Transcriptome-Based Identification of a Functional Fasciola hepatica Carboxylesterase B
Abstract
:1. Introduction
2. Results
2.1. Carboxylesterase Enzymatic Assays and Zymograms
2.2. F. hepatica Transcriptome
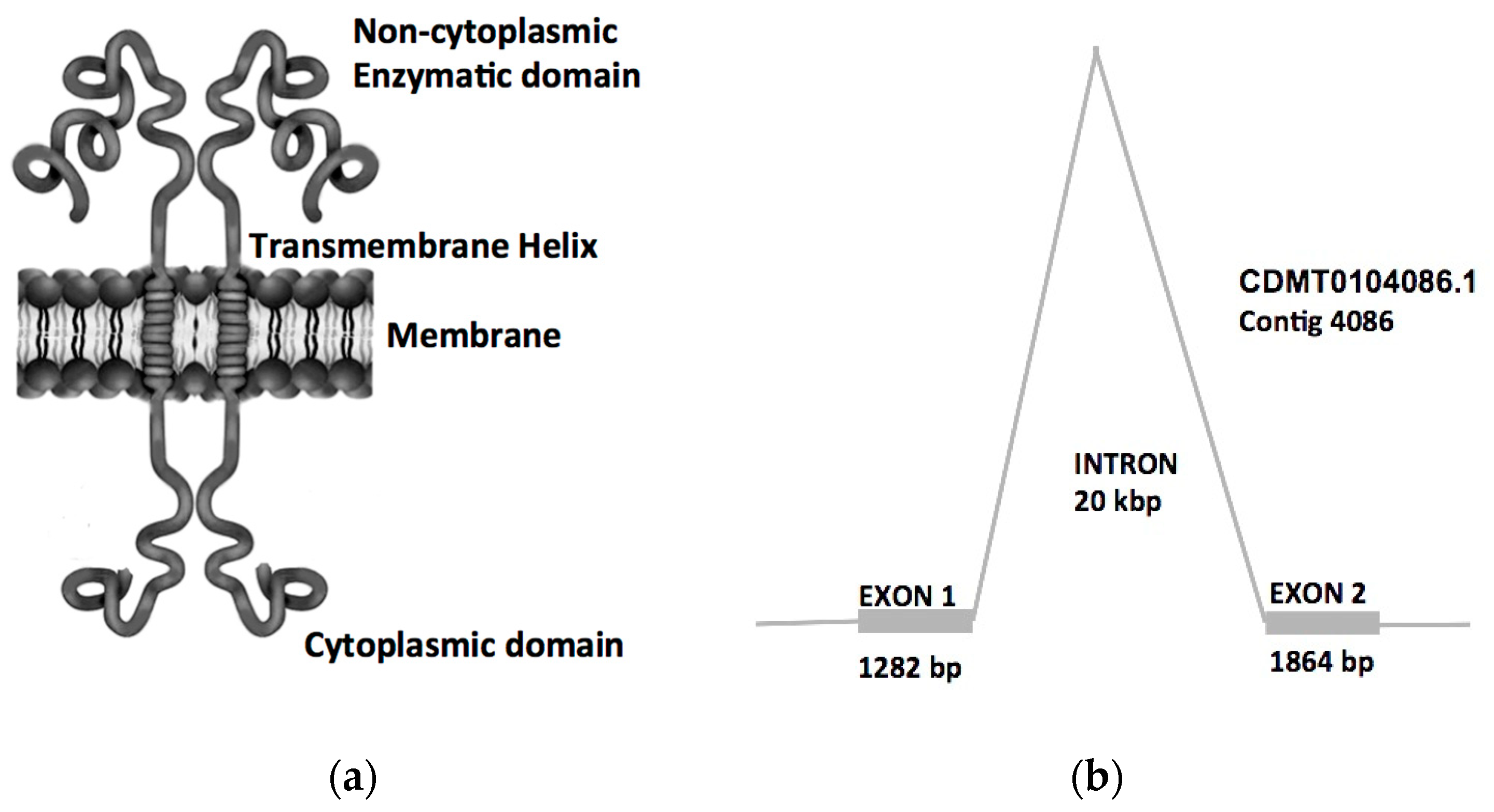
2.3. Bioinformatics Analysis
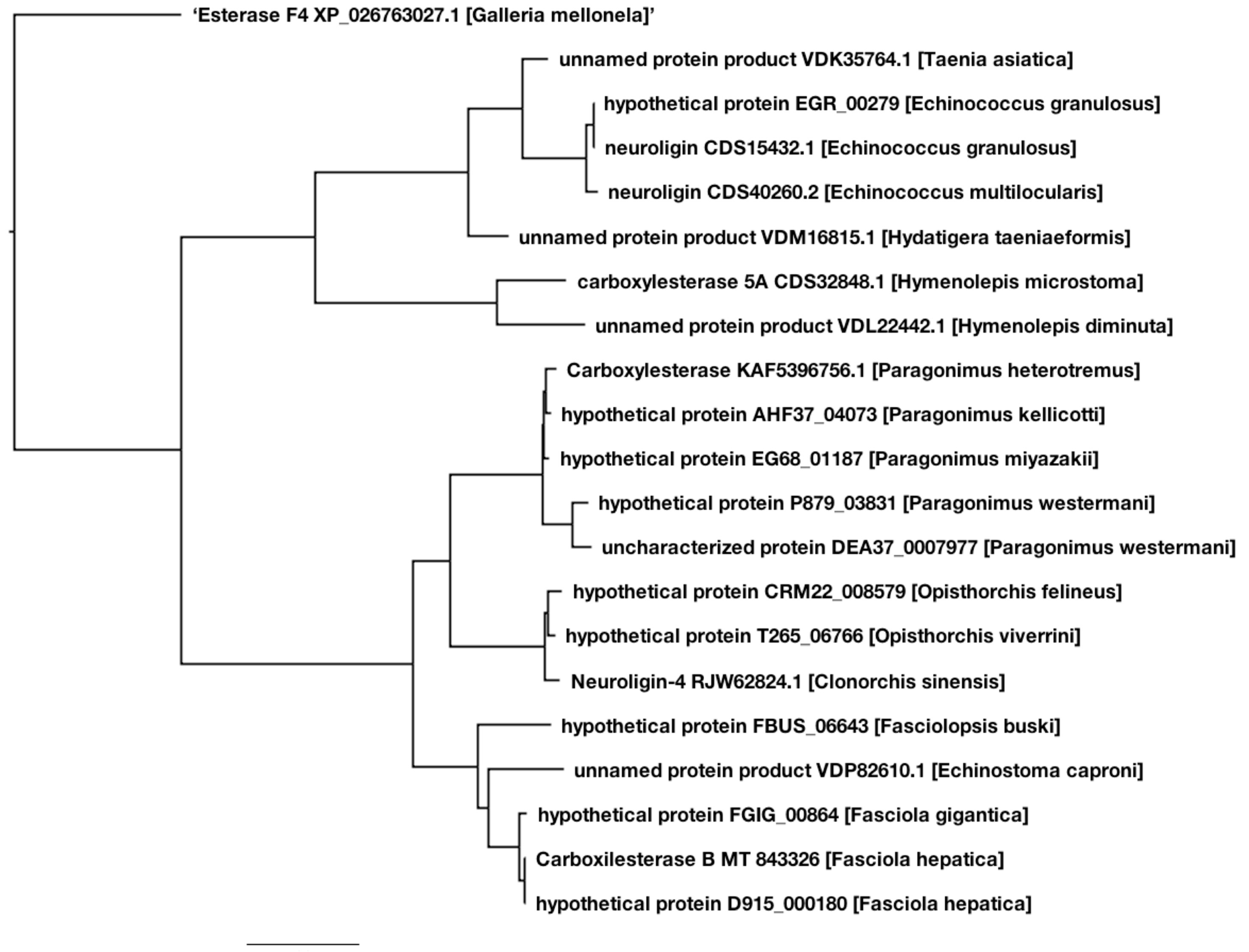
2.4. Phylogenetic Analysis
3. Discussion
4. Materials and Methods
4.1. Animals and the Parasite Strain
4.2. Enzyme Analysis
4.3. High throughput mRNA Sequencing
4.4. Phylogenetic Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Zumaquero-Ríos, J.L.; Sarracent-Pérez, J.; Rojas-García, R.; Rojas-Rivero, L.; Martínez-Tovilla, Y.; Valero, M.A.; Mas-Coma, S. Fascioliasis and Intestinal Parasitoses Affecting Schoolchildren in Atlixco, Puebla State, Mexico: Epidemiology and Treatment with Nitazoxanide. PLoS Negl. Trop. Dis. 2013, 7, e2553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bargues, M.D.; Artigas, P.; Khoubbane, M.; Mas-Coma, S. DNA Sequence Characterisation and Phylogeography of Lymnaea Cousini and Related Species, Vectors of Fascioliasis in Northern Andean Countries, with Description of L. meridensis n. Sp. (Gastropoda: Lymnaeidae). Parasites Vectors 2011, 4, 132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miranda-Miranda, E.; Cossio-Bayugar, R.; Aguilar Díaz, H.; Narváez Padilla, V.; Sachman-Ruiz, B.; Reynaud, E. Transcriptome Assembly Dataset of Anthelmintic Response in Fasciola hepatica. Data Brief 2021, 35, 106808. [Google Scholar] [CrossRef] [PubMed]
- Boyko, K.M.; Kryukova, M.V.; Petrovskaya, L.E.; Nikolaeva, A.Y.; Korzhenevsky, D.A.; Novototskaya-Vlasova, K.A.; Rivkina, E.M.; Dolgikh, D.A.; Kirpichnikov, M.P.; Popov, V.O. Crystal Structure of PMGL2 Esterase from the Hormone-Sensitive Lipase Family with GCSAG Motif around the Catalytic Serine. PLoS ONE 2020, 15, e0226838. [Google Scholar] [CrossRef]
- Boyko, K.M.; Kryukova, M.V.; Petrovskaya, L.E.; Kryukova, E.A.; Nikolaeva, A.Y.; Korzhenevsky, D.A.; Lomakina, G.Y.; Novototskaya-Vlasova, K.A.; Rivkina, E.M.; Dolgikh, D.A.; et al. Structural and Biochemical Characterization of a Cold-Active PMGL3 Esterase with Unusual Oligomeric Structure. Biomolecules 2021, 11, 57. [Google Scholar] [CrossRef] [PubMed]
- Scarcella, S.; Solana, M.V.; Fernandez, V.; Lamenza, P.; Ceballos, L.; Solana, H. Increase of Gluthatione S-Transferase, Carboxyl Esterase and Carbonyl Reductase in Fasciola hepatica Recovered from Triclabendazole Treated Sheep. Mol. Biochem. Parasitol. 2013, 191, 63–65. [Google Scholar] [CrossRef] [Green Version]
- Matoušková, P.; Vokřál, I.; Lamka, J.; Skálová, L. The Role of Xenobiotic-Metabolizing Enzymes in Anthelmintic Deactivation and Resistance in Helminths. Trends Parasitol. 2016, 32, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, M. Structure and Catalytic Properties of Carboxylesterase Isozymes Involved in Metabolic Activation of Prodrugs. Molecules 2008, 13, 412–431. [Google Scholar] [CrossRef] [Green Version]
- Parkinson, A. Biotransformation of xenobiotics. In Cassarett and Doulls Toxicology: The Basic Science of Poisons; McGraw-Hill Medical: New York, NY, USA, 1996; pp. 113–196. [Google Scholar]
- Kontogiannatos, D.; Swevers, L.; Maenaka, K.; Park, E.Y.; Iatrou, K.; Kourti, A. Functional Characterization of a Juvenile Hormone Esterase Related Gene in the Moth Sesamia nonagrioides through RNA Interference. PLoS ONE 2013, 8, e73834. [Google Scholar] [CrossRef] [Green Version]
- Istvan, E.S.; Mallari, J.P.; Corey, V.C.; Dharia, N.V.; Marshall, G.R.; Winzeler, E.A.; Goldberg, D.E. Esterase Mutation Is a Mechanism of Resistance to Antimalarial Compounds. Nat. Commun. 2017, 8, 14240. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.H.; Baptista, R.P.; Valenciano, A.L.; Zhou, B.; Kissinger, J.C.; Tumwebaze, P.K.; Rosenthal, P.J.; Cooper, R.A.; Yue, J.-M.; Cassera, M.B. Resistance to Some But Not Other Dimeric Lindenane Sesquiterpenoid Esters Is Mediated by Mutations in a Plasmodium falciparum Esterase. ACS Infect. Dis. 2020, 6, 2994–3003. [Google Scholar] [CrossRef] [PubMed]
- Dahan-Moss, Y.L.; Koekemoer, L.L. Analysis of Esterase Enzyme Activity in Adults of the Major Malaria Vector Anopheles funestus. Parasites Vectors 2016, 9, 110. [Google Scholar] [CrossRef] [Green Version]
- Schama, R.; Pedrini, N.; Juárez, M.P.; Nelson, D.R.; Torres, A.Q.; Valle, D.; Mesquita, R.D. Rhodnius Prolixus Supergene Families of Enzymes Potentially Associated with Insecticide Resistance. Insect Biochem. Mol. Biol. 2016, 69, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-L.; Huang, Y.; Lu, X.-P.; Jiang, X.-Z.; Smagghe, G.; Feng, Z.-J.; Yuan, G.-R.; Wei, D.; Wang, J.-J. Overexpression of Two α-Esterase Genes Mediates Metabolic Resistance to Malathion in the Oriental Fruit Fly, Bactrocera dorsalis (Hendel). Insect Mol. Biol. 2015, 24, 467–479. [Google Scholar] [CrossRef]
- Shin, D.; Smartt, C.T. Assessment of Esterase Gene Expression as a Risk Marker for Insecticide Resistance in Florida Culex nigripalpus (Diptera: Culicidae). J. Vector Ecol. 2016, 41, 63–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scarcella, S.; Miranda-Miranda, E.; Cossío-Bayúgar, R.; Ceballos, L.; Fernandez, V.; Solana, H. Increase of Carboxylesterase Activity in Fasciola hepatica Recovered from Triclabendazole Treated Sheep. Mol. Biochem. Parasitol. 2012, 185, 151–153. [Google Scholar] [CrossRef] [PubMed]
- Beesley, N.J.; Williams, D.J.L.; Paterson, S.; Hodgkinson, J. Fasciola hepatica Demonstrates High Levels of Genetic Diversity, a Lack of Population Structure and High Gene Flow: Possible Implications for Drug Resistance. Int. J. Parasitol. 2017, 47, 11–20. [Google Scholar] [CrossRef]
- Desper, R. Theoretical Foundation of the Balanced Minimum Evolution Method of Phylogenetic Inference and Its Relationship to Weighted Least-Squares Tree Fitting. Mol. Biol. Evol. 2003, 21, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Manchenko, G.P. Handbook of Detection of Enzymes on Electrophoretic Gels, 2nd ed.; CRC Press LLC: Boca Raton, FL, USA, 2003. [Google Scholar]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-Scale Gene Function Analysis with the PANTHER Classification System. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howe, K.L.; Bolt, B.J.; Cain, S.; Chan, J.; Chen, W.J.; Davis, P.; Done, J.; Down, T.; Gao, S.; Grove, C.; et al. WormBase 2016: Expanding to Enable Helminth Genomic Research. Nucleic Acids Res. 2016, 44, D774–D780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howe, K.L.; Bolt, B.J.; Shafie, M.; Kersey, P.; Berriman, M. WormBase ParaSite—A Comprehensive Resource for Helminth Genomics. Mol. Biochem. Parasitol. 2017, 215, 2–10. [Google Scholar] [CrossRef]
- Ericsson, C.; Franzén, B.; Nistér, M. Frozen Tissue Biobanks. Tissue Handling, Cryopreservation, Extraction, and Use for Proteomic Analysis. Acta Oncol. 2006, 45, 643–661. [Google Scholar] [CrossRef] [PubMed]
- Peterson, G.L. A Simplification of the Protein Assay Method of Lowry et al. Which Is More Generally Applicable. Anal. Biochem. 1977, 83, 346–356. [Google Scholar] [CrossRef]
- Bardi, L.; Dell’oro, V.; Delfini, C.; Marzona, M. A Rapid Spectrophotometric Method to Determine Esterase Activity of Yeast Cells in an Aqueous Medium. J. Inst. Brew. 1993, 99, 385–388. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001; ISBN 978-0-87969-577-4. [Google Scholar]
- Kelley, L.; Mezulis, S.; Yates, C.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 Web Portal for Protein Modeling, Prediction and Analysis | Nature Protocols. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boratyn, G.M.; Schäffer, A.A.; Agarwala, R.; Altschul, S.F.; Lipman, D.J.; Madden, T.L. Domain Enhanced Lookup Time Accelerated BLAST. Biol. Direct 2012, 7, 12. [Google Scholar] [CrossRef] [Green Version]

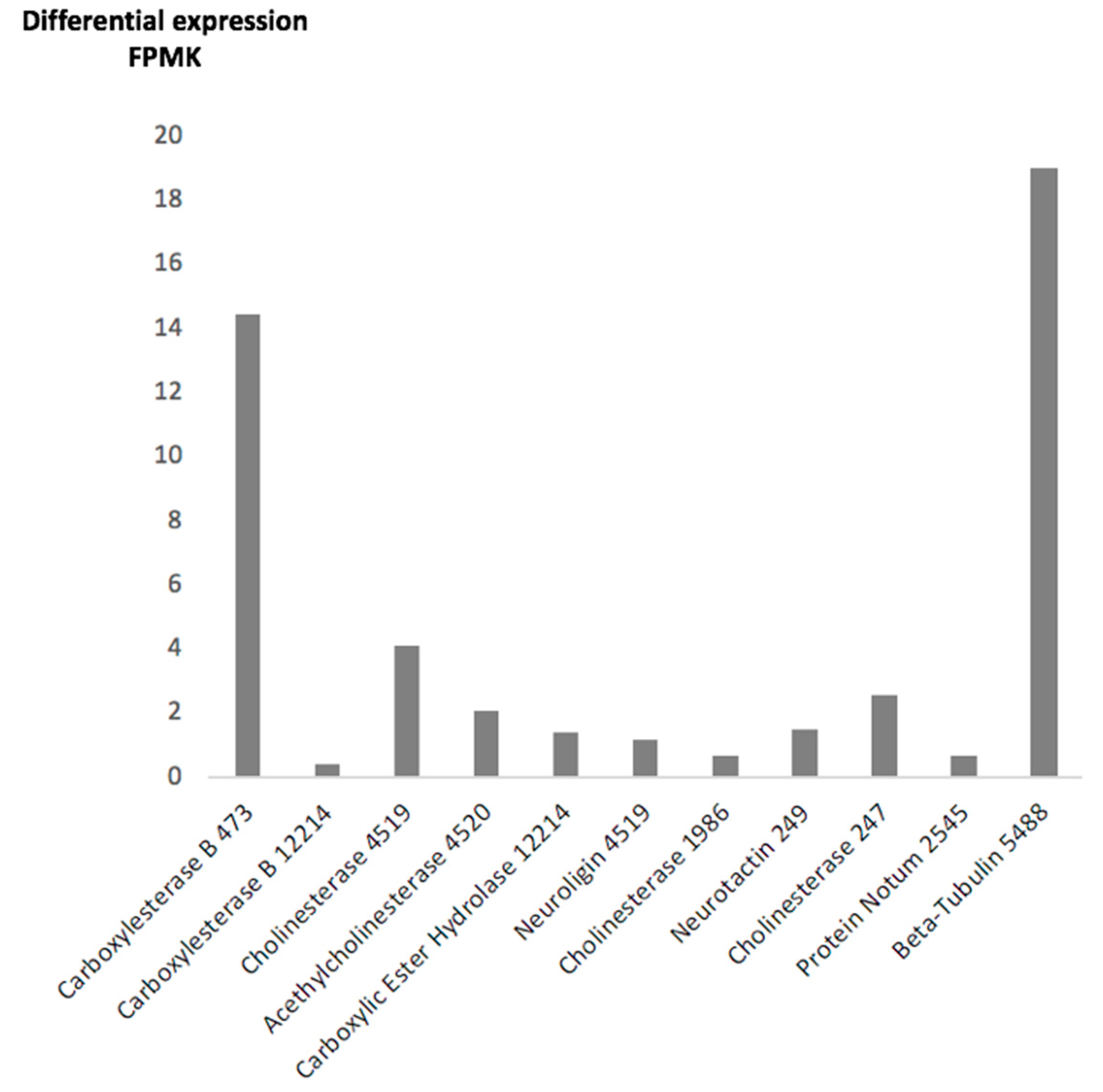
| Transcript SRR13076124 Spot No. | Size bp | AA | Molecular Mass | Expression FPKM | Description | Genome Identity UNIPROTKB/Description | Genome Identity GenBank/Description |
|---|---|---|---|---|---|---|---|
| 473 | 3146 | 735 | 83.5 | 14.45 | Carboxylesterase B | A0A4E0S0J7/Coesterase | D915-000180/Hypotetical |
| 12214 | 1837 | 424 | 25.41 | 0.42 | Carboxylesterase B | No match | KZ430433.1/ Juvenile hormone esterase |
| 4519 | 3482 | 697 | 78.8 | 4.11 | Cholinesterase | AOAA4E0RPG5/Neuroligin | THD27534.1/Cholinesterase |
| 4520 | 1816 | 383 | 44.0 | 2.1 | Acetylcholinesterase | AOAA4E0RPG/Carboxylic ester hydrolase | THD27534/Cholinesterase |
| 12214 | 1837 | 430 | 48.4 | 1.4 | Carboxylic Ester Hydrolase | A0A4E0S092 /Uncharacterized protein | D915_006009/Hypotetical |
| 4519 | 2094 | 697 | 78.8 | 1.2 | Neuroligin | D915_001711/Carboxylic ester hydrolase | THD27534/Cholinesterase |
| 1986 | 1424 | 502 | 56.9 | 0.7 | Cholinesterase | A0A2H1CBM8 Cholinesterase | PIS84875.1 /Neuroligin |
| 249 | 2085 | 694 | 79.7 | 1.5 | Neurotactin | A0A4E0RJZ9/Neurotactin | THD28606.1/Neurotactin |
| 247 | 1510 | 490 | 56.19 | 2.6 | Cholinesterase | A0A2H1CVT6/Cholinesterase | THD28606/Neurotactin |
| 2545 | 2304 | 606 | 69.2 | 0.7 | Uncharacterized | 0A2H1CVW9/Uncharacterized | THD28213.1/Protein Notum |
| No match | 1934 | 644 | 72.7 | No data | Cholinesterase | A0A2H1CPF2/Carboxylesterase | PIS89339/ Cholinesterase |
| No match | 1689 | 562 | 63.1 | No data | Carboxylesterase | No match | KZ429703.1/Carboxylesterase |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pedroza-Gómez, Y.J.; Cossio-Bayugar, R.; Aguilar-Díaz, H.; Scarcella, S.; Reynaud, E.; Sanchez-Carbente, M.d.R.; Narváez-Padilla, V.; Miranda-Miranda, E. Transcriptome-Based Identification of a Functional Fasciola hepatica Carboxylesterase B. Pathogens 2021, 10, 1454. https://doi.org/10.3390/pathogens10111454
Pedroza-Gómez YJ, Cossio-Bayugar R, Aguilar-Díaz H, Scarcella S, Reynaud E, Sanchez-Carbente MdR, Narváez-Padilla V, Miranda-Miranda E. Transcriptome-Based Identification of a Functional Fasciola hepatica Carboxylesterase B. Pathogens. 2021; 10(11):1454. https://doi.org/10.3390/pathogens10111454
Chicago/Turabian StylePedroza-Gómez, Yaretzi J., Raquel Cossio-Bayugar, Hugo Aguilar-Díaz, Silvana Scarcella, Enrique Reynaud, María del Rayo Sanchez-Carbente, Verónica Narváez-Padilla, and Estefan Miranda-Miranda. 2021. "Transcriptome-Based Identification of a Functional Fasciola hepatica Carboxylesterase B" Pathogens 10, no. 11: 1454. https://doi.org/10.3390/pathogens10111454
APA StylePedroza-Gómez, Y. J., Cossio-Bayugar, R., Aguilar-Díaz, H., Scarcella, S., Reynaud, E., Sanchez-Carbente, M. d. R., Narváez-Padilla, V., & Miranda-Miranda, E. (2021). Transcriptome-Based Identification of a Functional Fasciola hepatica Carboxylesterase B. Pathogens, 10(11), 1454. https://doi.org/10.3390/pathogens10111454








