Advances and Future Perspective on Detection Technology of Human Norovirus
Abstract
1. Introduction
2. Detection Techniques
2.1. Morphological Methods
2.2. Immunological Methods
2.3. Molecular Detection Methods
2.3.1. Isothermal Amplification
2.3.2. Thermal Cycling Amplification
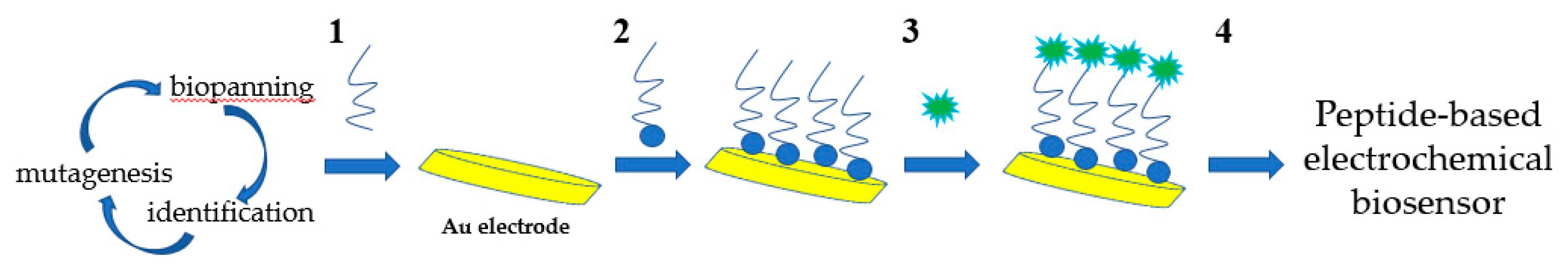
2.4. Biosensor
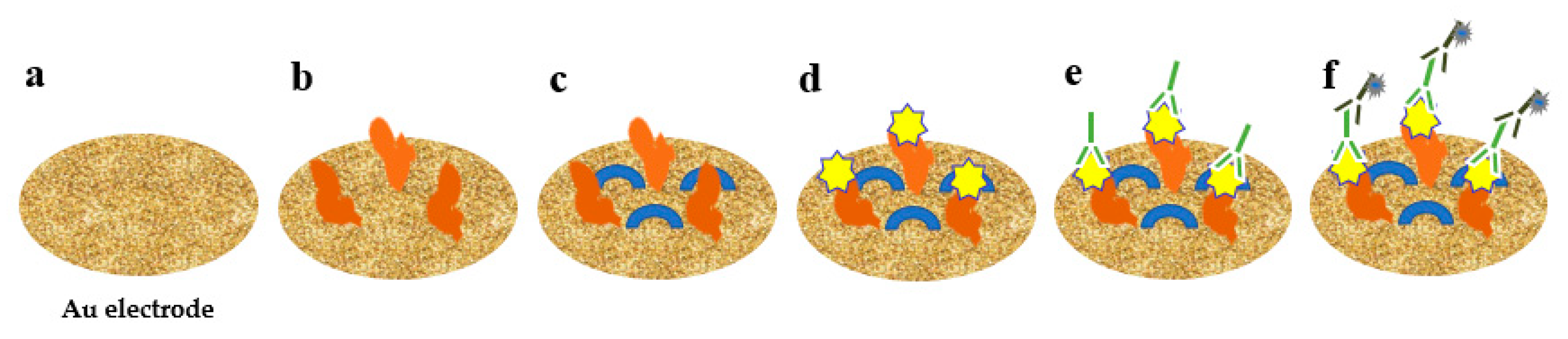
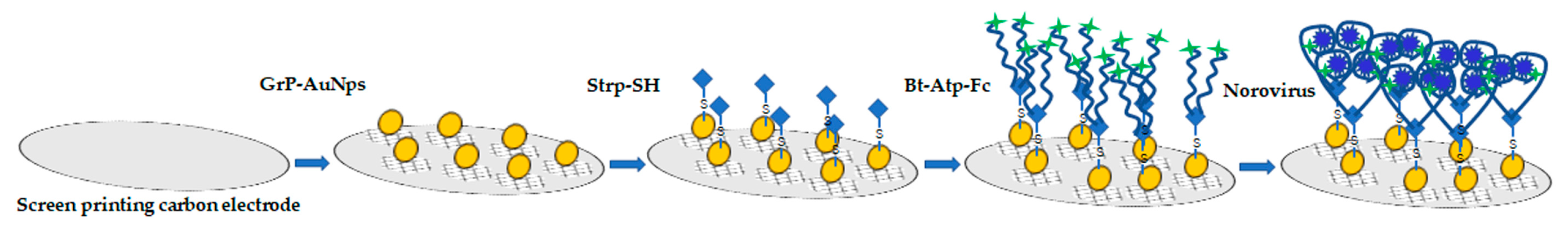
2.4.1. Electrochemical Biosensor
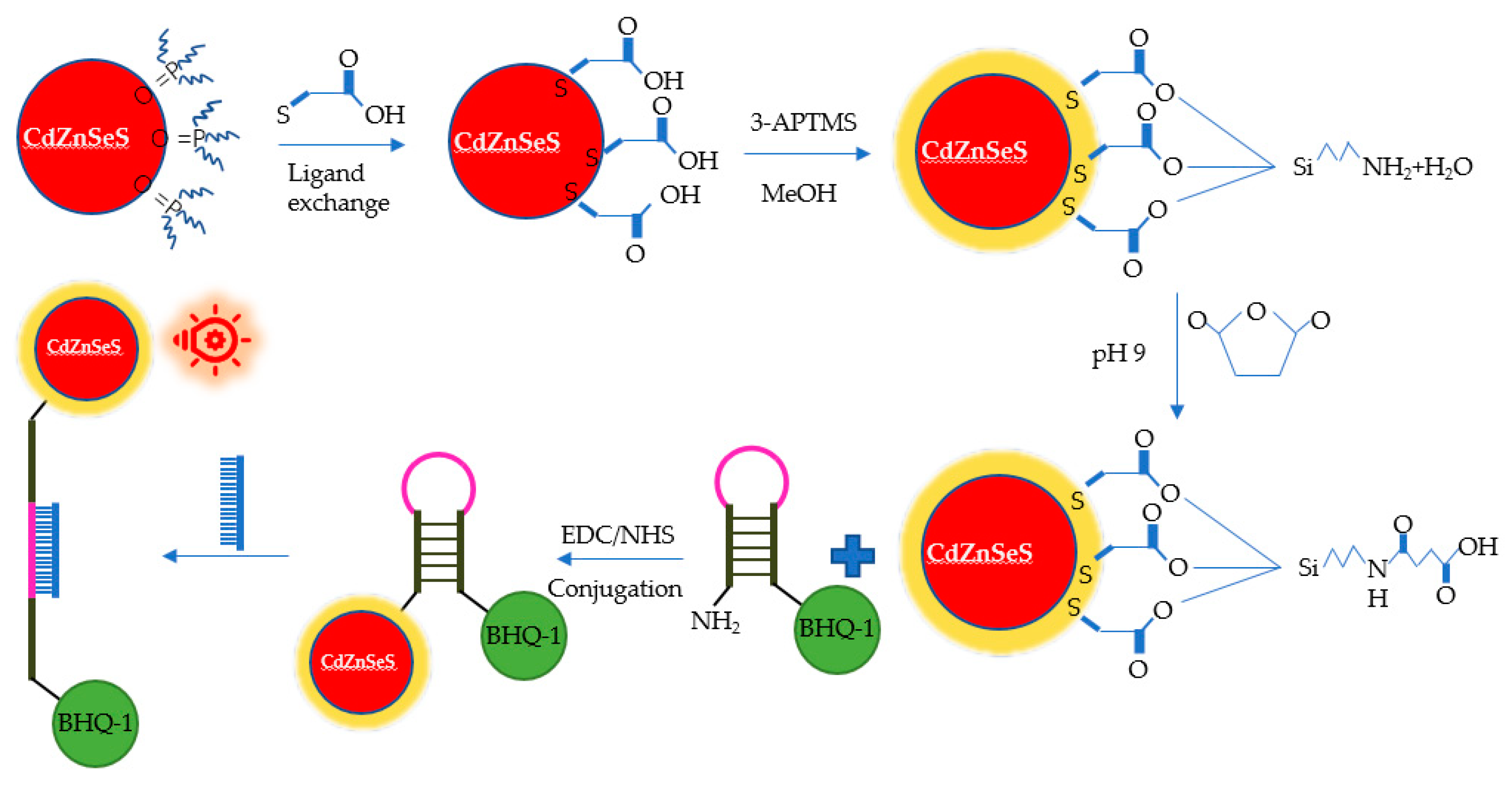
2.4.2. Optical Biosensor
3. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chhabra, P.; Graaf, M.D.; Parra, G.I.; Chan, C.W.; Vinjé, J. Updated classification of norovirus genogroups and genotypes. J. Gen. Virol. 2019, 100, 1393–1406. [Google Scholar] [CrossRef]
- Ahmed, S.M.; Hall, A.J.; Robinson, A.E.; Verhoef, L.; Premkumar, P.; Parashar, U.D.; Koopmans, M.; Lopman, B.A. Global prevalence of norovirus in cases of gastroenteritis: A systematic review and meta-analysis. Lancet Infect. Dis. 2014, 14, 725–730. [Google Scholar] [CrossRef]
- Bartsch, S.M.; Lopman, B.A.; Sachiko, O.; Hall, A.J.; Lee, B.Y.; Olson, D.R. Global economic burden of norovirus gastroenteritis. PLoS ONE 2016, 11, e0151219. [Google Scholar]
- Hemming, M.; Räsänen, S.; Huhti, L.; Paloniemi, M.; Salminen, M.; Vesikari, T. Major reduction of rotavirus, but not norovirus, gastroenteritis in children seen in hospital after the introduction of RotaTeq vaccine into the National Immunization Programme in Finland. Eur. J. Pediatr. 2013, 172, 739–746. [Google Scholar] [CrossRef]
- Hallowell, B.D.; Parashar, U.D.; Curns, A.; Degroote, N.P.; Tate, J.E. Trends in the laboratory detection of rotavirus before and after implementation of routine rotavirus vaccination-United States, 2000–2018. MMWR Morb. Mortal. Wkly. Rep. 2019, 68, 539–543. [Google Scholar] [CrossRef]
- Jonesteller, C.L.; Burnett, E.; Yen, C.; Tate, J.E.; Parashar, U.D. Effectiveness of rotavirus vaccination: A systematic review of the first decade of global postlicensure data, 2006–2016. Clin. Infect. Dis. 2017, 65, 840–850. [Google Scholar] [CrossRef]
- Zheng, D.P.; Ando, T.; Fankhauser, R.L.; Beard, R.S.; Glass, R.I.; Monroe, S.S. Norovirus classification and proposed strain nomenclature. Virology 2006, 346, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Liu, H.; Wilen, C.B.; Sychev, Z.E.; Desai, C.; Hykes, B.L., Jr.; Orchard, R.C.; McCune, B.T.; Kim, K.; Nice, T.J.; et al. A secreted viral nonstructural protein determines intestinal norovirus pathogenesis. Cell Host Microbe 2019, 25, 845–857. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wang, M.; Wang, K.; Estes, M.K. Sequence and genomic organization of Norwalk virus. Virology 1993, 195, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Prasad, B.V.; Hardy, M.E.; Dokland, T.; Bella, J.; Rossmann, M.G.; Estes, M.K. X-ray crystallographic structure of the Norwalk virus capsid. Science 1999, 286, 287–290. [Google Scholar] [CrossRef]
- Ford-Siltz, L.A.; Tohma, K.; Parra, G.I. Understanding the relationship between norovirus diversity and immunity. Gut Microbes 2021, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Vongpunsawad, S.; Venkataram Prasad, B.V.; Estes, M.K. Norwalk virus minor capsid protein VP2 associates within the VP1 shell domain. J. Virol. 2013, 87, 4818. [Google Scholar] [CrossRef]
- Atmar, R.L. Noroviruses: State of the art. Food Environ. Virol. 2010, 2, 117–126. [Google Scholar] [CrossRef]
- Reymão, T.K.A.; Fumian, T.M.; Justino, M.C.A.; Hernandez, J.M.; Bandeira, R.S.; Lucena, M.S.S.; Teixeira, D.M.; Farias, F.P.; Silva, L.D.; Linhares, A.C.; et al. Norovirus RNA in serum associated with increased fecal viral load in children: Detection, quantification and molecular analysis. PLoS ONE 2018, 13, e0199763. [Google Scholar] [CrossRef] [PubMed]
- Bozkurt, H.; Phan-Thien, K.Y.; Ogtrop, F.V.; Bell, T.; Mcconchie, R. Outbreaks, occurrence, and control of norovirus and hepatitis a virus contamination in berries: A review. Crit. Rev. Food Sci. Nutr. 2020, 61, 116–138. [Google Scholar] [CrossRef] [PubMed]
- Herman, K.M.; Hall, A.J.; Gould, L.H. Outbreaks attributed to fresh leafy vegetables, United States, 1973–2012. Epidemiol. Infect. 2015, 143, 3011–3021. [Google Scholar] [CrossRef]
- Gyawali, P.; Fletcher, G.C.; Mccoubery, D.J.; Hewitt, J. Norovirus in shellfish: An overview of post-harvest treatments and their challenges. Food Control 2019, 99, 171–179. [Google Scholar] [CrossRef]
- Knight, A.; Haines, J.; Stals, A.; Li, D.; Uyttendaele, M.; Knight, A.; Jaykus, L. A systematic review of human norovirus survival reveals a greater persistence of human norovirus RT-qPCR signals compared to those of cultivable surrogate viruses. Int. J. Food Microbiol. 2016, 216, 40–49. [Google Scholar] [CrossRef]
- Becker, B.; Dabisch-Ruthe, M.; Pfannebecker, J. Inactivation of murine norovirus on fruit and vegetable surfaces by vapor phase hydrogen peroxide. J. Food Prot. 2020, 83, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Barclay, L.; Park, G.W.; Vega, E.; Hall, A.; Parashar, U.; Vinjé, J.; Lopman, B. Infection control for norovirus. Clin. Microbiol. Infect. 2014, 20, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Cook, N.; Knight, A.; Richards, G.P. Persistence and elimination of human norovirus in food and on food contact surfaces: A critical review. J. Food Prot. 2016, 79, 1273–1294. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.K.; Watanabe, M.; Zhu, S.; Graves, C.L.; Keyes, L.R.; Grau, K.R.; Gonzalez-Hernandez, M.B.; Iovine, N.M.; Wobus, C.E.; Vinje, J.; et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 2014, 346, 755–759. [Google Scholar] [CrossRef]
- Jones, M.K.; Grau, K.R.; Costantini, V.; Kolawole, A.O.; De Graaf, M.; Freiden, P.; Graves, C.L.; Koopmans, M.; Wallet, S.M.; Tibbetts, S.A.; et al. Human norovirus culture in B cells. Nat. Protoc. 2015, 10, 1939–1947. [Google Scholar] [CrossRef] [PubMed]
- Ettayebi, K.; Crawford, S.E.; Murakami, K.; Broughman, J.R.; Karandikar, U.; Tenge, V.R.; Neill, F.H.; Blutt, S.E.; Zeng, X.; Qu, L.; et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 2016, 353, 1387–1393. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.Y.; Blutt, S.E.; Crawford, S.E.; Ettayebi, K.; Zeng, X.; Saxena, K.; Ramani, S.; Karandikar, U.C.; Zachos, N.C.; Estes, M.K. Human intestinal enteroids: New models to study gastrointestinal virus infections. Methods Mol. Biol. 2019, 1576, 229–247. [Google Scholar] [PubMed]
- Dycke, J.V.; Ny, A.; Conceição-Neto, N.; Maes, J.; Hosmillo, M.; Cuvry, A.; Goodfellow, I.; Nogueira, T.C.; Verbeken, E.; Matthijnssens, J.; et al. A robust human norovirus replication model in zebrafish larvae. PLoS Pathog. 2019, 15, e1008009. [Google Scholar]
- Thornhill, T.S.; Wyatt, R.G.; Kalica, A.R.; Dolin, R.; Chanock, R.M.; Kapikian, A.Z. Detection by immune electron microscopy of 26-to 27-nm viruslike particles associated with two family outbreaks of gastroenteritis. J. Infect. Dis. 1977, 135, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Riepenhoff-Talty, M.; Barrett, H.J.; Spada, B.A.; Ogra, P.L. Negative staining and immune electron microscopy as techniques for rapid diagnosis of viral agents. Ann. N. Y. Acad. Sci. 1983, 420, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Richards, A.F.; Lopman, B.; Gunn, A.; Curry, A.; Ellis, D.; Cotterill, H.; Ratcliffe, S.; Jenkins, M.; Appleton, H.; Gallimore, C.I.; et al. Evaluation of a commercial ELISA for detecting Norwalk-like virus antigen in faeces. J. Clin. Virol. 2003, 26, 109–115. [Google Scholar] [CrossRef]
- Kapikian, A.Z.; Greenberg, H.B.; Cline, W.L.; Kalica, A.R.; Wyatt, R.G.; James, H.D., Jr.; Lloyd, N.L.; Chanock, R.M.; Ryder, R.W.; Kim, H.W. Prevalence of antibody to the Norwalk agent by a newly developed immune adherence hemagglutination assay. J. Med. Virol. 1978, 2, 281–294. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, H.B.; Wyatt, R.G.; Valdesuso, J.; Kalica, A.R.; London, W.T.; Chanock, R.M.; Kapikian, A.Z. Solid-phase microtiter radioimmunoassay for detection of the Norwalk strain of acute nonbacterial, epidemic gastroenteritis virus and its antibodies. J. Med. Virol. 1978, 2, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Gary, G.W.; Kaplan, J.E.; Stine, S.E.; Anderson, L.J. Detection of Norwalk virus antibodies and antigen with a biotin-avidin immunoassay. J. Clin. Microbiol. 1985, 22, 274–278. [Google Scholar] [CrossRef]
- Herrmann, J.E.; Nowak, N.A.; Blacklow, N.R. Detection of Norwalk virus in stools by enzyme immunoassay. J. Med. Virol. 1985, 17, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wang, M.; Graham, D.Y.; Estes, M.K. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 1992, 66, 6527–6532. [Google Scholar] [CrossRef] [PubMed]
- Xi, J.N.; Graham, D.Y.; Wang, K.N.; Estes, M.K. Norwalk virus genome cloning and characterization. Science 1990, 250, 1580–1583. [Google Scholar] [CrossRef] [PubMed]
- Green, K.Y.; Lew, J.F.; Jiang, X.; Kapikian, A.Z.; Estes, M.K. Comparison of the reactivities of baculovirus-expressed recombinant Norwalk virus capsid antigen with those of the native Norwalk virus antigen in serologic assays and some epidemiologic observations. J. Clin. Microbiol. 1993, 31, 2185–2191. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, J.E.; Blacklow, N.R.; Matsui, S.M.; Lewis, T.L.; Estes, M.K.; Ball, J.K.; Brinker, J.P. Monoclonal antibodies for detection of Norwalk virus antigen in stools. J. Clin. Microbiol. 1995, 33, 2511–2513. [Google Scholar] [CrossRef]
- Okame, M.; Shiota, T.; Hansman, G.; Takagi, M.; Yagyu, F.; Takanashi, S.; Phan, T.G.; Shimizu, Y.; Kohno, H.; Okitsu, S.; et al. Anti-norovirus polyclonal antibody and its potential for development of an antigen-ELISA. J. Med. Virol. 2007, 79, 1180–1186. [Google Scholar] [CrossRef]
- Takanashi, S.; Okame, M.; Shiota, T.; Takagi, M.; Yagyu, F.; Tung, P.G.; Nishimura, S.; Katsumata, N.; Igarashi, T.; Okitsu, S.; et al. Development of a rapid immunochromatographic test for noroviruses genogroups I and II. J. Virol. Methods 2008, 148, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Lu, F.; Lyu, C.; Wu, Q.; Zhang, J.; Tian, P.; Xue, L.; Xu, T.; Wang, D. Broad-range and effective detection of human noroviruses by colloidal gold immunochromatographic assay based on the shell domain of the major capsid protein. BMC Microbiol. 2021, 21, 22. [Google Scholar] [CrossRef] [PubMed]
- Khamrin, P.; Nguyen, T.A.; Phan, T.G.; Satou, K.; Masuoka, Y.; Okitsu, S.; Maneekarn, N.; Nishio, O.; Ushijima, H. Evaluation of immunochromatography and commercial enzyme-linked immunosorbent assay for rapid detection of norovirus antigen in stool samples. J. Virol. Methods 2008, 147, 360–363. [Google Scholar] [CrossRef] [PubMed]
- Okitsu-Negishi, S.; Okame, M.; Shimizu, Y.; Phan, T.G.; Tomaru, T.; Kamijo, S.; Sato, T.; Yagyu, F.; Muller, W.E.G.; Ushijima, H. Detection of norovirus antigens from recombinant virus-like particles and stool samples by a commercial norovirus enzyme-linked immunosorbent assay kit. J. Clin. Microbiol. 2006, 44, 3784–3786. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kirby, A.; Gurgel, R.Q.; Dove, W.; Vieira, S.C.F.; Cunliffe, N.A.; Cuevas, L.E. An evaluation of the RIDASCREEN and IDEIA enzyme immunoassays and the RIDAQUICK immunochromatographic test for the detection of norovirus in faecal specimens. J. Clin. Virol. 2010, 49, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Hyun, J.; Kim, J.S.; Song, W.; Kang, H.J.; Lee, K.M. Evaluation of the SD Bioline Norovirus rapid immunochromatography test using fecal specimens from Korean gastroenteritis patients. J. Virol. Methods 2012, 186, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Jonckheere, S.; Botteldoorn, N.; Vandecandelaere, P.; Frans, J.; Laffut, W. Multicenter evaluation of the revised RIDA® QUICK test (N1402) for rapid detection of norovirus in a diagnostic laboratory setting. Diagn. Microbiol. Infect. Dis. 2017, 88, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Bruin, E.D.; Duizer, E.; Vennema, H.; Koopmans, M.P.G. Diagnosis of Norovirus outbreaks by commercial ELISA or RT-PCR. J. Virol. Methods 2006, 137, 259–264. [Google Scholar] [CrossRef]
- Ambert-Balay, K.; Pothier, P. Evaluation of 4 immunochromatographic tests for rapid detection of norovirus in faecal samples. J. Clin. Virol. 2013, 56, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Bruggink, L.D.; Catton, M.G.; Marshall, J.A. Evaluation of the Bioline Standard Diagnostics SD immunochromatographic norovirus detection kit using fecal specimens from Australian gastroenteritis incidents. Diagn. Microbiol. Infect. Dis. 2013, 76, 147–152. [Google Scholar] [CrossRef]
- Gray, J.J.; Kohli, E.; Ruggeri, F.M.; Vennema, H.; Sánchez-Fauquier, A.; Schreier, E.; Gallimore, C.I.; Iturriza-Gomara, M.; Giraudon, H.; Pothier, P.; et al. European multicenter evaluation of commercial enzyme immunoassays for detecting norovirus antigen in fecal samples. Clin. Vaccine Immunol. 2007, 14, 1349–1355. [Google Scholar] [CrossRef]
- Katayama, K.; Shirato-Horikoshi, H.; Kojima, S.; Kageyama, T.; Oka, T.; Hoshino, F.B.; Fukushi, S.; Shinohara, M.; Uchida, K.; Suzuki, Y.; et al. Phylogenetic analysis of the complete genome of 18 Norwalk-like viruses. Virology 2002, 299, 225–239. [Google Scholar] [CrossRef]
- Kojima, S.; Kageyama, T.; Fukushi, S.; Hoshino, F.B.; Shinohara, M.; Uchida, K.; Natori, K.; Takeda, N.; Katayama, K. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J. Virol. Methods 2002, 100, 107–114. [Google Scholar] [CrossRef]
- Fukuda, S.; Takao, S.; Kuwayama, M.; Shimazu, Y.; Miyazaki, K. Rapid detection of norovirus from fecal specimens by real-time reverse transcription-loop-mediated isothermal amplification assay. J. Clin. Microbiol. 2006, 44, 1376–1381. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Xu, Z.; Nie, K.; Ding, X.; Guan, L.; Wang, J.; Xian, Y.; Wu, X.; Ma, X. Visual detection of norovirus genogroup II by reverse transcription loop-mediated isothermal amplification with hydroxynaphthol blue dye. Food Environ. Virol. 2014, 6, 196–201. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.B.; Seo, D.J.; Oh, H.; Kingsley, D.H.; Choi, C. Development of one-step reverse transcription loop-mediated isothermal amplification for norovirus detection in oysters. Food Control 2017, 73, 1002–1009. [Google Scholar] [CrossRef]
- Greene, S.R.; Moe, C.L.; Jaykus, L.A.; Cronin, M.; Grosso, L.; Aarle, P.V. Evaluation of the NucliSens Basic Kit assay for detection of Norwalk virus RNA in stool specimens. J. Virol. Methods 2003, 108, 123–131. [Google Scholar] [CrossRef]
- Moore, C.; Clark, E.M.; Gallimore, C.I.; Corden, S.A.; Westmoreland, D. Evaluation of a broadly reactive nucleic acid sequence based amplification assay for the detection of noroviruses in faecal material. J. Clin. Virol. 2004, 29, 290–296. [Google Scholar] [CrossRef]
- Lamhoujeb, S.; Charest, H.; Fliss, I.; Ngazoa, S.; Jean, J. Real-time molecular beacon NASBA for rapid and sensitive detection of norovirus GII in clinical samples. Can. J. Microbiol. 2009, 55, 1375–1380. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.D.; Jaykus, L.A. Development of a recombinase polymerase amplification assay for detection of epidemic human noroviruses. Sci. Rep. 2017, 7, 40244. [Google Scholar] [CrossRef] [PubMed]
- Vinjé, J. Advances in laboratory methods for detection and typing of norovirus. J. Clin. Microbiol. 2015, 53, 373–381. [Google Scholar] [CrossRef]
- Kageyama, T.; Kojima, S.; Shinohara, M.; Uchida, K.; Fukushi, S.; Hoshino, F.B.; Takeda, N.; Katayama, K. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 2003, 41, 1548–1557. [Google Scholar] [CrossRef] [PubMed]
- Jothikumar, N.; Lowther, J.A.; Henshilwood, K.; Lees, D.N.; Hill, V.R.; Vinje, J. Rapid and sensitive detection of noroviruses by using TaqMan-based one-step reverse transcription-PCR assays and application to naturally contaminated shellfish samples. Appl. Environ. Microbiol. 2005, 71, 1870–1875. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhang, Z.; Wu, Q.; Tian, P.; Geng, H.; Xu, T.; Wang, D. Redesigned duplex RT-qPCR for the detection of GI and GII human noroviruses. Engineering 2020, 6, 442–448. [Google Scholar] [CrossRef]
- Gilpatrick, S.G.; Schwab, K.J.; Estes, M.K.; Atmar, R.L. Development of an immunomagnetic capture reverse transcription-PCR assay for the detection of Norwalk virus. J. Virol. Methods 2000, 90, 69–78. [Google Scholar] [CrossRef]
- Cannon, J.L.; Vinjé, J. Histo-blood group antigen assay for detecting noroviruses in water. Appl. Environ. Microbiol. 2008, 74, 6818–6819. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tian, P.; Engelbrektson, A.; Mandrell, R. Two-log increase in sensitivity for detection of norovirus in complex samples by concentration with porcine gastric mucin conjugated to magnetic beads. Appl. Environ. Microbiol. 2008, 74, 4271–4276. [Google Scholar] [CrossRef] [PubMed]
- Dancho, B.A.; Chen, H.; Kingsley, D.H. Discrimination between infectious and non-infectious human norovirus using porcine gastric mucin. Int. J. Food Microbiol. 2012, 155, 222–226. [Google Scholar] [CrossRef]
- Wang, D.; Xu, S.; Yang, D.; Young, G.M.; Tian, P. New in situ capture quantitative (real-time) reverse transcription-PCR method as an alternative approach for determining inactivation of Tulane Virus. Appl. Environ. Microbiol. 2014, 80, 2120–2124. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Tian, P. Inactivation conditions for human norovirus measured by an in situ capture-qRT-PCR method. Int. J. Food Microbiol. 2014, 172, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Tian, P.; Yang, D.; Shan, L.; Li, Q.; Liu, D.; Wang, D. Estimation of human norovirus infectivity from environmental water samples by in situ capture RT-qPCR method. Food Environ. Virol. 2018, 10, 29–38. [Google Scholar] [CrossRef]
- Zhou, Z.; Tian, Z.; Li, Q.; Tian, P.; Wu, Q.; Wang, D.; Shi, X. In situ capture RT-qPCR: A new simple and sensitive method to detect human norovirus in oysters. Front. Microbiol. 2017, 8, 554. [Google Scholar] [CrossRef] [PubMed]
- Batule, B.S.; Kim, S.U.; Mun, H.; Choi, C. Colorimetric detection of norovirus in oyster samples through DNAzyme as a signaling probe. J. Agric. Food Chem. 2018, 66, 3003–3008. [Google Scholar] [CrossRef]
- Polo, D.; Schaeffer, J.; Fournet, N.; Le Saux, J.; Parnaudeau, S.; McLeod, C.; Le Guyader, F.S. Digital PCR for quantifying norovirus in oysters implicated in outbreaks, France. Emerg. Infect. Dis. 2016, 22, 2189–2191. [Google Scholar] [CrossRef]
- Monteiro, S.; Santos, R. Nanofluidic digital PCR for the quantification of Norovirus for water quality assessment. PLoS ONE 2017, 12, e0179985. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, C.; Hoeper, D.; Maede, D.; Johne, R. Analysis of frozen strawberries involved in a large norovirus gastroenteritis outbreak using next generation sequencing and digital PCR. Food Microbiol. 2018, 76, 390–395. [Google Scholar] [CrossRef] [PubMed]
- Coudray-Meunier, C.; Fraisse, A.; Martin-Latil, S.; Guillier, L.; Delannoy, S.; Fach, P.; Perelle, S. A comparative study of digital RT-PCR and RT-qPCR for quantification of Hepatitis A virus and Norovirus in lettuce and water samples. Int. J. Food Microbiol. 2015, 201, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Suffredini, E.; Pepe, T.; Ventrone, I.; Croci, L. Norovirus detection in shellfish using two Real-Time RT-PCR methods. New Microbiol. 2011, 34, 9–16. [Google Scholar] [PubMed]
- Navidad, J.F.; Griswold, D.J.; Gradus, M.S.; Bhattacharyya, S. Evaluation of Luminex xTAG gastrointestinal pathogen analyte-specific reagents for high-throughput, simultaneous detection of bacteria, viruses, and parasites of clinical and public health importance. J. Clin. Microbiol. 2013, 51, 3018–3024. [Google Scholar] [CrossRef]
- Binnicker, M. Multiplex molecular panels for diagnosis of gastrointestinal infection: Performance, result interpretation, and cost-effectiveness. J. Clin. Microbiol. 2015, 53, 3723–3728. [Google Scholar] [CrossRef]
- Chhabra, P.; Gregoricus, N.; Weinberg, G.A.; Halasa, N.; Chappell, J.; Hassan, F.; Selvarangan, R.; Mijatovic-Rustempasic, S.; Ward, M.L.; Bowen, M.; et al. Comparison of three multiplex gastrointestinal platforms for the detection of gastroenteritis viruses. J. Clin. Virol. 2017, 95, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, R.; Cho, J.; Qiu, Y.Y.; Parsons, B.D.; Lee, B.E.; Chui, L.; Freedman, S.B.; Pang, X. High genetic variability of norovirus leads to diagnostic test challenges. J. Clin. Virol. 2017, 96, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Clark, L.C., Jr.; Lyons, C. Electrode systems for continuous monitoring in cardiovascular surgery. Ann. N. Y. Acad. Sci. 1962, 102, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Montagnana, M.; Caputo, M.; Giavarina, D.; Lippi, G. Overview on self-monitoring of blood glucose. Clin. Chim. Acta 2009, 402, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Lu, Y. Using personal glucose meters and functional DNA sensors to quantify a variety of analytical targets. Nat. Chem. 2011, 3, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Zhang, J.; Lu, Y. Transforming the blood glucose meter into a general healthcare meter for in vitro diagnostics in mobile health. Biotechnol. Adv. 2016, 34, 331–341. [Google Scholar] [CrossRef]
- Zeng, L.; Gong, J.; Rong, P.; Liu, C.; Chen, J. A portable and quantitative biosensor for cadmium detection using glucometer as the point-of-use device. Talanta 2019, 198, 412–416. [Google Scholar] [CrossRef] [PubMed]
- Vo-Dinh, T.; Cullum, B. Biosensors and biochips: Advances in biological and medical diagnostics. Fresenius J. Anal. Chem. 2000, 366, 540–551. [Google Scholar] [CrossRef]
- Justino, C.I.L.; Freitas, A.C.; Pereira, R.; Duarte, A.C.; Santos, T.A.P.R. Recent developments in recognition elements for chemical sensors and biosensors. TrAC Trend. Anal. Chem. 2015, 68, 2–17. [Google Scholar] [CrossRef]
- Saylan, Y.; Erdem, Z.; Nal, S.; Denizli, A. An alternative medical diagnosis method: Biosensors for virus detection. Biosensors 2019, 9, 65. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.A.; Kwon, J.; Kim, D.; Yang, S. A rapid, sensitive and selective electrochemical biosensor with concanavalin A for the preemptive detection of norovirus. Biosens. Bioelectron. 2015, 64, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Kitajima, M.; Mani, K.; Kanhere, E.; Whittle, A.J.; Triantafyllou, M.S.; Miao, J. Miniaturized Electrochemical Sensor Modified with Aptamers for Rapid Norovirus Detection. In Proceedings of the 2016 IEEE 11th Annual International Conference on Nano/Micro Engineered and Molecular Systems (NEMS 2016), Matsushima, Japan, 17–20 April 2016; pp. 587–590. [Google Scholar]
- Hwang, H.J.; Ryu, M.Y.; Park, C.Y.; Ahn, J.; Park, H.G.; Choi, C.; Ha, S.; Park, T.J.; Park, J.P. High sensitive and selective electrochemical biosensor: Label-free detection of human norovirus using affinity peptide as molecular binder. Biosens. Bioelectron. 2017, 87, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Chand, R.; Neethirajan, S. Microfluidic platform integrated with graphene-gold nano-composite aptasensor for one-step detection of norovirus. Biosens. Bioelectron. 2017, 98, 47–53. [Google Scholar] [CrossRef]
- Baek, S.H.; Kim, M.W.; Park, C.Y.; Choi, C.; Kailasa, S.K.; Park, J.P.; Park, T.J. Development of a rapid and sensitive electrochemical biosensor for detection of human norovirus via novel specific binding peptides. Biosens. Bioelectron. 2019, 123, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Yakes, B.J.; Papafragkou, E.; Conrad, S.M.; Neill, J.D.; Ridpath, J.F.; Burkhardt, W., 3rd; Kulka, M.; Degrasse, S.L. Surface plasmon resonance biosensor for detection of feline calicivirus, a surrogate for norovirus. Int. J. Food Microbiol. 2013, 162, 152–158. [Google Scholar] [CrossRef]
- Ashiba, H.; Sugiyama, Y.; Wang, X.; Shirato, H.; Higo-Moriguchi, K.; Taniguchi, K.; Ohki, Y.; Fujimaki, M. Detection of norovirus virus-like particles using a surface plasmon resonance-assisted fluoroimmunosensor optimized for quantum dot fluorescent labels. Biosens. Bioelectron. 2017, 93, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Fahmida, N.; Chowdhury, A.D.; Takemura, K.; Lee, J.; Adegoke, O.; Deo, V.K.; Abe, F.; Suzuki, T.; Park, E.Y. Single-step detection of norovirus tuning localized surface plasmon resonance-induced optical signal between gold nanoparticles and quantum dots. Biosens. Bioelectron. 2018, 122, 16–24. [Google Scholar]
- Han, K.N.; Choi, J.; Kwon, J. Three-dimensional paper-based slip device for one-step point-of-care testing. Sci. Rep. 2016, 6, 25710. [Google Scholar] [CrossRef] [PubMed]
- Adegoke, O.; Seo, M.; Kato, T.; Kawahito, S.; Park, E.Y. An ultrasensitive SiO2-encapsulated alloyed CdZnSeS quantum dot-molecular beacon nanobiosensor for norovirus. Biosens. Bioelectron. 2016, 86, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.; Breshears, L.E.; Perea, S.; Morrison, C.M.; Betancourt, W.Q.; Reynolds, K.A.; Yoon, J.Y. Smartphone-based paper microfluidic particulometry of norovirus from environmental water samples at the single copy level. ACS Omega 2019, 4, 11180–11188. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.R.; Takemeura, K.; Li, T.; Kitamoto, N.; Tanaka, T.; Suzuki, T.; Park, E.Y. Size-controlled preparation of peroxidase-like graphene-gold nanoparticle hybrids for the visible detection of norovirus-like particles. Biosens. Bioelectron. 2017, 87, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Ahn, S.; Chakkarapani, S.K.; Kang, S.H. Supersensitive detection of the norovirus immunoplasmon by 3D total internal reflection scattering defocus microscopy with wavelength-dependent transmission grating. ACS Sens. 2019, 4, 2515–2523. [Google Scholar] [CrossRef]
- Da Silva, E.T.S.G.; Souto, D.E.P.; Barragan, J.T.C.; Giarola, J.d.F.; de Moraes, A.C.M.; Kubota, L.T. Electrochemical biosensors in point-of-care devices: Recent advances and future trends. ChemElectroChem 2017, 4, 778–794. [Google Scholar] [CrossRef]
- Wang, R.; Wang, Y.; Lassiter, K.; Li, Y.; Hargis, B.; Tung, S.; Berghman, L.; Bottje, W. Interdigitated array microelectrode based impedance immunosensor for detection of avian influenza virus H5N1. Talanta 2009, 79, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Taleat, Z.; Khoshroo, A.; Mazloum-Ardakani, M. Screen-printed electrodes for biosensing: A review (2008–2013). Microchim. Acta 2014, 181, 865–891. [Google Scholar] [CrossRef]
- Alonso-Lomillo, M.A.; Domínguez-Renedo, O.; Arcos-Martínez, M.J. Screen-printed biosensors in microbiology; a review. Talanta 2010, 82, 1629–1636. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.; Madzivhandila, P.; Ntuli, L.; Bezuidenhout, P.; Zheng, H.; Land, K. Printed paper–based electrochemical sensors for low-cost point-of-need applications. Electrocatalysis 2019, 10, 342–351. [Google Scholar] [CrossRef]
- Shi, H.; Nie, K.; Dong, B.; Long, M.; Xu, H.; Liu, Z. Recent progress of microfluidic reactors for biomedical applications. Chem. Eng. J. 2019, 361, 635–650. [Google Scholar] [CrossRef]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef]
- Coltro, W.K.T.; Cheng, C.; Carrilho, E.; de Jesus, D.P. Recent advances in low-cost microfluidic platforms for diagnostic applications. Electrophoresis 2014, 35, 2309–2324. [Google Scholar] [CrossRef] [PubMed]
- Jans, H.; Huo, Q. Gold nanoparticle-enabled biological and chemical detection and analysis. Chem. Soc. Rev. 2012, 41, 2849–2866. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Wang, E. Synthesis and electrochemical applications of gold nanoparticles. Anal. Chim. Acta 2007, 598, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.C.; Chen, L.C.; Huang, C.H.; Ding, S.J.; Chang, C.C.; Chang, H.C. Development of the multi-functionalized gold nanoparticles with electrochemical-based immunoassay for protein A detection. J. Electroanal. Chem. 2008, 619, 39–45. [Google Scholar] [CrossRef]
- Xiao, T.; Huang, J.; Wang, D.; Meng, T.; Yang, X. Au and Au-Based nanomaterials: Synthesis and recent progress in electrochemical sensor applications. Talanta 2020, 206, 120210. [Google Scholar] [CrossRef] [PubMed]
- Ambartsumyan, O.; Gribanyov, D.; Kukushkin, V.; Kopylov, A.; Zavyalova, E. SERS-Based Biosensors for Virus Determination with Oligonucleotides as Recognition Elements. Int. J. Mol. Sci. 2020, 21, 3373. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; Wang, R.; Hargis, B.; Lu, H.; Li, Y. A SPR aptasensor for detection of avian influenza virus H5N1. Sensors 2012, 12, 12506–12518. [Google Scholar] [CrossRef]
- Riedel, T.; Rodriguez-Emmenegger, C.; Andres, D.L.S.P.; Bědajánková, A.; Jinoch, P.; Boltovets, P.M.; Brynda, E. Diagnosis of Epstein-Barr virus infection in clinical serum samples by an SPR biosensor assay. Biosens. Bioelectron. 2014, 55, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Lee, G.; Ko, H.; Chang, Y.W.; Kang, M.; Pyun, J. Development of SPR biosensor for the detection of human hepatitis B virus using plasma-treated parylene-N film. Biosens. Bioelectron. 2014, 56, 286–294. [Google Scholar] [CrossRef]
- Omar, N.A.S.; Fen, Y.W.; Abdullah, J.; Sadrolhosseini, A.R.; Kamil, Y.M.; Fauzi, N.I.M.; Hashim, H.S.; Mahdi, M.A. Quantitative and selective surface plasmon resonance response based on a reduced graphene oxide–polyamidoamine nanocomposite for detection of dengue virus E-proteins. Nanomaterials 2020, 10, 569. [Google Scholar] [CrossRef]
- Shang, J.; Piskarev, V.E.; Xia, M.; Huang, P.; Jiang, X.; Likhosherstov, L.M.; Novikova, O.S.; Newburg, D.S.; Ratner, D.M. Identifying human milk glycans that inhibit norovirus binding using surface plasmon resonance. Glycobiology 2013, 23, 1491–1498. [Google Scholar] [CrossRef]
- Miroslav, P. Overview of piezoelectric biosensors, immunosensors and DNA sensors and their applications. Materials 2018, 11, 448. [Google Scholar]
- Saylan, Y.; Akgnüllü, S.; Yavuz, H.; Ünal, S.; Denizli, A. Molecularly imprinted polymer based sensors for medical applications. Sensors 2019, 19, 1279. [Google Scholar] [CrossRef]
- Li, D.; Wang, J.; Wang, R.; Li, Y.; Abi-Ghanem, D.; Berghman, L.; Hargis, B.; Lu, H. A nanobeads amplified QCM immunosensor for the detection of avian influenza virus H5N1. Biosens. Bioelectron. 2011, 26, 4146–4154. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Li, Y. Hydrogel based QCM aptasensor for detection of avian influenza virus. Biosens. Bioelectron. 2013, 42, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, L.; Callaway, Z.T.; Lu, H.; Huang, T.J.; Li, Y. A nanowell-based QCM aptasensor for rapid and sensitive detection of avian influenza virus. Sens. Actuators B Chem. 2017, 240, 934–940. [Google Scholar] [CrossRef]
- Yao, C.; Zhu, T.; Tang, J.; Wu, R.; Chen, Q.; Chen, M.; Zhang, B.; Huang, J.; Fu, W. Hybridization assay of hepatitis B virus by QCM peptide nucleic acid biosensor. Biosens. Bioelectron. 2008, 23, 879–885. [Google Scholar] [CrossRef] [PubMed]
- Giamblanco, N.; Conoci, S.; Russo, D.; Marletta, G. Single-step label-free hepatitis B virus detection by a piezoelectric biosensor. RSC Adv. 2015, 5, 38152–38158. [Google Scholar] [CrossRef]

| Biosensor | Bioreceptor | Signal 1 | Target | LOD (with Linear Range) 2 | Reference |
|---|---|---|---|---|---|
| Electrochemistry | Concanavalin A | CV | NoV GII.4 subtype | 35 genomic copies/mL (102–106 genomic copies/mL) | [89] |
| MNV-specific aptamer | SWV | MHV-1 2 | (0–1.0 × 104 PFU/mL) | [90] | |
| Noro-1 affinity peptide | QCM/CV/EIS | rP2/ NoV GII.4 subtype | rP2 (99.8 nM) GII.4 (7.8 genomic copies/mL) | [91] | |
| 81-bases-long aptamer | DPV | GII VLPs | 100 pM (100 pM–3.5 nM) | [92] | |
| NoroBP-nonFoul (FlexL)2 peptide | EIS | HuNoV GII.4 subtype | 1.7 genomic copies/mL (0–105 genomic copies/mL) | [93] | |
| Optics | FCV antibody | SPR | FCV 3 | ≈104 TCID50 FCV/mL | [94] |
| Anti-HuNoV GII.4 monoclonal antibody (12A11) | SPR | GII.4 VLPs | 0.01 ng/mL | [95] | |
| Anti-norovirus antibody (NS14) | LSPR | NoV GII VLPs/NoV | 12.1 × 10−15 g/mL (10−14–10−9 genomic copies/mL) 95.0 genomic copies/mL (102–105 genomic copies/mL) | [96] | |
| Anti-norovirus GII.4 antibody (for capture) Anti-norovirus GII.4 capsid protein VP1 antibody (for detection) | The assay results could be visualised by the naked eye | NoV GII.4 subtype | 9.5 × 104 genomic copies/mL (1.58 × 105–7.9 × 107 genomic copies/mL) | [97] | |
| Molecular beacon probes (contained 20 bp complementary to NV genogroup II RNA) | UV–vis absorption and fluorescence emission measurements | NoV GII RNA (in human serum) NoV GII RNA (in buffer) | In human serum: 8.2 genomic copies/mL In buffer: 9.3 genomic copies/mL | [98] | |
| Anti-norovirus capsid protein VP1 polyclonal antibody | Benchtop fluorescence microscope | NoV GII (diluted in deionised water) NoV GII (diluted in reclaimed wastewater) | In deionised water: 1 genomic copies/μL In reclaimed wastewater: 10 genomic copies/μL | [99] | |
| Anti-norovirus antibody (NS14) | Colorimetric detection (OD450 nm) | HuNoV GII.4 VLP | 92.7 pg/mL (100 pg/mL to 10 μg/mL) | [100] | |
| Monoclonal capture antibody (C01875M) Monoclonal detection antibody (C01874M) | 3D Total internal reflection-scattering defocused imaging with wavelength-dependent transmission grating | NoV GI capsid protein VP1 | 820 yM (820 yM–92.45 pM) | [101] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, N.; Pan, G.; Liu, P.; Rong, S.; Gao, Z.; Li, Q. Advances and Future Perspective on Detection Technology of Human Norovirus. Pathogens 2021, 10, 1383. https://doi.org/10.3390/pathogens10111383
Wang N, Pan G, Liu P, Rong S, Gao Z, Li Q. Advances and Future Perspective on Detection Technology of Human Norovirus. Pathogens. 2021; 10(11):1383. https://doi.org/10.3390/pathogens10111383
Chicago/Turabian StyleWang, Nan, Guiying Pan, Ping Liu, Shaofeng Rong, Zhiyong Gao, and Qianqian Li. 2021. "Advances and Future Perspective on Detection Technology of Human Norovirus" Pathogens 10, no. 11: 1383. https://doi.org/10.3390/pathogens10111383
APA StyleWang, N., Pan, G., Liu, P., Rong, S., Gao, Z., & Li, Q. (2021). Advances and Future Perspective on Detection Technology of Human Norovirus. Pathogens, 10(11), 1383. https://doi.org/10.3390/pathogens10111383






