Active Human and Porcine Serum Induce Competence for Genetic Transformation in the Emerging Zoonotic Pathogen Streptococcus suis
Abstract
1. Introduction
2. Methods
2.1. Bacterial Strains, Plasmids, and Culture Conditions
2.2. RNA-Seq
2.3. Standard Assay for DNA Transformation
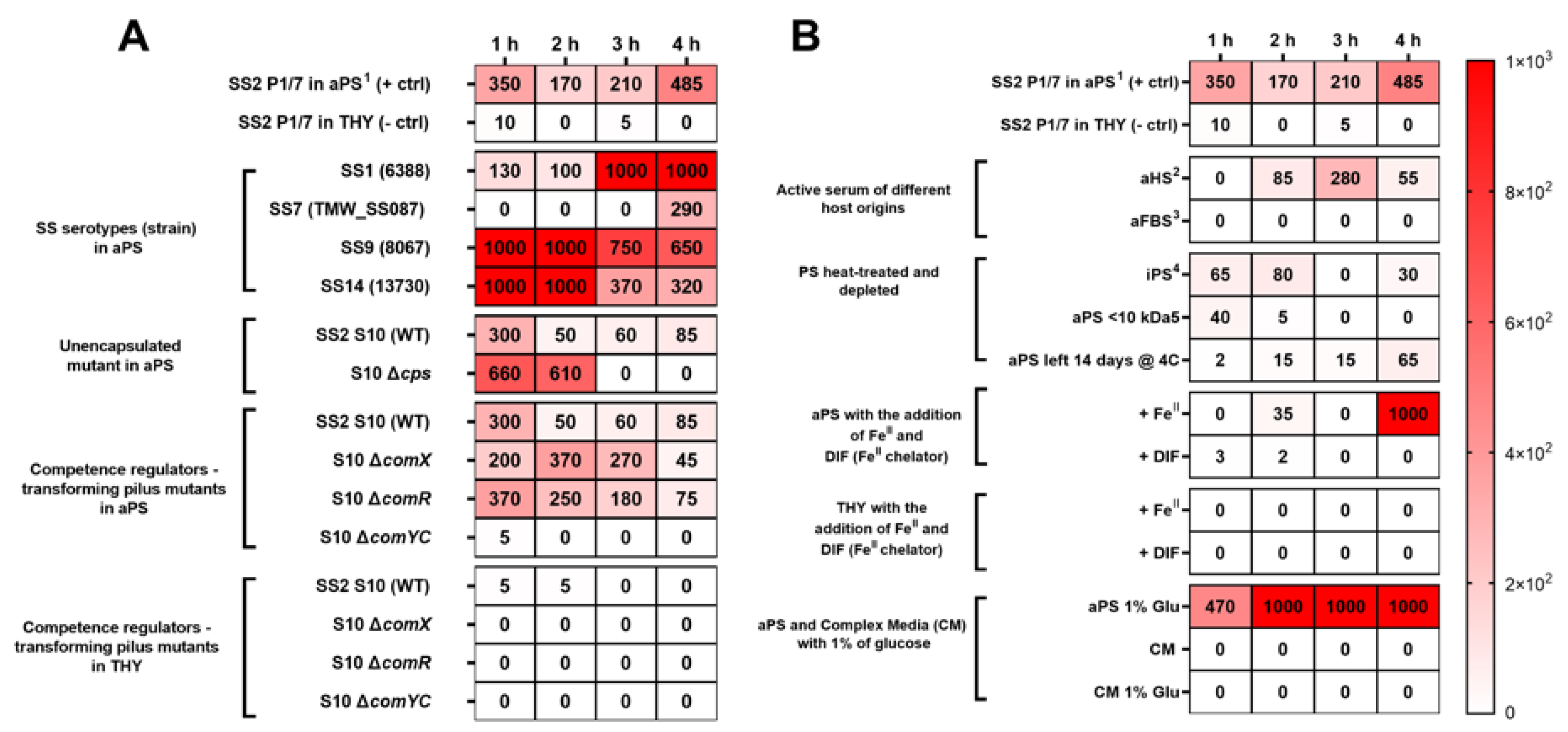
3. Results and Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethical Considerations
References
- Blokesch, M. Natural competence for transformation. Curr. Biol. 2016, 26, R1126–R1130. [Google Scholar] [CrossRef] [PubMed]
- Marks, L.R.; Reddinger, R.M.; Hakansson, A.P. High Levels of Genetic Recombination during Nasopharyngeal Carriage and Biofilm Formation in Streptococcus pneumoniae. MBio 2012, 3, 276–301. [Google Scholar] [CrossRef] [PubMed]
- Ochman, H.; Lawrence, J.G.; Groisman, E.A. Lateral gene transfer and the nature of bacterial innovation. Nature 2000, 405, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, L.; Boutry, C.; de Frahan, M.H.; Delplace, B.; Fremaux, C.; Horvath, P.; Boyaval, P.; Hols, P. A Novel Pheromone Quorum-Sensing System Controls the Development of Natural Competence in Streptococcus thermophilus and Streptococcus salivarius. J. Bacteriol. 2010, 192, 1444–1454. [Google Scholar] [CrossRef]
- Claverys, J.-P.; Prudhomme, M.; Martin, B. Induction of Competence Regulons as a General Response to Stress in Gram-Positive Bacteria. Annu. Rev. Microbiol. 2006, 60, 451–475. [Google Scholar] [CrossRef]
- Fontaine, L.; Wahl, A.; Fléchard, M.; Mignolet, J.; Hols, P. Regulation of competence for natural transformation in streptococci. Infect. Genet. Evol. 2015, 33, 343–360. [Google Scholar] [CrossRef]
- Bjedov, I. Stress-Induced Mutagenesis in Bacteria. Science 2003, 300, 1404–1409. [Google Scholar] [CrossRef]
- Goyette-Desjardins, G.; Auger, J.-P.; Xu, J.; Segura, M.; Gottschalk, M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent—an update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microbes Infect. 2014, 3, e45. [Google Scholar] [CrossRef]
- Ferrando, M.L.; de Greeff, A.; van Rooijen, W.J.M.; Stockhofe-Zurwieden, N.; Nielsen, J.; Wichgers Schreur, P.J.; Pannekoek, Y.; Heuvelink, A.; van der Ende, A.; Smith, H.; et al. Host-pathogen Interaction at the Intestinal Mucosa Correlates with Zoonotic Potential of Streptococcus suis. J. Infect. Dis. 2015, 212, 95–105. [Google Scholar] [CrossRef]
- Holden, M.T.G.; Hauser, H.; Sanders, M.; Ngo, T.H.; Cherevach, I.; Cronin, A.; Goodhead, I.; Mungall, K.; Quail, M.A.; Price, C.; et al. Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS ONE 2009, 4, e6072. [Google Scholar] [CrossRef]
- Weinert, L.A.; Chaudhuri, R.R.; Wang, J.; Peters, S.E.; Corander, J.; Jombart, T.; Baig, A.; Howell, K.J.; Vehkala, M.; Välimäki, N.; et al. Genomic signatures of human and animal disease in the zoonotic pathogen Streptococcus suis. Nat. Commun. 2015, 6, 6740. [Google Scholar] [CrossRef] [PubMed]
- Zaccaria, E.; van Baarlen, P.; de Greeff, A.; Morrison, D.A.; Smith, H.; Wells, J.M. Control of Competence for DNA Transformation in Streptococcus suis by Genetically Transferable Pherotypes. PLoS ONE 2014, 9, e99394. [Google Scholar] [CrossRef] [PubMed]
- Berends, E.T.M.; Kuipers, A.; Ravesloot, M.M.; Urbanus, R.T.; Rooijakkers, S.H.M. Bacteria under stress by complement and coagulation. FEMS Microbiol. Rev. 2014, 38, 1146–1171. [Google Scholar] [CrossRef] [PubMed]
- The M.R. Sequencing Enabled with Kit, I.R.-Z.P. rRNA D. Microbial RNA Sequencing Enabled with the Illumina Ribo-ZeroTM Plus rRNA Depletion Kit. Available online: https://www.illumina.com/content/dam/illumina-marketing/documents/products/technotes/ribo-zero-plus-data-concordance-tech-note-1270-2020-003.pdf (accessed on 20 January 2021).
- Ferrando, M.L.; Fuentes, S.; De Greeff, A.; Smith, H.; Wells, J.M. ApuA, a multifunctional α-glucan-degrading enzyme of Streptococcus suis, mediates adhesion to porcine epithelium and mucus. Microbiology 2010, 156, 2818–2828. [Google Scholar] [CrossRef]
- Trappetti, C.; Potter, A.J.; Paton, A.W.; Oggioni, M.R.; Paton, J.C. LuxS Mediates Iron-Dependent Biofilm Formation, Competence, and Fratricide in Streptococcus pneumoniae. Infect. Immun. 2011, 79, 4550–4558. [Google Scholar] [CrossRef]
- Segura, M.; Fittipaldi, N.; Calzas, C.; Gottschalk, M. Critical Streptococcus suis Virulence Factors: Are They All Really Critical? Trends Microbiol. 2017, 25, 585–599. [Google Scholar] [CrossRef]
- Luke, N.R.; Howlett, A.J.; Shao, J.; Campagnari, A.A. Expression of Type IV Pili by Moraxella catarrhalis Is Essential for Natural Competence and Is Affected by Iron Limitation. Infect. Immun. 2004, 72, 6262–6270. [Google Scholar] [CrossRef]
- Huang, K.; Zhang, Q.; Song, Y.; Zhang, Z.; Zhang, A.; Xiao, J.; Jin, M. Characterization of spectinomycin resistance in Streptococcus suis Leads to two novel insights into drug resistance formation and dissemination mechanism. Antimicrob. Agents Chemother. 2016, 60, 6390–6392. [Google Scholar] [CrossRef]
- Zaccaria, E.; Wels, M.; van Baarlen, P.; Wells, J.M. Temporal Regulation of the Transformasome and Competence Development in Streptococcus suis. Front. Microbiol. 2016, 7, 1922. [Google Scholar] [CrossRef]
- Chaguza, C.; Cornick, J.E.; Everett, D.B. Mechanisms and impact of genetic recombination in the evolution of Streptococcus pneumoniae. Comput. Struct. Biotechnol. J. 2015, 13, 241–247. [Google Scholar] [CrossRef]
- van der Hee, B.; Madsen, O.; Vervoort, J.; Smidt, H.; Wells, J.M. Congruence of Transcription Programs in Adult Stem Cell-Derived Jejunum Organoids and Original Tissue During Long-Term Culture. Front. Cell Dev. Biol. 2020, 8, 375. [Google Scholar] [CrossRef] [PubMed]
- Benis, N.; Wells, J.M.; Smits, M.A.; Kar, S.K.; van der Hee, B.; dos Santos, V.A.P.M.; Suarez-Diez, M.; Schokker, D. High-level integration of murine intestinal transcriptomics data highlights the importance of the complement system in mucosal homeostasis. BMC Genomics 2019, 20, 1028. [Google Scholar] [CrossRef] [PubMed]
- Seele, J.; Beineke, A.; Hillermann, L.-M.; Jaschok-Kentner, B.; von Pawel-Rammingen, U.; Valentin-Weigand, P.; Baums, C.G. The immunoglobulin M-degrading enzyme of Streptococcus suis, Ide Ssuis, is involved in complement evasion. Vet. Res. 2015, 46, 45. [Google Scholar] [CrossRef] [PubMed]
- Vaillancourt, K.; Bonifait, L.; Grignon, L.; Frenette, M.; Gottschalk, M.; Grenier, D. Identification and characterization of a new cell surface protein possessing factor H-binding activity in the swine pathogen and zoonotic agent Streptococcus suis. J. Med. Microbiol. 2013, 62, 1073–1080. [Google Scholar] [CrossRef]
- Siegele, D.A. Universal Stress Proteins in Escherichia coli. J. Bacteriol. 2005, 187, 6253–6254. [Google Scholar] [CrossRef]
| SS2 P1/7 Locus Tag | Protein | Functions | aPS vs. THY (Log2 Fold Expression Change) |
|---|---|---|---|
| SSU0049 | ComR | Transcriptional activator of ComX | −2.42 |
| SSU0016 | ComX | Master transcriptional regulator of the transformasome | −0.59 |
| SSU0061 | CinA | DNA binding and homologous recombination | −0.31 |
| SSU0062 | RecA | −0.70 | |
| SSU0924 | RadC | −0.52 | |
| SSU1083 | CoiA | 0.66 | |
| SSU0126 | ComYA | Transformasome apparatus | 4.02 |
| SSU0127 | ComYB | 4.72 | |
| SSU0128 | ComYC | 5.48 | |
| SSU0129 | ComYD | 4.91 | |
| SSU0130 | ComYE | 3.05 | |
| SSU0131 | ComYF | 4.20 | |
| SSU0132 | ComYG | 4.26 | |
| SSU0133 | ComYH | −2.51 | |
| SSU0144 | SsbB | ssDNA binding and protection | −1.34 |
| SSU0610 | ComEA | dsDNA receptor and channel | 2.07 |
| SSU0611 | ComEC | 2.33 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrando, M.L.; Gussak, A.; Mentink, S.; Gutierrez, M.F.; van Baarlen, P.; Wells, J.M. Active Human and Porcine Serum Induce Competence for Genetic Transformation in the Emerging Zoonotic Pathogen Streptococcus suis. Pathogens 2021, 10, 156. https://doi.org/10.3390/pathogens10020156
Ferrando ML, Gussak A, Mentink S, Gutierrez MF, van Baarlen P, Wells JM. Active Human and Porcine Serum Induce Competence for Genetic Transformation in the Emerging Zoonotic Pathogen Streptococcus suis. Pathogens. 2021; 10(2):156. https://doi.org/10.3390/pathogens10020156
Chicago/Turabian StyleFerrando, Maria Laura, Alex Gussak, Saskia Mentink, Marcela Fernandez Gutierrez, Peter van Baarlen, and Jerry Mark Wells. 2021. "Active Human and Porcine Serum Induce Competence for Genetic Transformation in the Emerging Zoonotic Pathogen Streptococcus suis" Pathogens 10, no. 2: 156. https://doi.org/10.3390/pathogens10020156
APA StyleFerrando, M. L., Gussak, A., Mentink, S., Gutierrez, M. F., van Baarlen, P., & Wells, J. M. (2021). Active Human and Porcine Serum Induce Competence for Genetic Transformation in the Emerging Zoonotic Pathogen Streptococcus suis. Pathogens, 10(2), 156. https://doi.org/10.3390/pathogens10020156





