Prevalence and Phylogenetic Analysis of Microsporidium Enterocytozoon bieneusi in Diarrheal Patients
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Sample Collection
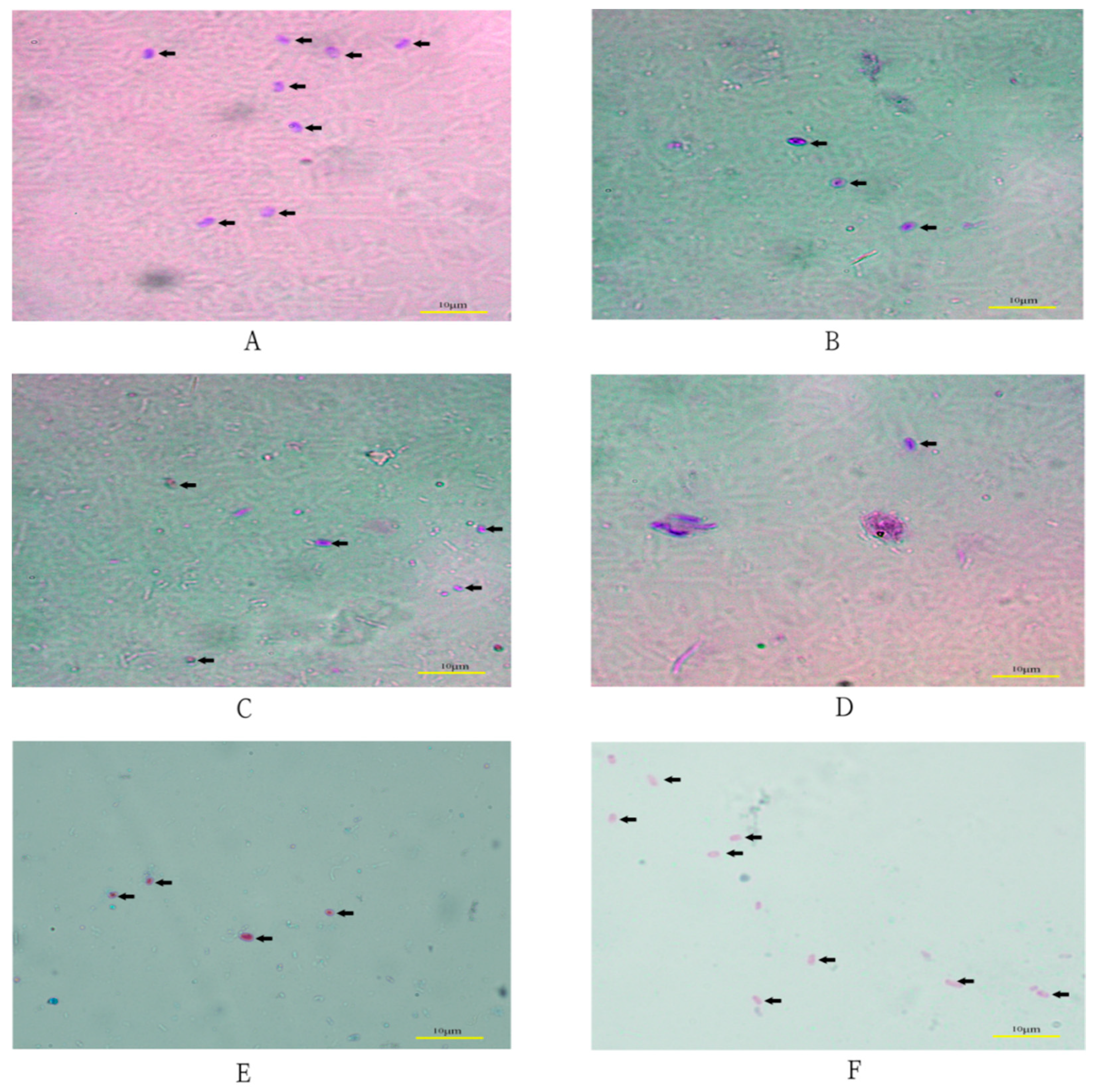
4.3. Microscopic Examination
4.4. DNA Extraction and Separation
4.5. PCR Amplification
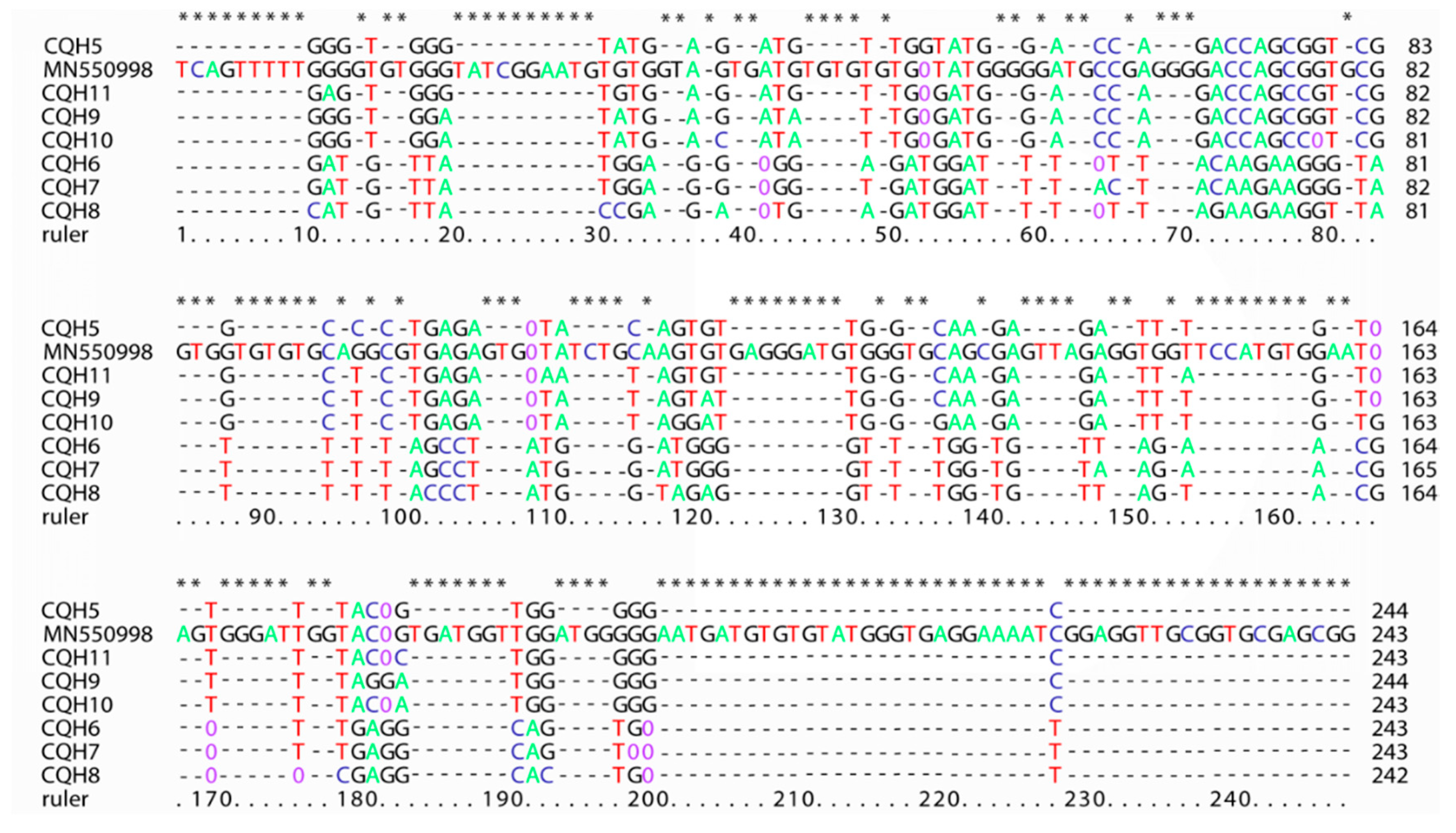
4.6. Sequences Difference and Phylogenetic Analysis
4.7. Statistical Analysis
4.8. Nucleotide Sequence Accession Numbers
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Santín, M.; Fayer, R. Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res. Vet. Sci. 2011, 90, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Ramanan, P.; Pritt, B.S. Extraintestinal microsporidiosis. J. Clin. Microbiol. 2014, 52, 3839–3844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghoyounchi, R.; Ahmadpour, E.; Spotin, A.; Mahami-Oskouei, M.; Rezamand, A.; Aminisani, N.; Ghojazadeh, M.; Berahmat, R.; Mikaeili-Galeh, T. Microsporidiosis in Iran: A systematic review and meta-analysis. Asian Pac. J. Trop. Med. 2017, 10, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Mathis, A.; Weber, R.; Deplazes, P. Reviews: July 2005, Volume 18, Issue 3 Author’s Correction: Zoonotic Potential of the Microsporidia. Clin. Microbiol. Rev. 2005, 18, 423–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwak, D.; Seo, M.G. Genetic analysis of zoonotic gastrointestinal protozoa and microsporidia in shelter cats in South Korea. Pathogens 2020, 9, 894. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.S.; Wang, R.J.; Fan, X.C.; Liu, T.L.; Zhang, L.X.; Zhao, G.H. Prevalence and genotypes of Enterocytozoon bieneusi in China. Acta Trop. 2018, 183, 142–152. [Google Scholar] [CrossRef]
- Khanduja, S.; Ghoshal, U.; Agarwal, V.; Pant, P.; Ghoshal, U.C. Identification and genotyping of Enterocytozoon bieneusi amonghuman immunodeficiency virus infected patients. J. Infect. Public Health 2017, 10, 31–40. [Google Scholar] [CrossRef] [Green Version]
- Desportes, I.; Le Charpentier, Y.; Galian, A.; Bernard, F.; Cochand-Priollet, B.; Lavergne, A.; Ravisse, P.; Modigliani, R. Occurrence of a new microsporidan: Enterocytozoon bieneusi n. g., n. sp., in the Enterocytes of human patient with AIDS. J. Eukaryot. Microbiol. 1985, 32, 250–254. [Google Scholar]
- Zhang, X.; Wang, Z.; Su, Y.; Liang, X.; Sun, X.; Peng, S.; Lu, H.; Jiang, N.; Yin, J.; Xiang, M.; et al. Identification and genotyping of Enterocytozoon bieneusi in China. J. Clin. Microbiol. 2011, 49, 2006–2008. [Google Scholar] [CrossRef] [Green Version]
- Santín, M.; Fayer, R. Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: A consensus. J. Eukaryot. Microbiol. 2009, 56, 34–38. [Google Scholar] [CrossRef]
- Qiu, L.; Xia, W.; Li, W.; Ping, J.; Ding, S.; Liu, H. The prevalence of microsporidia in China: A systematic review and meta-analysis. Sci. Rep. 2019, 9, 3174. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, H.; Zhao, X.; Zhang, L.; Zhang, G.; Guo, M.; Liu, L.; Feng, Y.; Xiao, L. Zoonotic cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. J. Clin. Microbiol. 2013, 51, 557–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.; Li, D.; Chang, Y.; Wu, Y.; Guo, Z.; Jia, L.; Xu, J.; Li, J.; Qi, M.; Wang, R.; et al. Molecular characterization of three intestinal protozoans in hospitalized children with different disease backgrounds in Zhengzhou, central China. Parasites Vectors 2019, 12, 543. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Yu, F.; Zhao, A.; Zhang, Y.; Wei, Z.; Li, D.; Zhang, L. Unusual dominant genotype NIA1 of Enterocytozoon bieneusi in children in Southern Xinjiang, China. PLoS Negl. Trop. Dis. 2020, 14, e0008293. [Google Scholar] [CrossRef]
- Yang, J.; Song, M.; Wan, Q.; Li, Y.; Lu, Y.; Jiang, Y.; Tao, W.; Li, W. Enterocytozoon bieneusi genotypes in children in northeast China and assessment of risk of zoonotic transmission. J. Clin. Microbiol. 2014, 52, 4363–4367. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Fan, Y.; Koehler, A.V.; Ma, G.; Li, T.; Hu, M.; Gasser, R.B. First survey of Cryptosporidium, Giardia and Enterocytozoon in diarrhoeic children from Wuhan, China. Infect. Genet. Evol. 2017, 51, 127–131. [Google Scholar] [CrossRef]
- Liu, H.; Shen, Y.; Yin, J.; Yuan, Z.; Jiang, Y.; Xu, Y.; Pan, W.; Hu, Y.; Cao, J. Prevalence and genetic characterization of Cryptosporidium, Enterocytozoon, Giardia and Cyclospora in diarrheal outpatients in china. BMC Infect. Dis. 2014, 14, 25. [Google Scholar] [CrossRef] [Green Version]
- Ding, S.; Huang, W.; Qin, Q.; Tang, J.; Liu, H. Genotype Identification and Phylogenetic Analysis of Enterocytozoon bieneusi Isolates from Stool Samples of Diarrheic Children. J. Parasitol. 2018, 104, 297–301. [Google Scholar] [CrossRef]
- Liu, H.; Jiang, Z.; Yuan, Z.; Yin, J.; Wang, Z.; Yu, B.; Zhou, D.; Shen, Y.; Cao, J. Infection by and genotype characteristics of Enterocytozoon bieneusi in HIV/AIDS patients from Guangxi Zhuang autonomous region, China. BMC Infect. Dis. 2017, 17, 1–8. [Google Scholar] [CrossRef]
- Didier, E.S.; Weiss, L.M. Microsporidiosis: Not just in AIDS patients. Curr. Opin. Infect. Dis. 2011, 24, 490–495. [Google Scholar] [CrossRef]
- Zhang, W.; Ren, G.; Zhao, W.; Yang, Z.; Shen, Y.; Sun, Y.; Liu, A.; Cao, J. Genotyping of Enterocytozoon bieneusi and subtyping of Blastocystis in cancer patients: Relationship to diarrhea and assessment of zoonotic transmission. Front. Microbiol. 2017, 8, 1835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prasertbun, R.; Mori, H.; Sukthana, Y.; Popruk, S.; Kusolsuk, T.; Hagiwara, K.; Mahittikorn, A. Enterocytozoon bieneusi and Cryptosporidium: A cross-sectional study conducted throughout Thailand. BMC Infect. Dis. 2019, 19, 808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henriques-Gil, N.; Haro, M.; Izquierdo, F.; Fenoy, S.; del Aguila, C. Phylogenetic approach to the variability of the microsporidian Enterocytozoon bieneusi and its implications for inter- and intrahost transmission. Appl. Environ. Microbiol. 2010, 76, 3333–3342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palareti, G.; Legnani, C.; Cosmi, B.; Antonucci, E.; Erba, N.; Poli, D.; Testa, S.; Tosetto, A. Comparison between different D-Dimer cutoff values to assess the individual risk of recurrent venous thromboembolism: Analysis of results obtained in the DULCIS study. Int. J. Lab. Hematol. 2016, 38, 42–49. [Google Scholar] [CrossRef]
- Dengjel, B.M.; Zahler, M.; Hermanns, W.; Heinritzi, K.; Spillmann, T.; Thomschke, A.; Loscher, T.; Gothe, R.; Rinder, H. Zoonotic Potential of Enterocytozoon bieneusi. J. Clin. Microbiol. 2001, 39, 4495–4499. [Google Scholar] [CrossRef] [Green Version]
- Reetz, J.; Rinder, H.; Thomschke, A.; Manke, H.; Schwebs, M.; Bruderek, A. First detection of the microsporidium Enterocytozoon bieneusi in non-mammalian hosts (chickens). Int. J. Parasitol. 2002, 32, 785–787. [Google Scholar] [CrossRef]
- Sulaiman, I.M.; Bern, C.; Gilman, R.; Cama, V.; Kawai, V.; Vargas, D.; Ticona, E.; Vivar, A.; Xiao, L. A Molecular Biologic Study of Enterocytozoon bieneusi in HIV-Infected Patients in Lima, Peru. J. Eukaryot. Microbiol. 2003, 50, 591–596. [Google Scholar] [CrossRef]
- Sulaiman, I.M.; Fayer, R.; Yang, C.; Santin, M.; Matos, O.; Xiao, L. Molecular characterization of Enterocytozoon bieneusi in cattle indicates that only some isolates have zoonotic potential. Parasitol. Res. 2004, 92, 328–334. [Google Scholar] [CrossRef]
- Santín, M.; Trout, J.M.; Fayer, R. Enterocytozoon bieneusi genotypes in dairy cattle in the eastern United States. Parasitol. Res. 2005, 97, 535–538. [Google Scholar] [CrossRef]
- Sulaiman, I.M.; Fayer, R.; Lal, A.A.; Trout, J.M.; Schaefer, F.W.; Xiao, L. Molecular characterization of microsporidia indicates that wild mammals harbor host-adapted Enterocytozoon spp. as well as human-pathogenic Enterocytozoon bieneusi. Appl. Environ. Microbiol. 2003, 69, 4495–4501. [Google Scholar] [CrossRef] [Green Version]
- Yakoob, J.; Abbas, Z.; Beg, M.A.; Jafri, W.; Naz, S.; Khalid, A.; Khan, R. Microsporidial infections due to Encephalitozoon intestinalis in non-HIV-infected patients with chronic diarrhoea. Epidemiol. Infect. 2012, 140, 1773–1779. [Google Scholar] [CrossRef] [PubMed]
- Karim, M.R.; Dong, H.; Li, T.; Yu, F.; Li, D.; Zhang, L.; Li, J.; Wang, R.; Li, S.; Li, X.; et al. Predomination and new genotypes of Enterocytozoon bieneusi in captive nonhuman primates in zoos in China: High genetic diversity and zoonotic significance. PLoS ONE 2015, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; Jiang, J.; Cai, Y.N.; Wang, C.F.; Xu, P.; Yang, G.L.; Zhao, Q. Molecular characterization of Enterocytozoon bieneusi in domestic rabbits (Oryctolagus cuniculus) in northeastern China. Korean J. Parasitol. 2016, 54, 81–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Lin, Y.; Li, Q.; Zhang, S.; Wei, T.; Wan, Q.; Jiang, Y.; Li, W. Widespread presence of human-pathogenic Enterocytozoon bieneusi genotype D in farmed foxes (Vulpes vulpes) and raccoon dogs (Nyctereutes procyonoides) in China: First identification and zoonotic concern. Parasitol. Res. 2015, 114, 4341–4348. [Google Scholar] [CrossRef]
- Amer, S.; Kim, S.; Han, J.I.; Na, K.J. Prevalence and genotypes of Enterocytozoon bieneusi in wildlife in Korea: A public health concern. Parasites Vectors 2019, 12, 1–7. [Google Scholar] [CrossRef]
- Yue, D.M.; Ma, J.G.; Li, F.C.; Hou, J.L.; Zheng, W.B.; Quan, Z.; Zhang, X.X.; Zhu, X.Q. Occurrence of Enterocytozoon bieneusi in donkeys (Equus asinus) in China: A public health concern. Front. Microbiol. 2017, 8, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Deng, L.; Li, W.; Yu, X.; Gong, C.; Liu, X.; Zhong, Z.; Xie, N.; Lei, S.; Yu, J.; Fu, H.; et al. Correction: First report of the human-pathogenic Enterocytozoon bieneusi from red-bellied tree squirrels (Callosciurus erythraeus) in Sichuan, China. PLoS ONE 2016, 11, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Cama, V.; Feng, Y.; Gilman, R.H.; Bern, C.; Zhang, X.; Xiao, L. Population genetic analysis of Enterocytozoon bieneusi in humans. Int. J. Parasitol. 2012, 42, 287–293. [Google Scholar] [CrossRef]
- Didier, E.S.; Orenstein, J.M.; Aldras, A.; Bertucci, D.; Rogers, L.B.; Janney, F.A. Comparison of three staining methods for detecting microsporidia in fluids. J. Clin. Microbiol. 1995, 33, 3138–3145. [Google Scholar] [CrossRef] [Green Version]
- Hawash, Y. DNA extraction from protozoan oocysts/cysts in feces for diagnostic PCR. Korean J. Parasitol. 2014, 52, 263–271. [Google Scholar] [CrossRef]
- Mirsepasi, H.; Persson, S.; Struve, C.; Andersen, L.O.B.; Petersen, A.M.; Krogfelt, K.A. Microbial diversity in fecal samples depends on DNA extraction method: EasyMag DNA extraction compared to QIAamp DNA stool mini kit extraction. BMC Res. Notes 2014, 7, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menu, E.; Mary, C.; Toga, I.; Raoult, D.; Ranque, S.; Bittar, F. Evaluation of two DNA extraction methods for the PCR-based detection of eukaryotic enteric pathogens in fecal samples. BMC Res. Notes 2018, 11, 4–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buckholt, M.A.; Lee, J.H.; Tzipori, S. Prevalence of Enterocytozoon bieneusi in swine: An 18-month survey at a slaughterhouse in Massachusetts. Appl. Environ. Microbiol. 2002, 68, 2595–2599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]

| Locations | Chongqing | Suizhou, Hubei | Qingzhou, Shandong | |||
|---|---|---|---|---|---|---|
| Adult | Children | Adult | Adult | Children | ||
| Number of samples | 68 | 64 | 20 | 17 | 27 | |
| Positive a (%) | microscope | 5 (7.35%) | 7 (10.94%) | 2 (10%) | 1 (5.88%) | 4 (14.81%) |
| PCR | 5 (7.35%) | 9 (14.06%) | 1 (5%) | 0 | 5 (18.52%) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zang, M.; Li, J.; Tang, C.; Ding, S.; Huang, W.; Qin, Q.; Liu, H. Prevalence and Phylogenetic Analysis of Microsporidium Enterocytozoon bieneusi in Diarrheal Patients. Pathogens 2021, 10, 128. https://doi.org/10.3390/pathogens10020128
Zang M, Li J, Tang C, Ding S, Huang W, Qin Q, Liu H. Prevalence and Phylogenetic Analysis of Microsporidium Enterocytozoon bieneusi in Diarrheal Patients. Pathogens. 2021; 10(2):128. https://doi.org/10.3390/pathogens10020128
Chicago/Turabian StyleZang, Manman, Jinjin Li, Chun Tang, Songtao Ding, Wei Huang, Qizhong Qin, and Handeng Liu. 2021. "Prevalence and Phylogenetic Analysis of Microsporidium Enterocytozoon bieneusi in Diarrheal Patients" Pathogens 10, no. 2: 128. https://doi.org/10.3390/pathogens10020128





