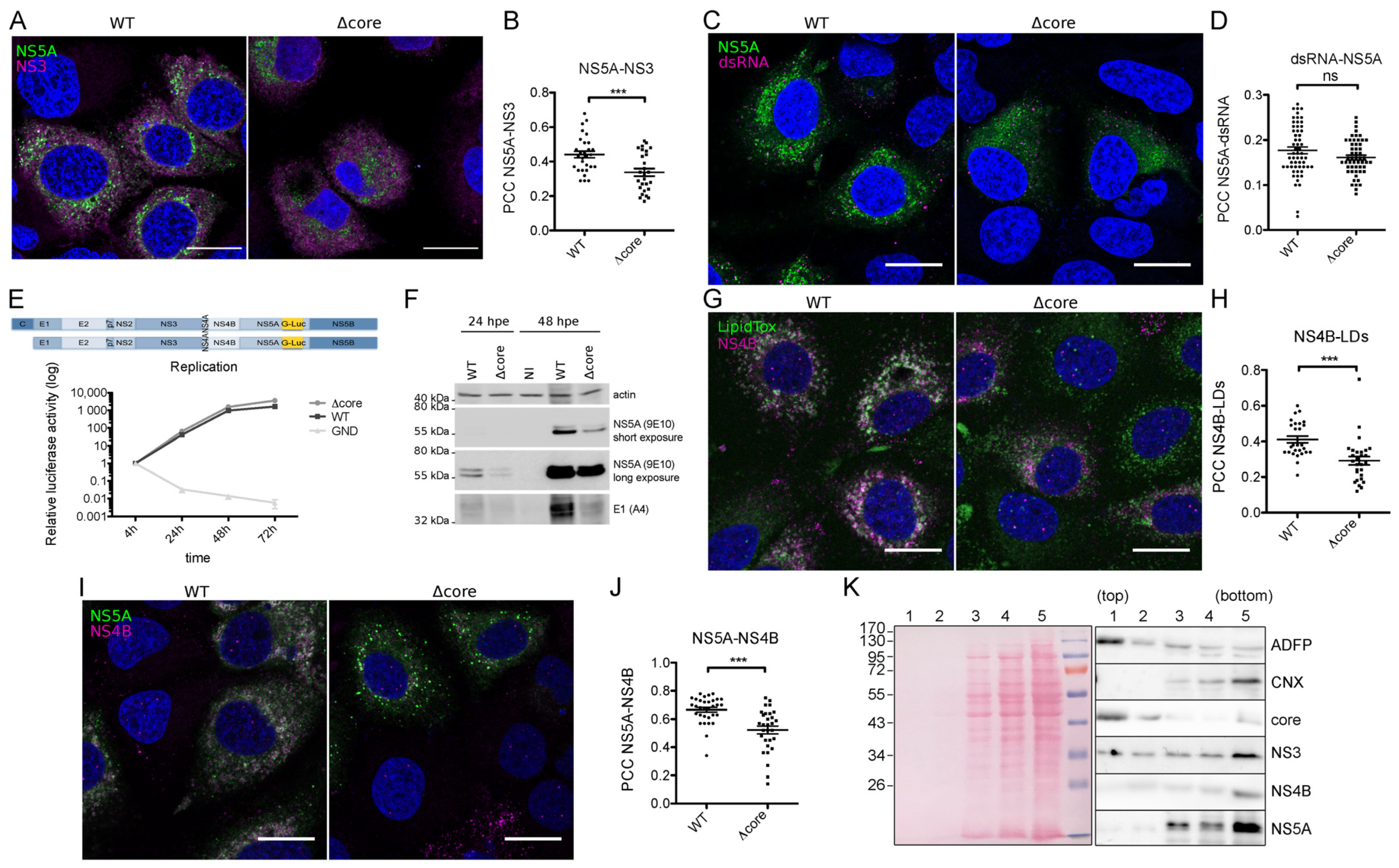
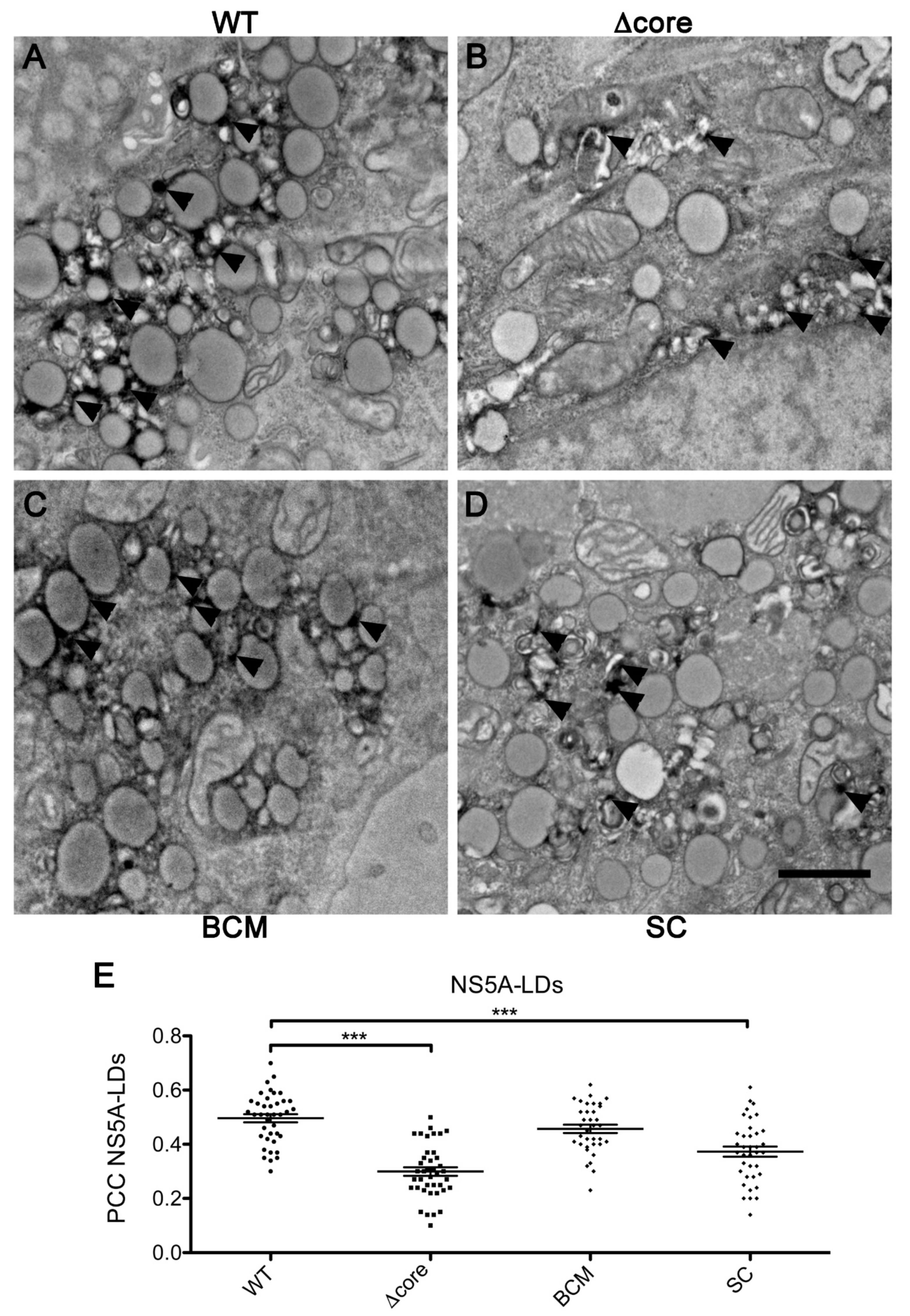
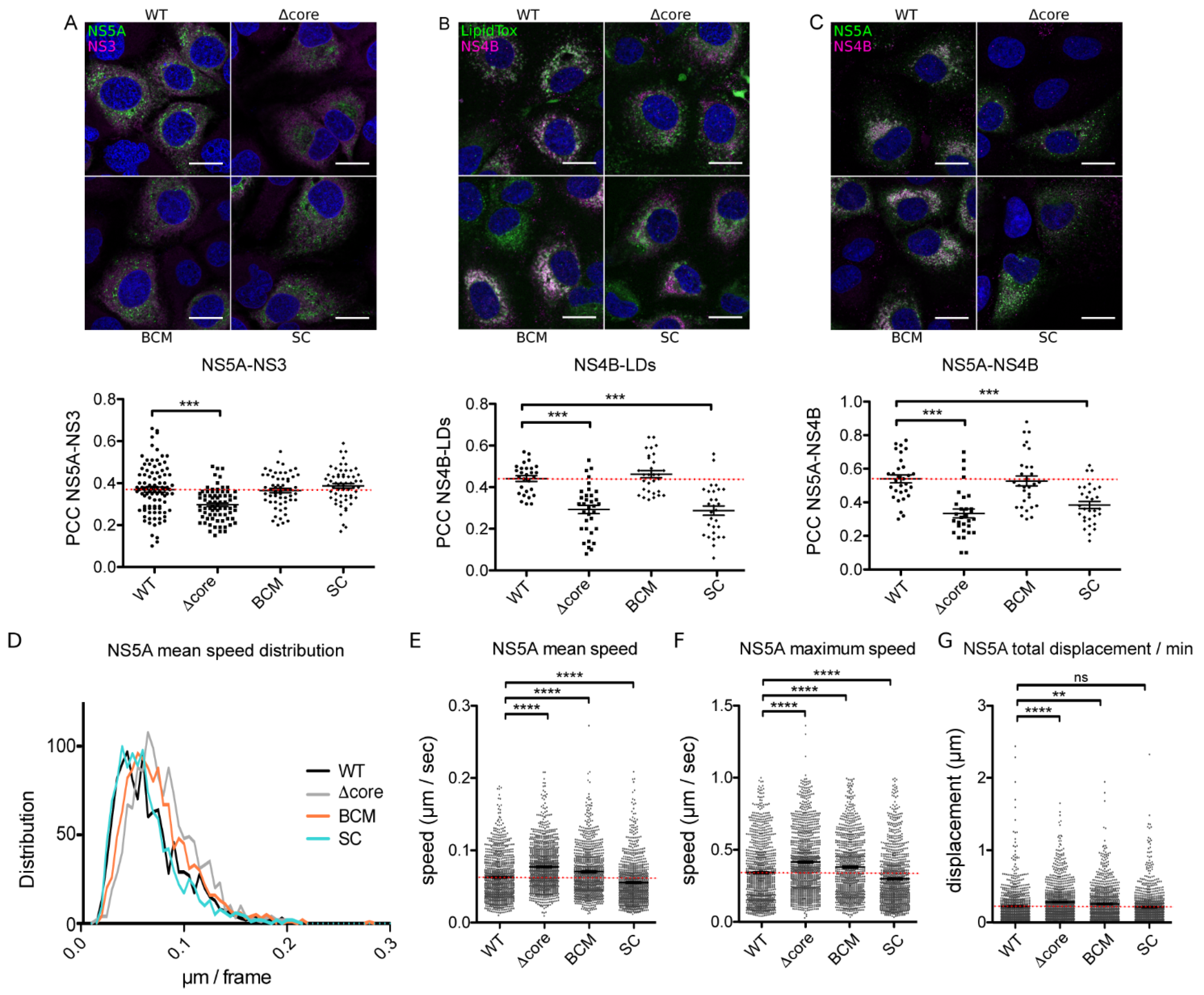
2. Results
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Primers
4.3. Plasmids
4.4. RNA Transcription and Electroporation
4.5. Antibodies and Reagents
4.6. Videomicroscopy
4.7. Immunofluorescence
4.8. APEX2 Labeling and Transmission Electron Microscopy (TEM)
4.9. Purification of Lipid Droplets
4.10. Quantification and Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Hepatitis Programme Global Hepatitis Report; World Health Organization: Geneva, Switzerland, 2017; ISBN 978-92-4-156545-5. [Google Scholar]
- Choo, Q.L.; Richman, K.H.; Han, J.H.; Berger, K.; Lee, C.; Dong, C.; Gallegos, C.; Coit, D.; Medina-Selby, R.; Barr, P.J. Genetic Organization and Diversity of the Hepatitis C Virus. Proc. Natl. Acad. Sci. USA 1991, 88, 2451–2455. [Google Scholar] [CrossRef]
- Neddermann, P.; Tomei, L.; Steinkuhler, C.; Gallinari, P.; Tramontano, A.; De Francesco, R. The Nonstructural Proteins of the Hepatitis C Virus: Structure and Functions. Biol. Chem. 1997, 378, 469–476. [Google Scholar]
- Suzuki, T.; Ishii, K.; Aizaki, H.; Wakita, T. Hepatitis C Viral Life Cycle. Adv. Drug Deliv. Rev. 2007, 59, 1200–1212. [Google Scholar] [CrossRef]
- Lohmann, V.; Korner, F.; Koch, J.; Herian, U.; Theilmann, L.; Bartenschlager, R. Replication of Subgenomic Hepatitis C Virus RNAs in a Hepatoma Cell Line. Science 1999, 285, 110–113. [Google Scholar] [CrossRef] [PubMed]
- Lohmann, V.; Körner, F.; Herian, U.; Bartenschlager, R. Biochemical Properties of Hepatitis C Virus NS5B RNA-Dependent RNA Polymerase and Identification of Amino Acid Sequence Motifs Essential for Enzymatic Activity. J. Virol. 1997, 71, 8416–8428. [Google Scholar] [CrossRef]
- Jones, C.T.; Murray, C.L.; Eastman, D.K.; Tassello, J.; Rice, C.M. Hepatitis C Virus p7 and NS2 Proteins Are Essential for Production of Infectious Virus. J. Virol. 2007, 81, 8374–8383. [Google Scholar] [CrossRef] [PubMed]
- Popescu, C.-I.; Callens, N.; Trinel, D.; Roingeard, P.; Moradpour, D.; Descamps, V.; Duverlie, G.; Penin, F.; Héliot, L.; Rouillé, Y.; et al. NS2 Protein of Hepatitis C Virus Interacts with Structural and Non-Structural Proteins towards Virus Assembly. PLoS Pathog. 2011, 7, e1001278. [Google Scholar] [CrossRef] [PubMed]
- Romero-Brey, I.; Merz, A.; Chiramel, A.; Lee, J.Y.; Chlanda, P.; Haselman, U.; Santarella-Mellwig, R.; Habermann, A.; Hoppe, S.; Kallis, S.; et al. Three-Dimensional Architecture and Biogenesis of Membrane Structures Associated with Hepatitis C Virus Replication. PLoS Pathog. 2012, 8, e1003056. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Aizaki, H.; He, J.-W.; Lai, M.M.C. Interactions between Viral Nonstructural Proteins and Host Protein hVAP-33 Mediate the Formation of Hepatitis C Virus RNA Replication Complex on Lipid Raft. J. Virol. 2004, 78, 3480–3488. [Google Scholar] [CrossRef]
- Egger, D.; Wolk, B.; Gosert, R.; Bianchi, L.; Blum, H.E.; Moradpour, D.; Bienz, K. Expression of Hepatitis C Virus Proteins Induces Distinct Membrane Alterations Including a Candidate Viral Replication Complex. J. Virol. 2002, 76, 5974–5984. [Google Scholar] [CrossRef]
- Paul, D.; Romero-Brey, I.; Gouttenoire, J.; Stoitsova, S.; Krijnse-Locker, J.; Moradpour, D.; Bartenschlager, R. NS4B Self-Interaction through Conserved C-Terminal Elements Is Required for the Establishment of Functional Hepatitis C Virus Replication Complexes. J. Virol. 2011, 85, 6963–6976. [Google Scholar] [CrossRef]
- Popescu, C.-I.; Riva, L.; Vlaicu, O.; Farhat, R.; Rouillé, Y.; Dubuisson, J. Hepatitis C Virus Life Cycle and Lipid Metabolism. Biology 2014, 3, 892–921. [Google Scholar] [CrossRef]
- Miyanari, Y.; Atsuzawa, K.; Usuda, N.; Watashi, K.; Hishiki, T.; Zayas, M.; Bartenschlager, R.; Wakita, T.; Hijikata, M.; Shimotohno, K. The Lipid Droplet Is an Important Organelle for Hepatitis C Virus Production. Nat. Cell Biol. 2007, 9, 1089–1097. [Google Scholar] [CrossRef]
- Ross-Thriepland, D.; Harris, M. Hepatitis C Virus NS5A: Enigmatic but Still Promiscuous 10 Years On! J. Gen. Virol. 2015, 96, 727–738. [Google Scholar] [CrossRef]
- Biswas, A.; Treadaway, J.; Tellinghuisen, T.L. Interaction between Nonstructural Proteins NS4B and NS5A Is Essential for Proper NS5A Localization and Hepatitis C Virus RNA Replication. J. Virol. 2016, 90, 7205–7218. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.L.; Belyaeva, T.; Stonehouse, N.J.; Pearson, A.R.; Harris, M. All Three Domains of the Hepatitis C Virus Nonstructural NS5A Protein Contribute to RNA Binding. J. Virol. 2010, 84, 9267–9277. [Google Scholar] [CrossRef] [PubMed]
- Masaki, T.; Suzuki, R.; Murakami, K.; Aizaki, H.; Ishii, K.; Murayama, A.; Date, T.; Matsuura, Y.; Miyamura, T.; Wakita, T.; et al. Interaction of Hepatitis C Virus Nonstructural Protein 5A with Core Protein Is Critical for the Production of Infectious Virus Particles. J. Virol. 2008, 82, 7964–7976. [Google Scholar] [CrossRef] [PubMed]
- Zayas, M.; Long, G.; Madan, V.; Bartenschlager, R. Coordination of Hepatitis C Virus Assembly by Distinct Regulatory Regions in Nonstructural Protein 5A. PLoS Pathog. 2016, 12, e1005376. [Google Scholar] [CrossRef]
- Appel, N.; Zayas, M.; Miller, S.; Krijnse-Locker, J.; Schaller, T.; Friebe, P.; Kallis, S.; Engel, U.; Bartenschlager, R. Essential Role of Domain III of Nonstructural Protein 5A for Hepatitis C Virus Infectious Particle Assembly. PLoS Pathog. 2008, 4, e1000035. [Google Scholar] [CrossRef] [PubMed]
- Tellinghuisen, T.L.; Foss, K.L.; Treadaway, J. Regulation of Hepatitis C Virion Production via Phosphorylation of the NS5A Protein. PLoS Pathog. 2008, 4, e1000032. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Goonawardane, N.; Stewart, H.; Harris, M. A Role for Domain I of the Hepatitis C Virus NS5A Protein in Virus Assembly. PLoS Pathog. 2018, 14, e1006834. [Google Scholar] [CrossRef] [PubMed]
- Alsaleh, K.; Delavalle, P.-Y.; Pillez, A.; Duverlie, G.; Descamps, V.; Rouille, Y.; Dubuisson, J.; Wychowski, C. Identification of Basic Amino Acids at the N-Terminal End of the Core Protein That Are Crucial for Hepatitis C Virus Infectivity. J. Virol. 2010, 84, 12515–12528. [Google Scholar] [CrossRef] [PubMed]
- Gawlik, K.; Gallay, P.A. HCV Core Protein and Virus Assembly: What We Know without Structures. Immunol. Res. 2014, 60, 1–10. [Google Scholar] [CrossRef]
- Camus, G.; Herker, E.; Modi, A.A.; Haas, J.T.; Ramage, H.R.; Farese, R.V.; Ott, M. Diacylglycerol Acyltransferase-1 Localizes Hepatitis C Virus NS5A Protein to Lipid Droplets and Enhances NS5A Interaction with the Viral Capsid Core. J. Biol. Chem. 2013, 288, 9915–9923. [Google Scholar] [CrossRef]
- Gottwein, J.M.; Jensen, T.B.; Mathiesen, C.K.; Meuleman, P.; Serre, S.B.N.; Lademann, J.B.; Ghanem, L.; Scheel, T.K.H.; Leroux-Roels, G.; Bukh, J. Development and Application of Hepatitis C Reporter Viruses with Genotype 1 to 7 Core-Nonstructural Protein 2 (NS2) Expressing Fluorescent Proteins or Luciferase in Modified JFH1 NS5A. J. Virol. 2011, 85, 8913–8928. [Google Scholar] [CrossRef]
- Shulla, A.; Randall, G. Spatiotemporal Analysis of Hepatitis C Virus Infection. PLoS Pathog. 2015, 11, e1004758. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.M.; Patel, A.H.; Targett-Adams, P.; McLauchlan, J. The Hepatitis C Virus NS4B Protein Can Trans-Complement Viral RNA Replication and Modulates Production of Infectious Virus. J. Virol. 2009, 83, 2163–2177. [Google Scholar] [CrossRef]
- Piccininni, S.; Varaklioti, A.; Nardelli, M.; Dave, B.; Raney, K.D.; McCarthy, J.E.G. Modulation of the Hepatitis C Virus RNA-Dependent RNA Polymerase Activity by the Non-Structural (NS) 3 Helicase and the NS4B Membrane Protein. J. Biol. Chem. 2002, 277, 45670–45679. [Google Scholar] [CrossRef]
- Tanaka, T.; Kuroda, K.; Ikeda, M.; Wakita, T.; Kato, N.; Makishima, M. Hepatitis C Virus NS4B Targets Lipid Droplets through Hydrophobic Residues in the Amphipathic Helices. J. Lipid Res. 2013, 54, 881–892. [Google Scholar] [CrossRef]
- Han, Q.; Manna, D.; Belton, K.; Cole, R.; Konan, K.V. Modulation of Hepatitis C Virus Genome Encapsidation by Nonstructural Protein 4B. J. Virol. 2013, 87, 7409–7422. [Google Scholar] [CrossRef] [PubMed]
- Gouttenoire, J.; Montserret, R.; Paul, D.; Castillo, R.; Meister, S.; Bartenschlager, R.; Penin, F.; Moradpour, D. Aminoterminal Amphipathic α-Helix AH1 of Hepatitis C Virus Nonstructural Protein 4B Possesses a Dual Role in RNA Replication and Virus Production. PLoS Pathog. 2014, 10, e1004501. [Google Scholar] [CrossRef]
- Lam, S.S.; Martell, J.D.; Kamer, K.J.; Deerinck, T.J.; Ellisman, M.H.; Mootha, V.K.; Ting, A.Y. Directed Evolution of APEX2 for Electron Microscopy and Proximity Labeling. Nat. Methods 2014, 12, 51–54. [Google Scholar] [CrossRef] [PubMed]
- Eyre, N.S.; Hampton-Smith, R.J.; Aloia, A.L.; Eddes, J.S.; Simpson, K.J.; Hoffmann, P.; Beard, M.R. Phosphorylation of NS5A Serine-235 Is Essential to Hepatitis C Virus RNA Replication and Normal Replication Compartment Formation. Virology 2016, 491, 27–44. [Google Scholar] [CrossRef]
- Boulant, S.; Douglas, M.W.; Moody, L.; Budkowska, A.; Targett-Adams, P.; McLauchlan, J. Hepatitis C Virus Core Protein Induces Lipid Droplet Redistribution in a Microtubule- and Dynein-Dependent Manner. Traffic 2008, 9, 1268–1282. [Google Scholar] [CrossRef] [PubMed]
- Lyn, R.K.; Kennedy, D.C.; Stolow, A.; Ridsdale, A.; Pezacki, J.P. Dynamics of Lipid Droplets Induced by the Hepatitis C Virus Core Protein. Biochem. Biophys. Res. Commun. 2010, 399, 518–524. [Google Scholar] [CrossRef]
- Lyn, R.K.; Hope, G.; Sherratt, A.R.; McLauchlan, J.; Pezacki, J.P. Bidirectional Lipid Droplet Velocities Are Controlled by Differential Binding Strengths of HCV Core DII Protein. PLoS ONE 2013, 8, e78065. [Google Scholar] [CrossRef][Green Version]
- Wölk, B.; Büchele, B.; Moradpour, D.; Rice, C.M. A Dynamic View of Hepatitis C Virus Replication Complexes. J. Virol. 2008, 82, 10519–10531. [Google Scholar] [CrossRef] [PubMed]
- Eyre, N.S.; Fiches, G.N.; Aloia, A.L.; Helbig, K.J.; McCartney, E.M.; McErlean, C.S.P.; Li, K.; Aggarwal, A.; Turville, S.G.; Beard, M.R. Dynamic Imaging of the Hepatitis C Virus NS5A Protein during a Productive Infection. J. Virol. 2014, 88, 3636–3652. [Google Scholar] [CrossRef] [PubMed]
- Vlaicu, O.; Selescu, T.; Pastrama, F.; Munteanu, C.; Riva, L.; Dubuisson, J.; Rouille, Y.; Popescu, C.-I. Novel Replicons and Trans -Encapsidation Systems for Hepatitis C Virus Proteins Live Imaging and Virus-Host Interaction Proteomics. J. Virol. Methods 2017, 246, 42–50. [Google Scholar] [CrossRef]
- Roohvand, F.; Maillard, P.; Lavergne, J.-P.; Boulant, S.; Walic, M.; Andréo, U.; Goueslain, L.; Helle, F.; Mallet, A.; McLauchlan, J.; et al. Initiation of Hepatitis C Virus Infection Requires the Dynamic Microtubule Network: Role of the viral nucleocapsid protein. J. Biol. Chem. 2009, 284, 13778–13791. [Google Scholar] [CrossRef]
- Fiches, G.N.; Eyre, N.S.; Aloia, A.L.; Van Der Hoek, K.; Betz-Stablein, B.; Luciani, F.; Chopra, A.; Beard, M.R. HCV RNA Traffic and Association with NS5A in Living Cells. Virology 2016, 493, 60–74. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Cortese, M.; Haselmann, U.; Tabata, K.; Romero-Brey, I.; Funaya, C.; Schieber, N.L.; Qiang, Y.; Bartenschlager, M.; Kallis, S.; et al. Spatiotemporal Coupling of the Hepatitis C Virus Replication Cycle by Creating a Lipid Droplet- Proximal Membranous Replication Compartment. Cell Rep. 2019, 27, 3602–3617.e5. [Google Scholar] [CrossRef]
- Dimitrova, M.; Imbert, I.; Kieny, M.P.; Schuster, C. Protein-Protein Interactions between Hepatitis C Virus Nonstructural Proteins. J. Virol. 2003, 77, 5401–5414. [Google Scholar] [CrossRef]
- Benureau, Y.; Warter, L.; Malcolm, B.A.; Martin, A. A Comparative Analysis of the Substrate Permissiveness of HCV and GBV-B NS3/4A Proteases Reveals Genetic Evidence for an Interaction with NS4B Protein during Genome Replication. Virology 2010, 406, 228–240. [Google Scholar] [CrossRef]
- Einav, S.; Gerber, D.; Bryson, P.D.; Sklan, E.H.; Elazar, M.; Maerkl, S.J.; Glenn, J.S.; Quake, S.R. Discovery of a Hepatitis C Target and Its Pharmacological Inhibitors by Microfluidic Affinity Analysis. Nat. Biotechnol. 2008, 26, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Quinkert, D.; Bartenschlager, R.; Lohmann, V. Quantitative Analysis of the Hepatitis C Virus Replication Complex. J. Virol. 2005, 79, 13594–13605. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, H.; Taketa, K.; Miyano, K.; Yamane, T.; Sato, J. Growth of Human Hepatoma Cells Lines with Differentiated Functions in Chemically Defined Medium. Cancer Res. 1982, 42, 3858–3863. [Google Scholar] [PubMed]
- Wakita, T.; Pietschmann, T.; Kato, T.; Date, T.; Miyamoto, M.; Zhao, Z.; Murthy, K.; Habermann, A.; Krausslich, H.-G.; Mizokami, M.; et al. Production of Infectious Hepatitis C Virus in Tissue Culture from a Cloned Viral Genome. Nat. Med. 2005, 11, 791–796. [Google Scholar] [CrossRef]
- Goueslain, L.; Alsaleh, K.; Horellou, P.; Roingeard, P.; Descamps, V.; Duverlie, G.; Ciczora, Y.; Wychowski, C.; Dubuisson, J.; Rouille, Y. Identification of GBF1 as a Cellular Factor Required for Hepatitis C Virus RNA Replication. J. Virol. 2010, 84, 773–787. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Date, T.; Miyamoto, M.; Furusaka, A.; Tokushige, K.; Mizokami, M.; Wakita, T. Efficient Replication of the Genotype 2a Hepatitis C Virus Subgenomic Replicon. Gastroenterology 2003, 125, 1808–1817. [Google Scholar] [CrossRef]
- Macdonald, A.; Crowder, K.; Street, A.; McCormick, C.; Saksela, K.; Harris, M. The Hepatitis C Virus Non-Structural NS5A Protein Inhibits Activating Protein–1 Function by Perturbing Ras-ERK Pathway Signaling. J. Biol. Chem. 2003, 278, 17775–17784. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Tinevez, J.-Y.; Perry, N.; Schindelin, J.; Hoopes, G.M.; Reynolds, G.D.; Laplantine, E.; Bednarek, S.Y.; Shorte, S.L.; Eliceiri, K.W. TrackMate: An Open and Extensible Platform for Single-Particle Tracking. Methods 2017, 115, 80–90. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riva, L.; Spriet, C.; Barois, N.; Popescu, C.-I.; Dubuisson, J.; Rouillé, Y. Comparative Analysis of Hepatitis C Virus NS5A Dynamics and Localization in Assembly-Deficient Mutants. Pathogens 2021, 10, 172. https://doi.org/10.3390/pathogens10020172
Riva L, Spriet C, Barois N, Popescu C-I, Dubuisson J, Rouillé Y. Comparative Analysis of Hepatitis C Virus NS5A Dynamics and Localization in Assembly-Deficient Mutants. Pathogens. 2021; 10(2):172. https://doi.org/10.3390/pathogens10020172
Chicago/Turabian StyleRiva, Laura, Corentin Spriet, Nicolas Barois, Costin-Ioan Popescu, Jean Dubuisson, and Yves Rouillé. 2021. "Comparative Analysis of Hepatitis C Virus NS5A Dynamics and Localization in Assembly-Deficient Mutants" Pathogens 10, no. 2: 172. https://doi.org/10.3390/pathogens10020172
APA StyleRiva, L., Spriet, C., Barois, N., Popescu, C.-I., Dubuisson, J., & Rouillé, Y. (2021). Comparative Analysis of Hepatitis C Virus NS5A Dynamics and Localization in Assembly-Deficient Mutants. Pathogens, 10(2), 172. https://doi.org/10.3390/pathogens10020172





