Identification of Rice Seed-Derived Fusarium spp. and Development of LAMP Assay against Fusarium fujikuroi
Abstract
1. Introduction
2. Materials and Methods
2.1. Seed Samples, Fungal Isolation, and Preliminary Identification
2.2. Morphological Identification
2.3. DNA Extraction, PCR Amplification, and Sequencing
2.4. Phylogenetic Analysis
2.5. Pathogenicity Tests
2.6. LAMP Primer Design
2.7. LAMP Reaction and Optimization of Its Condition
2.8. Specificity and Sensitivity of LAMP Reaction
2.9. Detection of Rice Seed Samples
3. Results
3.1. Isolation and Identification Using Morphology and TEF1-α Gene Sequences
3.2. Diversity of Fusarium spp. on Rice Seeds
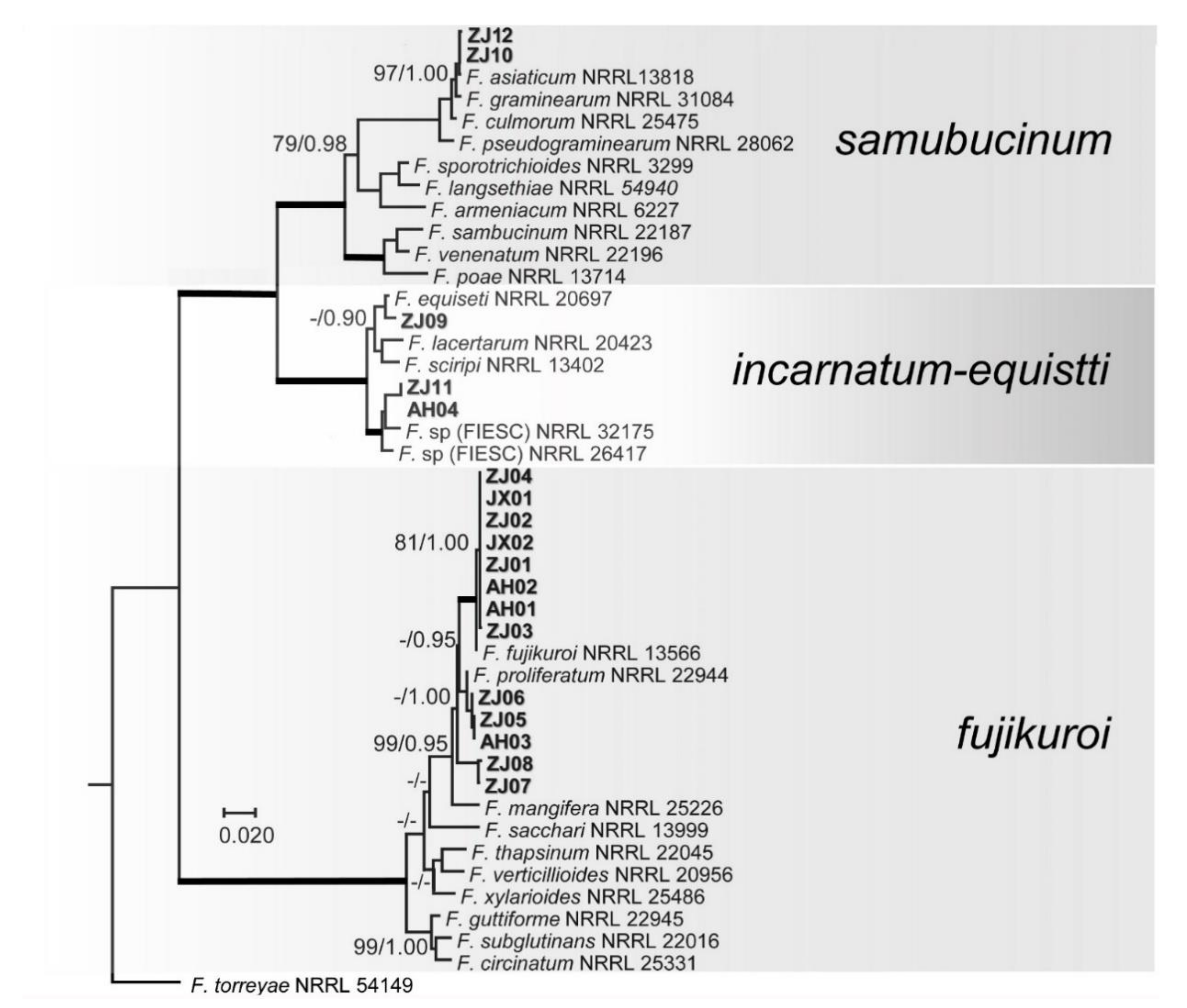
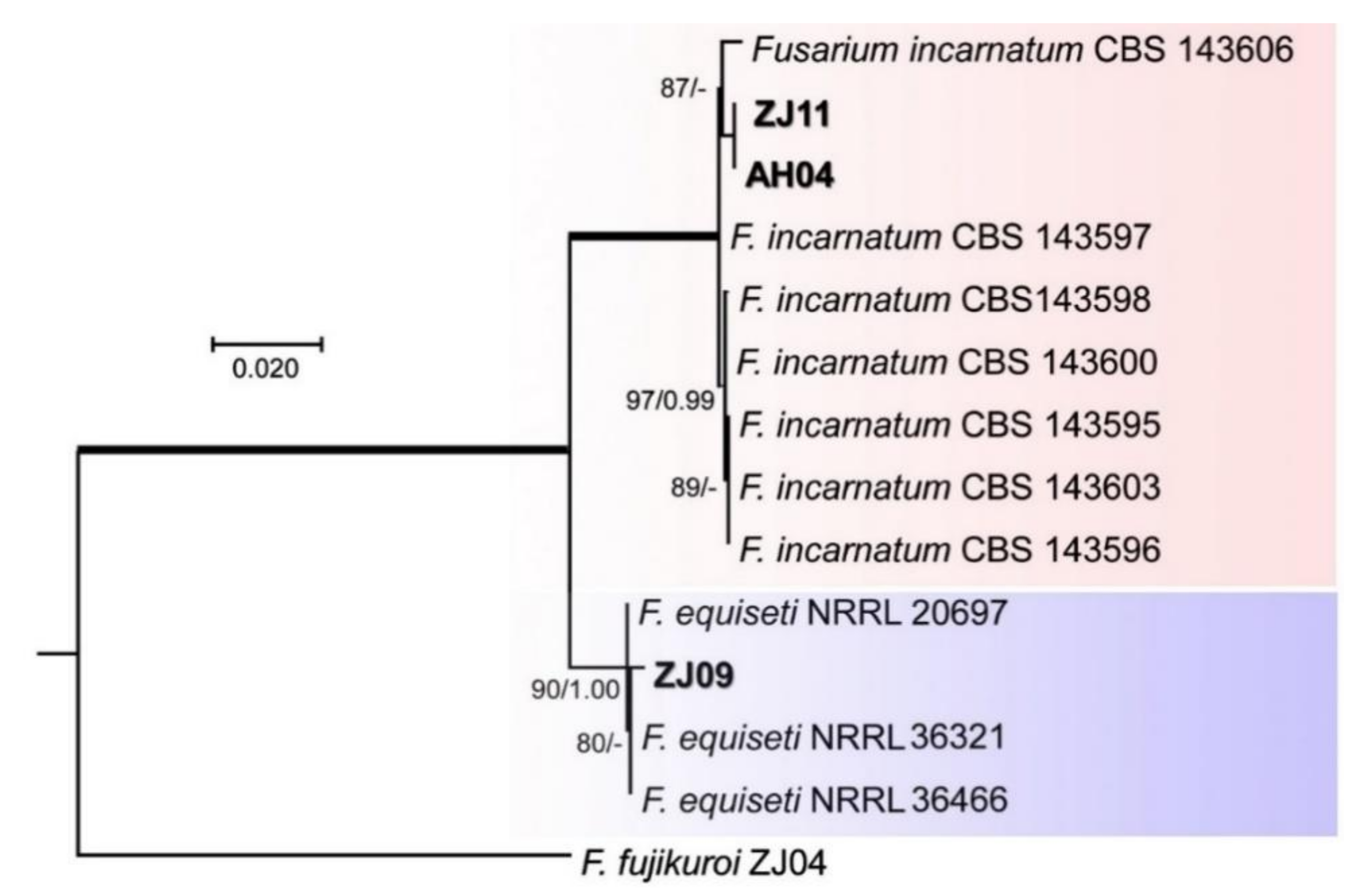
3.3. Phylogenetic Studies
3.4. Pathogenicity of Fusarium spp.
3.5. LAMP Primers
3.6. LAMP Reaction and Optimization of Its Conditions
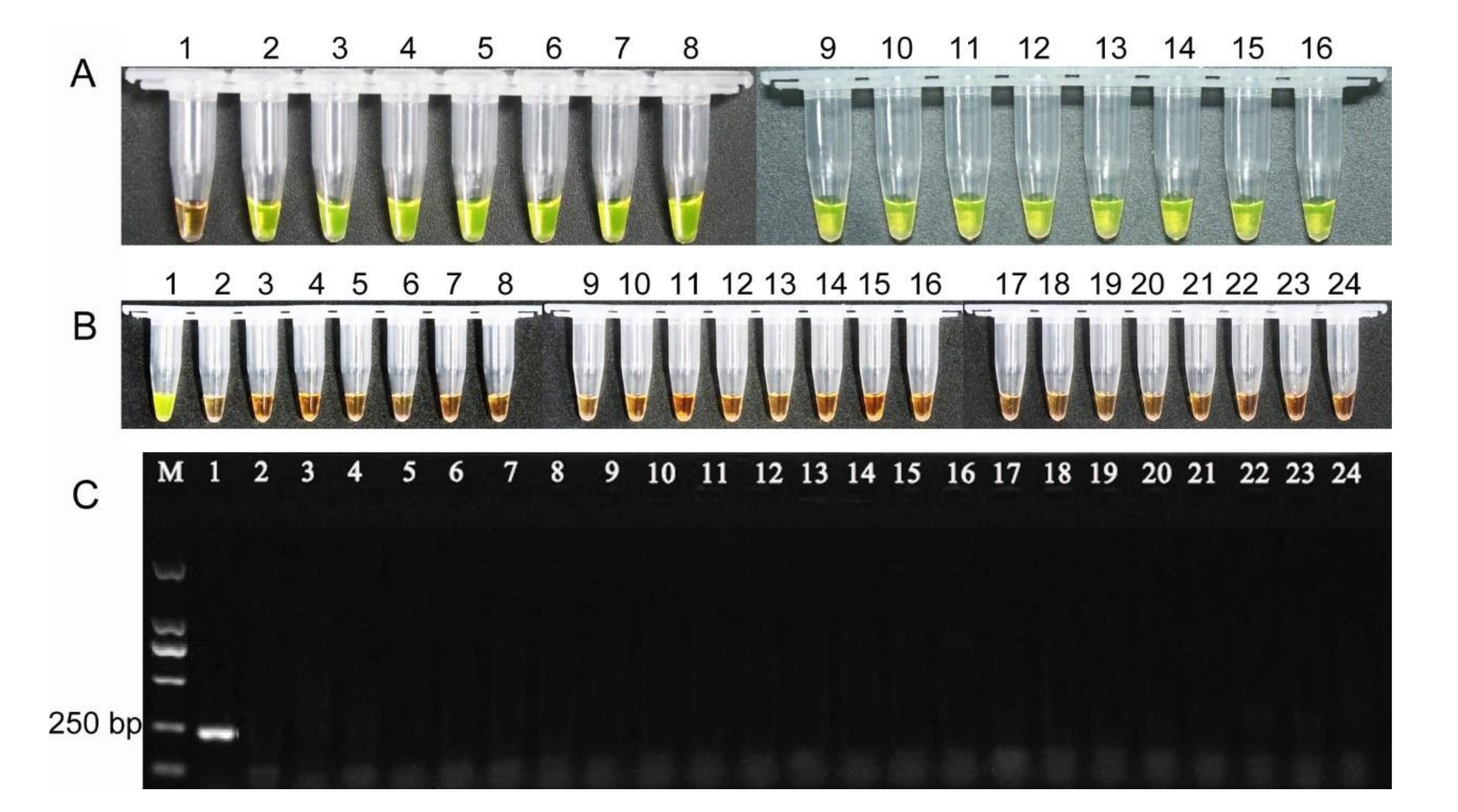
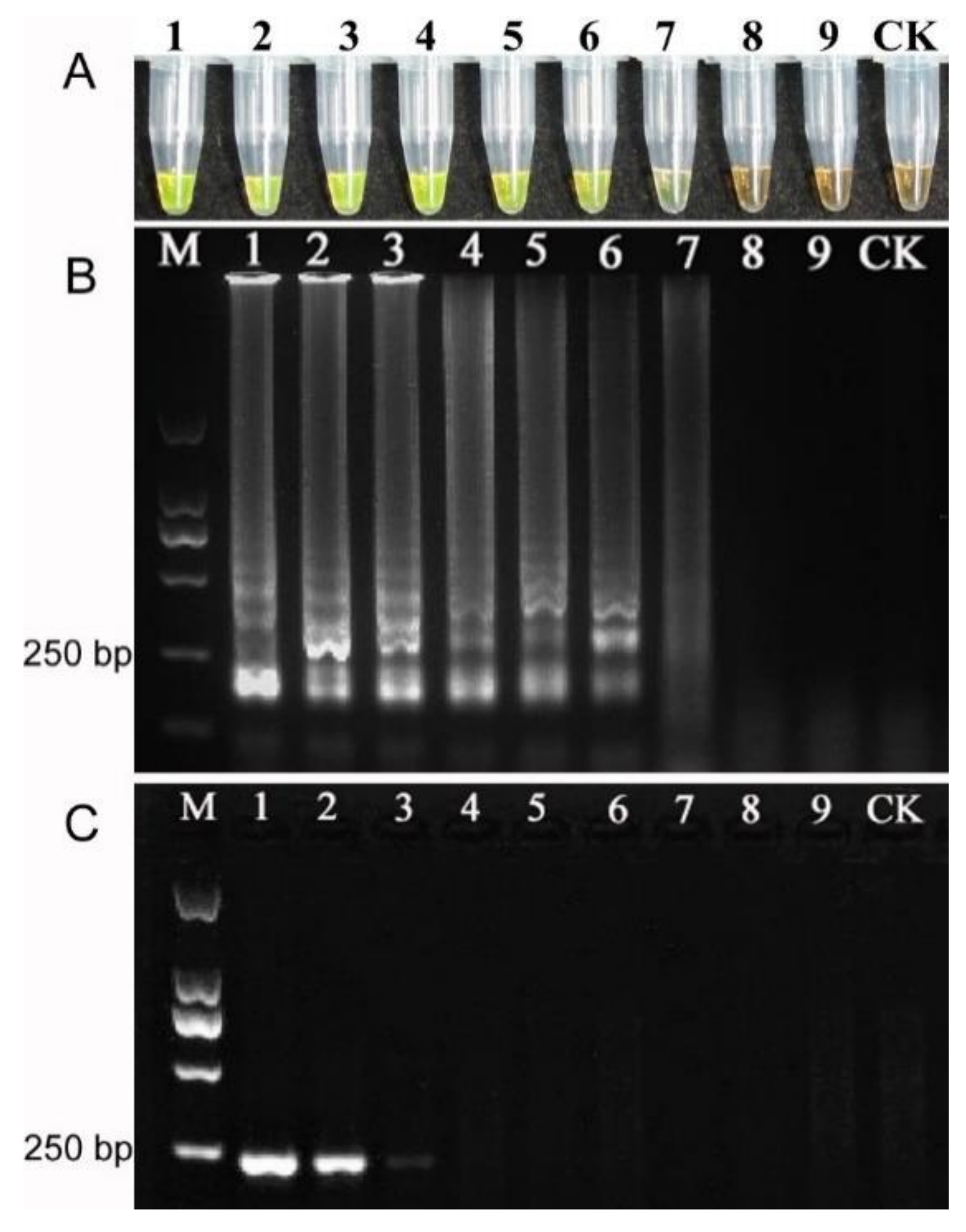
3.7. Specificity and Sensitivity of LAMP Reaction
3.8. Detection of Rice Seed Samples
4. Discussion
4.1. Diversity of Fusarium spp. on Rice Seeds
4.2. Identification of Fusarium spp.
4.3. Pathogenicity of Fusarium spp.
4.4. Application of LAMP as Diagnostic Tool
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahmed, T.; Shahid, M.; Noman, M.; Niazi, M.B.K.; Mahmood, F.; Manzoor, I.; Zhang, Y.; Li, B.; Yang, Y.; Yan, C. Silver Nanoparticles Synthesized by Using Bacillus cereus SZT1 Ameliorated the Damage of Bacterial Leaf Blight Pathogen in Rice. Pathogens 2020, 9, 160. [Google Scholar] [CrossRef] [PubMed]
- Amoah, B.K.; Rezanoor, H.N.; Nicholson, P.; Macdonald, M.V. Variation in the Fusarium section Liseola: Pathogenicity and genetic studies of isolates of Fusarium moniliforme Sheldon from different hosts in Ghana. Plant Pathol. 1995, 44, 563–572. [Google Scholar] [CrossRef]
- Nakamura, T.; Mitsuoka, K.; Sugano, M.; Tomita, K.; Murayama, T. Effects of Auxin and Gibberellin on Conidial Germination and Elongation of Young Hyphae in Gibberella fujikuroi and Penicillium notatum. Plant Cell Physiol. 1985, 26, 1433–1437. [Google Scholar] [CrossRef]
- Matic, S.; Gullino, M.L.; Spadaro, D. The puzzle of bakanae disease through interactions between Fusarium fujikuroi and rice. Front. Biosci. 2017, 9, 333–344. [Google Scholar] [CrossRef]
- Bashyal, B.M.; Aggarwal, R.; Banerjee, S.; Gupta, S.; Sharma, S. Pathogenicity, ecology and genetic diversity of the Fusarium spp. Associated with an emerging bakanae disease of rice (Oryza sativa L.) in India. In Microbial Diversity and Biotechnology in Food Security; Kharwar, R.N., Upadhyay, R., Eds.; Springer India: New Delhi, India, 2014; pp. 307–314. [Google Scholar]
- Zhang, S.Y.; Dai, D.J.; Wang, H.D.; Zhang, C.Q. One-step loop-mediated isothermal amplification (LAMP) for the rapid and sensitive detection of Fusarium fujikuroi in bakanae disease through NRPS31, an important gene in the gibberellic acid bio-synthesis. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- O’Donnell, K.; Rooney, A.P.; Proctor, R.H.; Brown, D.W.; McCormick, S.P.; Ward, T.J.; Frandsen, R.J.N.; Lysøe, E.; Rehner, S.A.; Aoki, T.; et al. Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genet. Biol. 2013, 52, 20–31. [Google Scholar] [CrossRef]
- Wulff, E.G.; Sørensen, J.L.; Lübeck, M.; Nielsen, K.F.; Thrane, U.; Torp, J. Fusarium spp. associated with rice Bakanae: Ecology, genetic diversity, pathogenicity and toxigenicity. Environ. Microbiol. 2010, 12, 649–657. [Google Scholar] [CrossRef]
- Choi, H.-W.; Hong, S.; Lee, Y.K.; Kim, W.G.; Chun, S. Taxonomy of Fusarium fujikuroi species complex associated with bakanae on rice in Korea. Australas. Plant Pathol. 2018, 47, 23–34. [Google Scholar] [CrossRef]
- Nicolli, C.P.; Haidukowski, M.; Susca, A.; Gomes, L.B.; Logrieco, A.; Stea, G.; Del Ponte, E.M.; Moretti, A.; Pfenning, L.H. Fusarium fujikuroi species complex in Brazilian rice: Unveiling increased phylogenetic diversity and toxigenic potential. Int. J. Food Microbiol. 2020, 330, 108667. [Google Scholar] [CrossRef]
- Li, M.; Li, T.; Duan, Y.; Yang, Y.; Wu, J.; Zhao, D.; Xiao, X.; Pan, X.; Chen, W.; Wang, J.; et al. Evaluation of Phenamacril and Ipconazole for Control of Rice Bakanae Disease Caused by Fusarium fujikuroi. Plant Dis. 2018, 102, 1234–1239. [Google Scholar] [CrossRef]
- Proctor, R.H.; Plattner, R.D.; Brown, D.W.; Seo, J.-A.; Lee, Y.-W. Discontinuous distribution of fumonisin biosynthetic genes in the Gibberella fujikuroi species complex. Mycol. Res. 2004, 108, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Tudzynski, B. Gibberellin biosynthesis in fungi: Genes, enzymes, evolution, and impact on biotechnology. Appl. Microbiol. Biotechnol. 2005, 66, 597–611. [Google Scholar] [CrossRef] [PubMed]
- Malonek, S.; Bömke, C.; Bornberg-Bauer, E.; Rojas, M.C.; Hedden, P.; Hopkins, P.; Tudzynski, B. Distribution of gibberellin biosynthetic genes and gibberellin production in the Gibberella fujikuroi species complex. Phytochemistry 2005, 66, 1296–1311. [Google Scholar] [CrossRef] [PubMed]
- Malonek, S.; Rojas, M.C.; Hedden, P.; Hopkins, P.; Tudzynski, B. Restoration of Gibberellin Production in Fusarium proliferatum by Functional Complementation of Enzymatic Blocks. Appl. Environ. Microbiol. 2005, 71, 6014–6025. [Google Scholar] [CrossRef] [PubMed]
- Kokkonen, M.; Ojala, L.; Parikka, P.; Jestoi, M. Mycotoxin production of selected Fusarium species at different culture conditions. Int. J. Food Microbiol. 2010, 143, 17–25. [Google Scholar] [CrossRef]
- Bömke, C.; Tudzynski, B. Diversity, regulation, and evolution of the gibberellin biosynthetic pathway in fungi compared to plants and bacteria. Phytochemistry 2009, 70, 1876–1893. [Google Scholar] [CrossRef]
- Tudzynski, B.; Mihlan, M.; Rojas, M.C.; Linnemannstöns, P.; Gaskin, P.; Hedden, P. Characterization of the final two genes of the gibberellin biosynthesis gene cluster of Gibberella fujikuroi: Des and P450-3 encode GA4 desaturase and the 13-hydroxylase, respectively. J. Biol. Chem. 2003, 278, 28635–28643. [Google Scholar] [CrossRef]
- Gomes, L.B.; Ward, T.J.; Badiale-Furlong, E.; Del Ponte, E.M. Species composition, toxigenic potential and pathogenicity of Fusarium graminearum species complex isolates from southern Brazilian rice. Plant Pathol. 2015, 64, 980–987. [Google Scholar] [CrossRef]
- Avila, C.F.; Moreira, G.M.; Nicolli, C.P.; Gomes, L.B.; Abreu, L.M.; Pfenning, L.H.; Haidukowski, M.; Moretti, A.; Logrieco, A.; Del Ponte, E.M. Fusarium incarnatum-equiseti species complex associated with Brazilian rice: Phylogeny, morphology and toxigenic potential. Int. J. Food Microbiol. 2019, 306, 108267. [Google Scholar] [CrossRef]
- Edwards, S.G.; O’Callaghan, J.; Dobson, A.D.W. PCR-based detection and quantification of mycotoxigenic fungi. Mycol. Res. 2002, 106, 1005–1025. [Google Scholar] [CrossRef]
- Aggarwal, R.; Gupta, S.; Banerjee, S.; Singh, V.B. Development of a SCAR marker for detection of Bipolaris sorokiniana causing spot blotch of wheat. Can. J. Microbiol. 2011, 57, 934–942. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, G.A.; Matić, S.; Ortu, G.; Garibaldi, A.; Spadaro, D.; Gullino, M.L. Development and Validation of a TaqMan Real-Time PCR Assay for the Specific Detection and Quantification of Fusarium fujikuroi in Rice Plants and Seeds. Phytopathology 2017, 107, 885–892. [Google Scholar] [CrossRef] [PubMed]
- Rong, Z.; Yuan, Y.; Ye, W.; Wang, X.; Zheng, X. Rapid diagnosis of rice bakanae caused by Fusarium fujikuroi and F. proliferatum using loop-mediated isothermal amplification assays. J. Phytopathol. 2018, 166, 283–290. [Google Scholar] [CrossRef]
- Sunani, S.K.; Bashyal, B.M.; Rawat, K.; Manjunatha, C.; Sharma, S.; Prakash, G.; Krishnan, S.G.; Singh, A.K.; Aggarwal, R. Development of PCR and loop mediated isothermal amplification assay for the detection of bakanae pathogen Fusarium fujikuroi. Eur. J. Plant Pathol. 2019, 154, 715–725. [Google Scholar] [CrossRef]
- Lee, J.; Chang, I.-Y.; Kim, H.; Yun, S.-H.; Leslie, J.F.; Lee, Y.-W. Genetic Diversity and Fitness of Fusarium graminearum Populations from Rice in Korea. Appl. Environ. Microbiol. 2009, 75, 3289–3295. [Google Scholar] [CrossRef]
- Nash, G.A.; Snyder, W.C. Quantitative enumerations by plate counts of propagules of bean root rot Fusarium in field soils. Phytopathology 1962, 52, 567–572. [Google Scholar]
- Leslie, J.F.; Summerell, B.A. The Fusarium Laboratory Manual; Blackwell Publishing: Ames, IA, USA, 2006. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef]
- O’Donnell, K.; Sutton, D.A.; Rinaldi, M.G.; Sarver, B.A.J.; Balajee, S.A.; Schroers, H.-J.; Summerbell, R.C.; Robert, V.A.R.G.; Crous, P.W.; Zhang, N.; et al. Internet-Accessible DNA Sequence Database for Identifying Fusaria from Human and Animal Infections. J. Clin. Microbiol. 2010, 48, 3708–3718. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Katoh, K.; Kuma, K.I.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Silvestro, D.; Michalak, I. raxmlGUI: A graphical front-end for RAxML. Org. Divers. Evol. 2012, 12, 335–337. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-L.; Ojaghian, M.R.; Zhang, J.-Z.; Zhu, S.-J. A new species of Scopulariopsis and its synergistic effect on pathogenicity of Verticillium dahliae on cotton plants. Microbiol. Res. 2017, 201, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Starkey, D.E.; Ward, T.J.; Aoki, T.; Gale, L.R.; Kistler, H.C.; Geiser, D.M.; Suga, H.; Tóth, B.; Varga, J.; O’Donnell, K. Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genet. Biol. 2007, 44, 1191–1204. [Google Scholar] [CrossRef]
- Burgess, L.W.; Nelson, P.E.; Toussoun, T.A.; Marasas, W.F.O. Fusarium scirpi: Emended Description and Notes on Geographic Distribution. Mycologia 1985, 77, 212. [Google Scholar] [CrossRef]
- Pascoe, I.G. Fusarium morphology I: Identification and characterization of a third conidial type, the mesoconidium. Mycotaxon 1990, 37, 121–160. [Google Scholar]
- O’Donnell, K.; Ward, T.J.; Geiser, D.M.; Kistler, H.C.; Aoki, T. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genet. Biol. 2004, 41, 600–623. [Google Scholar] [CrossRef]
- Amatulli, M.T.; Spadaro, D.; Gullino, M.L.; Garibaldi, A. Molecular identification of Fusarium spp. associated with bakanae disease of rice in Italy and assessment of their pathogenicity. Plant Pathol. 2010, 59, 839–844. [Google Scholar] [CrossRef]
- Skovgaard, K.; Rosendahl, S.; O’Donnell, K.; Nirenberg, H.I. Fusarium commune is a new species identified by morphological and molecular phylogenetic data. Mycologia 2003, 95, 630–636. [Google Scholar] [CrossRef] [PubMed]
- Moreira, G.M.; Nicolli, C.P.; Gomes, L.B.; Ogoshi, C.; Scheuermann, K.K.; Silva-Lobo, V.L.; Schurt, D.A.; Ritieni, A.; Moretti, A.; Pfenning, L.H.; et al. Nationwide survey reveals high diversity of Fusarium species and related mycotoxins in Brazilian rice: 2014 and 2015 harvests. Food Control. 2020, 113, 107171. [Google Scholar] [CrossRef]
- Maryani, N.; Lombard, L.; Poerba, Y.S.; Subandiyah, S.; Crous, P.W.; Kema, G.H.J. Phylogeny and genetic diversity of the banana Fusarium wilt pathogen Fusarium oxysporum f. sp. cubense in the Indonesian centre of origin. Stud. Mycol. 2019, 92, 155–194. [Google Scholar] [CrossRef] [PubMed]
- Prà, M.D.; Tonti, S.; Pancaldi, D.; Nipoti, P.; Alberti, I. First Report of Fusarium andiyazi Associated with Rice Bakanae in Italy. Plant Dis. 2010, 94, 1070. [Google Scholar] [CrossRef] [PubMed]
- Jahan, Q.S.A.; Meon, S.; Jaafar, H.; Ahmad, Z.A. Characterization of Fusarium proliferatum through species specific primers and its virulence on rice seeds. Int. J. Agric. Biol. 2013, 15, 649–656. [Google Scholar]
- Jeon, Y.-A.; Yu, S.-H.; Lee, Y.Y.; Park, H.-J.; Lee, S.; Sung, J.S.; Kim, Y.-G.; Lee, H.-S. Incidence, Molecular Characteristics and Pathogenicity of Gibberella fujikuroi Species Complex Associated with Rice Seeds from Asian Countries. Mycobiology 2013, 41, 225–233. [Google Scholar] [CrossRef]
- Dhingra, O.D.; Muchovej, J.J. Pod rot, seed rot, and root rot of snap bean and dry bean caused by Fusarium semitectum. Plant Dis. Report. 1979, 63, 84–87. [Google Scholar]
- Abbas, H.K.; Boyette, C.D.; Hoagland, R.E. Phytotoxicity of Fusarium, other fungal isolates, and of the phytotoxins fumonisin, fusaric acid, and moniliformin to jimsonweed. Phytoprotection 1995, 76, 17–25. [Google Scholar] [CrossRef]
- Izzati, M.Z.N.A.; Azmi, A.R.; Baharuddin, S. Secondary metabolite profiles and mating populations of Fusarium species in section Liseola associated with bakanae disease of rice. Malays. J. Microbiol. 2008, 4, 6–13. [Google Scholar] [CrossRef]
- Goswami, R.S.; Dong, Y.; Punja, Z.K. Host range and mycotoxin production by Fusarium equiseti isolates originating from ginseng fields1. Can. J. Plant Pathol. 2008, 30, 155–160. [Google Scholar] [CrossRef]
- Dong, F.; Xing, Y.J.; Lee, Y.W.; Mokoena, M.P.; Olaniran, A.O.; Xu, J.H.; Shi, J.R. Occurrence of Fusarium mycotoxins and toxigenic Fusarium species in freshly harvested rice in Jiangsu, China. World Mycotoxin J. 2020, 13, 201–212. [Google Scholar] [CrossRef]
- Desjardins, A.E.; McCormick, S.P.; Appell, M. Structure−Activity Relationships of Trichothecene Toxins in an Arabidopsis thaliana Leaf Assay. J. Agric. Food Chem. 2007, 55, 6487–6492. [Google Scholar] [CrossRef] [PubMed]
- Muhitch, M.J.; McCormick, S.P.; Alexander, N.J.; Hohn, T.M. Transgenic expression of the TRI101 or PDR5 gene increases resistance of tobacco to the phytotoxic effects of the trichothecene 4,15-diacetoxyscirpenol. Plant Sci. 2000, 157, 201–207. [Google Scholar] [CrossRef]
- Wiemann, P.; Sieber, C.M.K.; Von Bargen, K.W.; Studt, L.; Niehaus, E.-M.; Espino, J.J.; Huß, K.; Michielse, C.B.; Albermann, S.; Wagner, D.; et al. Deciphering the Cryptic Genome: Genome-wide Analyses of the Rice Pathogen Fusarium fujikuroi Reveal Complex Regulation of Secondary Metabolism and Novel Metabolites. PLoS Pathog. 2013, 9, e1003475. [Google Scholar] [CrossRef] [PubMed]
- Han, J.W.; Ahn, S.H.; Park, S.H.; Wang, S.Y.; Bae, G.U.; Seo, D.W.; Kwon, H.K.; Hong, S.; Lee, H.Y.; Lee, Y.W.; et al. Apicidin, a histone deacetylase inhibitor, inhibits proliferation of tumor cells via induction of p21WAF1/Cip1 and gelsolin. Cancer Res. 2000, 60, 6068–6074. [Google Scholar] [PubMed]
- Jin, J.-M.; Lee, S.; Lee, J.; Baek, S.-R.; Kim, J.-C.; Yun, S.-H.; Park, S.-Y.; Kang, S.; Lee, Y.-W. Functional characterization and manipulation of the apicidin biosynthetic pathway in Fusarium semitectum. Mol. Microbiol. 2010, 76, 456–466. [Google Scholar] [CrossRef] [PubMed]





| Species | Isolates | Origin | GenBank Access NO. | |||
|---|---|---|---|---|---|---|
| TEF1-α RPB1 RPB2 ITS | ||||||
| Fusarium fujikuroi | ZJ01 | Zhejiang China | MT560637 | MT560601 | MT560616 | |
| Fusarium fujikuroi | ZJ02 | Zhejiang China | MT560638 | MT560602 | MT560617 | |
| Fusarium fujikuroi | ZJ03 | Zhejiang China | MT560639 | MT560603 | MT560618 | |
| Fusarium fujikuroi | ZJ04 | Zhejiang China | MT560640 | MT560604 | MT560619 | |
| Fusarium fujikuroi | AH01 | Anhui China | MT560650 | MT560611 | MT560628 | |
| Fusarium fujikuroi | AH02 | Anhui China | MT560651 | MT560612 | MT560629 | |
| Fusarium fujikuroi | JX01 | Jiangxi China | MT560654 | MT560614 | MT560632 | |
| Fusarium fujikuroi | JX02 | Jiangxi China | MT560655 | MT560615 | MT560633 | |
| Fusarium proliferatum | ZJ05 | Zhejiang China | MT560641 | MT560605 | MT560620 | |
| Fusarium proliferatum | ZJ06 | Zhejiang China | MT560642 | MT560606 | MT560621 | |
| Fusarium proliferatum | AH03 | Anhui China | MT560652 | MT560613 | MT560630 | |
| Fusarium andiyazi | ZJ07 | Zhejiang China | MT560643 | MT560607 | MT560622 | |
| Fusarium andiyazi | ZJ08 | Zhejiang China | MT560644 | MT560608 | MT560623 | |
| Fusarium asiaticum | ZJ10 | Zhejiang China | MT560646 | MT560609 | MT560625 | |
| Fusarium asiaticum | ZJ12 | Zhejiang China | MT560648 | MT560610 | MT560627 | |
| Fusarium equiseti | ZJ09 | Zhejiang China | MT560645 | MT560624 | MT560634 | |
| Fusarium incarnatum | ZJ11 | Zhejiang China | MT560647 | MT560626 | MT560635 | |
| Fusarium incarnatum | AH04 | Anhui China | MT560653 | MT560631 | MT560636 | |
| Fusarium commune | ZJG17 | Zhejiang China | MT560649 | |||
| Fusarium andiyazi | CBS 119857 | Netherlands | KP662901 | LT996189 | KT154004 | |
| Fusarium_fujikuroi | NRRL 13566 | Taiwan | AF160279 | JX171456 | JX171570 | |
| Fusarium proliferatum | NRRL 22944 | Germany | AF160280 | JX171504 | HM068352 | |
| Fusarium_mangifera | NRRL 25226 | India | AF160281 | JX171509 | JX171622 | |
| Fusarium_sacchari | NRRL 13999 | India | AF160278 | JX171466 | JX171580 | |
| Fusarium_thapsinum | NRRL 22045 | Natal, South Africa | AF160270 | JX171487 | JX171600 | |
| Fusarium_subglutinans | NRRL 22016 | IL-USA | AF160289 | JX171486 | JX171599 | |
| Fusarium_circinatum | NRRL 25331 | CA-USA | AF160295 | JX171510 | JX171623 | |
| Fusarium_ananatum | NRRL 22945 | England | AF160297 | JX171505 | JX171618 | |
| Fusarium fujikuroi | NRRL 43610 | Iowa | HM347123 | HM347184 | EF470199 | |
| Fusarium verticillioides | NRRL43608 | Minnesota | HM347122 | HM347183 | EF470197 | |
| Fusarium proliferatum | NRRL43617 | Colorado | HM347124 | HM347185 | EF470206 | |
| Fusarium equiseti | InaCC F963 | Indonesia | LS479445 | LS479875 | LS479859 | |
| Fusarium asiaticum | NRRL 13818 | Japan | AF212451 | JX171459 | JX171573 | |
| Fusarium graminearum | NRRL 31084 | MI-USA | AY452957 | JX171531 | JX171644 | |
| Fusarium culmorum | NRRL 25475 | Denmark | KY873384 | JX171515 | JX171628 | |
| Fusarium pseudograminearum | NRRL 28062 | USA | AF212468 | JX171524 | JX171637 | |
| Fusarium sporotrichioides | NRRL 25479 | Germany | HM744652 | HM347144 | HQ154441 | |
| Fusarium graminearum | NRRL 43641 | Missouri | GQ505430 | HM347192 | GQ505494 | |
| Fusarium brachygibbosum | NRRL 34033 | Texas unknown | GQ505418 | HM347172 | GQ505482 | |
| Fusarium dimerum | NRRL36140 | Zhejiang China | HM347133 | HM347203 | HM347218 | |
| Fusarium incarnatum | CBS143596 | Moghan-Ardabil Iran | LT970779 | LT970751 | LT970815 | |
| Fusarium incarnatum | CBS143595 | Moghan-Ardabil Iran | LT970778 | LT970750 | LT970814 | |
| Fusarium incarnatum | CBS143598 | Moghan-Ardabil Iran | LT970780 | LT970752 | LT970816 | |
| Fusarium incarnatum | CBS143603 | Moghan-Ardabil Iran | LT970782 | LT970754 | LT970818 | |
| Fusarium incarnatum | CBS143600 | Moghan-Ardabil Iran | LT970781 | LT970753 | LT970817 | |
| Fusarium incarnatum | CBS143606 | Moghan-Ardabil Iran | LT970783 | LT970755 | LT970819 | |
| Fusarium incarnatum | CBS143597 | Moghan-Ardabil Iran | LT970784 | LT970756 | LT970820 | |
| Fusarium equiseti | NRRL 20697 | Chile | GQ505594 | GQ505772 | GQ505683 | |
| Fusarium equiseti | NRRL 36321 | Netherlands | GQ505647 | GQ505825 | GQ505736 | |
| Fusarium equiseti | NRRL 36466 | Denmark | GQ505653 | GQ505831 | GQ505742 | |
| Fusarium_sporotrichioides | NRRL 3299 | WI-USA | JX171444 | JX171558 | ||
| Fusarium_langsethiae | NRRL 54940 | Norway | JX171550 | JX171662 | ||
| Fusarium_armeniacum | NRRL 6227 | MO-USA | JX171446 | JX171560 | ||
| Fusarium_sambucinum | NRRL 22187 | England | JX171493 | JX171606 | ||
| Fusarium_venenatum | NRRL 22196 | Germany | JX171494 | JX171607 | ||
| Fusarium_poae | NRRL 13714 | Manitoba, Canada | JX171458 | JX171572 | ||
| Fusarium_sciripi | NRRL 13402 | NSW, Australia | JX171566 | JX171452 | ||
| Fusarium_lacertarum | NRRL 20423 | India | JX171467 | JX171581 | ||
| Fusarium_equiseti | NRRL 20697 | Chile | JX171481 | JX171595 | ||
| Fusarium_sp._FIESC_26a | NRRL 26417 | Cuba | JX171522 | JX171635 | ||
| Fusarium_sp._FIESC_15a | NRRL 32175 | TX-USA | JX171532 | JX171645 | ||
| Fusarium_verticillioides | NRRL 20956 | CA-USA | JX171485 | JX171598 | ||
| Fusarium_xylarioides | NRRL 25486 | Ivory Coast | JX171517 | JX171630 | ||
| Fusarium torreyae | NRRL 54149 | FL-USA | JX171548 | JX171660 | ||
| Primer Type | Primer Sequence (5′-3′) | Length |
|---|---|---|
| F3 | TTGACAAAGTTCGGTGCC | 18 |
| B3 | TCTGACTTGATTCACAGATG | 20 |
| FIP (F1c + F2) | GCTACCAAGGTAAGGTATCTGCTAAGTTGTTTGTCGGCTGATC | 43 |
| BIP (B1c + B2) | GCTTCACTGGCCTTGGAGTCATAAGGTTTATGGAGACGCAC | 41 |
| LoopF | GGCGTTCATACCACGACCT | 19 |
| LoopB | GTACGGTATCACCGTTGCATT | 21 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, H.; Wu, N.; Jin, S.; Ahmed, T.; Wang, H.; Li, B.; Wu, X.; Bao, Y.; Liu, F.; Zhang, J.-Z. Identification of Rice Seed-Derived Fusarium spp. and Development of LAMP Assay against Fusarium fujikuroi. Pathogens 2021, 10, 1. https://doi.org/10.3390/pathogens10010001
Jiang H, Wu N, Jin S, Ahmed T, Wang H, Li B, Wu X, Bao Y, Liu F, Zhang J-Z. Identification of Rice Seed-Derived Fusarium spp. and Development of LAMP Assay against Fusarium fujikuroi. Pathogens. 2021; 10(1):1. https://doi.org/10.3390/pathogens10010001
Chicago/Turabian StyleJiang, Hubiao, Na Wu, Shaomin Jin, Temoor Ahmed, Hui Wang, Bin Li, Xiaobi Wu, Yidan Bao, Fei Liu, and Jing-Ze Zhang. 2021. "Identification of Rice Seed-Derived Fusarium spp. and Development of LAMP Assay against Fusarium fujikuroi" Pathogens 10, no. 1: 1. https://doi.org/10.3390/pathogens10010001
APA StyleJiang, H., Wu, N., Jin, S., Ahmed, T., Wang, H., Li, B., Wu, X., Bao, Y., Liu, F., & Zhang, J.-Z. (2021). Identification of Rice Seed-Derived Fusarium spp. and Development of LAMP Assay against Fusarium fujikuroi. Pathogens, 10(1), 1. https://doi.org/10.3390/pathogens10010001







