Epigenetic Mechanisms of Plant Adaptation to Cadmium and Heavy Metal Stress
Abstract
1. Introduction
2. Understanding Heavy Metal and Metalloid Stress
3. Sources and Types of Heavy Metals and Metalloids in the Environment
4. Impact of Heavy Metal and Metalloid Stress on Plant Growth and Development
5. Epigenetic Mechanisms in Plant Resilience to Heavy Metal Stress
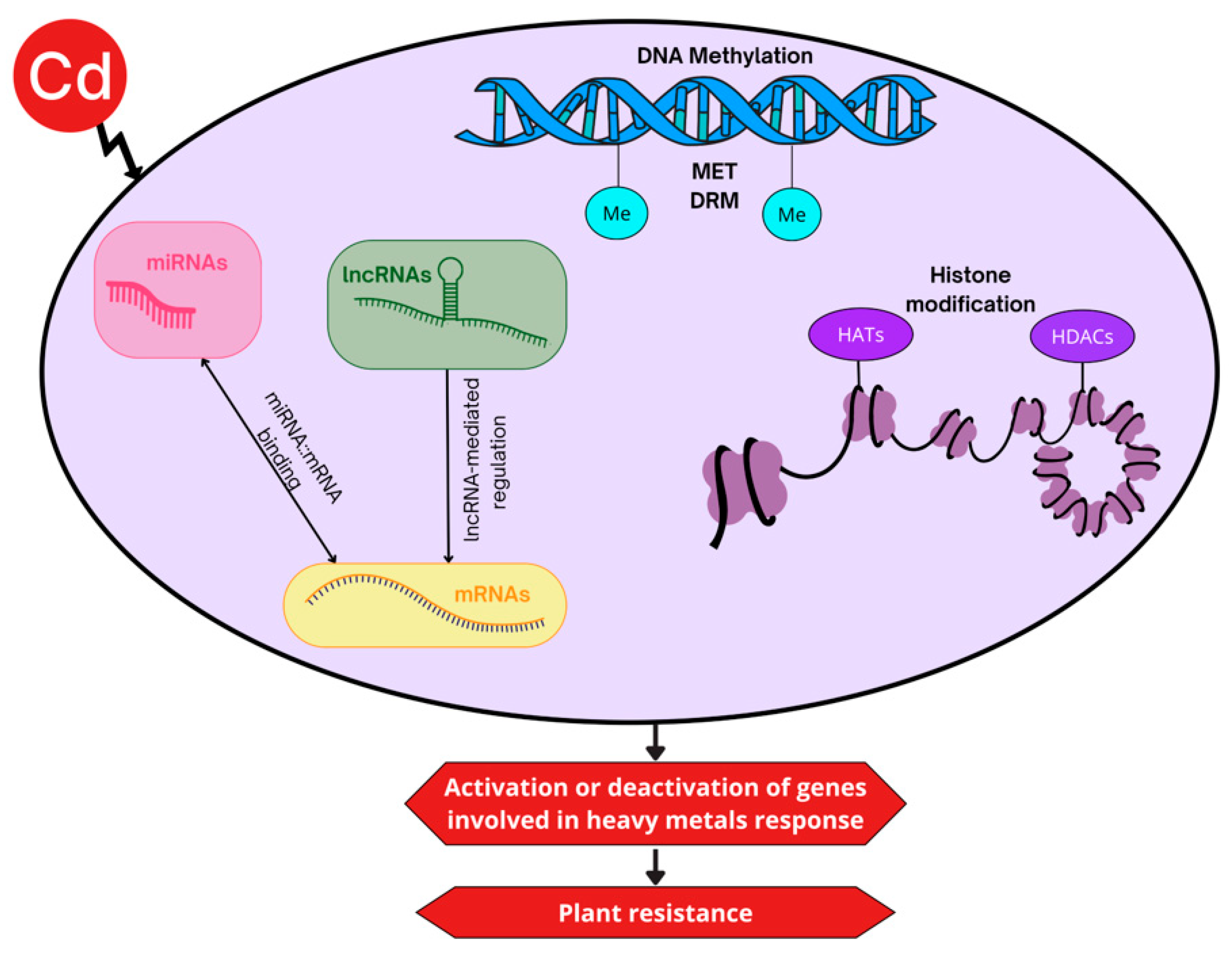
5.1. DNA Methylation
5.2. Histone Modifications
5.3. Non-Coding RNAs
5.4. Stress Memory and Transgenerational Epigenetic Inheritance
5.5. Epigenome Editing and Applications in Crop Improvement
6. Transgenerational Epigenetic Inheritance and Stress Memory
7. Chromatin Remodeling Under Cadmium Stress
8. The Role of DNA Methylation in Cadmium Stress Response
9. Impact of Cadmium Stress on Histone Acetylation
10. MicroRNAs and Their Role in Cadmium Tolerance
| Species | Findings | Citation |
|---|---|---|
| Nicotiana tabacum (Guiyan 1 vs. Yunyan 2) | High-throughput sequencing identified 72 known and 14 novel miRNAs differentially expressed. Twenty-eight known and five novel miRNAs were linked to tolerance. Cd accumulation was higher in Yunyan 2, but this cultivar showed smaller reductions in height and chlorophyll content, and a lower integrated stress score, suggesting cultivar-specific miRNA regulation. | [133] |
| Oryza sativa | Transgenic overexpression of miR390 elevated miR390 levels and reduced its target OsSRK transcript, confirming its role in cleaving a stress-responsive receptor kinase. | [129] |
| Triticum aestivum | qRT-PCR and psRNATarget analysis revealed organ-specific miRNA changes. Downregulation of miR398 increased CSD (Cu/Zn-SOD) but also H2O2, highlighting miR398’s role in balancing ROS detoxification. | [134] |
| Triticum aestivum (L17 vs. H17) | miRNA-seq and transcriptomics in low-Cd (L17) and high-Cd (H17) cultivars identified inversely regulated miR9664-3p and tea-miR159a. Thirty-two TaHMA genes were identified, with miRNA-mediated regulation of HMAs (e.g., TaHMA2;1) implicated in Cd sequestration and tolerance. | [137] |
11. Long Non-Coding RNAs (lncRNAs) in Heavy Metal Stress Response
| Species | Findings | Citation |
|---|---|---|
| Oryza sativa Indica “Huanghuazhan” | Hydroponic Cd and As exposure alters > 3300 lncRNAs; e.g., MSTRG.24054.4 acts in cis on nearby detoxification genes under combined stress. 10 µM CdCl2 + 250 mg/L melatonin reduces shoot Cd by 30%, improves K+/Ca2+ uptake, boosts photosynthesis, lowers MDA; transcriptome shows 2510 DE transcripts including six lncRNAs interacting with mRNAs for cell-wall enzymes and photosynthetic proteins, underpinning melatonin-mediated Cd tolerance. | [151] |
| Hordeum vulgare (Tibetan wild barley) | Al treatment induces 268 lncRNAs forming cis-acting lncRNA–mRNA pairs enriched in peroxisome and diterpenoid-biosynthesis pathways, implicating transcriptional/chromatin-level regulation of Al tolerance genes. | [152] |
| Hordeum vulgare (ZN8 vs. W6nk2) | Identified 9937 novel lncRNAs under 5 µM Cd; 5758 cis- and 4159 trans-acting pairs; eight lncRNAs act as miRNA mimics (e.g., targeting HvGAMYB), with virus-induced silencing of HvGAMYB causing Cd hypersensitivity and disrupted photosynthetic/antioxidant gene expression. | [154] |
| Hordeum vulgare (hull-less, X178 vs. X38) | RNA-Seq revealed 8299 lncRNAs (1884 cis, 3428 trans) under Cd stress; 26 lncRNAs and 150 mRNAs linked to tolerance; 12 lncRNAs form 18 lncRNA–mRNA pairs modulating DJ-1, EDR, PHT, ABC transporters; qRT-PCR validated candidates in genotype X178. | [155] |
| Cajanus cajan (pigeon pea) | Two modules enhance Al tolerance: TF CcNFYB3 → CcMATE35 increases citrate efflux; lncRNA CcLTCS → CcCS boosts citrate synthesis; co-overexpression synergistically improves Al detoxification and root health. | [156] |
| Triticum aestivum (wheat) | Cd stress induces >10,000 novel lncRNAs; 69 cis-regulate genes for Cd transport, photosynthesis, antioxidant defense; overexpression of lncRNA37228 (targets PSII D1) in Arabidopsis enhances Cd tolerance, linking PSII family to wheat Cd response. | [158] |
| Triticum aestivum (wheat) | TalncRNA18313 is Cd-inducible in leaves; heterologous expression in Arabidopsis lowers MDA and raises CAT, SOD, peroxidase activities; RNA-seq of overexpressors under Cd stress identifies 370 DE genes enriched in transcriptional regulation and antioxidative defense pathways. | [159] |
| Sorghum bicolor (sweet sorghum, H18 vs. L69) | lncRNA-seq in roots identified 1988 lncRNAs; 52 and 69 DE in H18 and L69 under Cd; 65 lncRNAs target 117 miRNAs, 1888 cis-genes; lncRNA 15962 sequesters sbi-miR5565e to derepress cell-wall genes; overexpression of four lncRNAs upregulates their cis-targets (including SbYS1); miRNA inhibition of lncRNA 11558 decreases SbYS1, confirming positive regulation. | [160] |
12. Future Perspectives: Epigenome Editing and Crop Improvement for Heavy Metals Resistance
13. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Abhinandan, K.; Skori, L.; Stanic, M.; Hickerson, N.M.; Jamshed, M.; Samuel, M.A. Abiotic Stress Signaling in Wheat–an Inclusive Overview of Hormonal Interactions during Abiotic Stress Responses in Wheat. Front. Plant Sci. 2018, 9, 734. [Google Scholar] [CrossRef] [PubMed]
- Devi, E.L.; Kumar, S.; Singh, T.B.; Sharma, S.K.; Beemrote, A.; Devi, C.P.; Chongtham, S.; Singh, C.H.; Yumlembam, R.A.; Haribhushan, A.; et al. Adaptation Strategies and Defence Mechanisms of Plants during Environmental Stress. In Medicinal Plants and Environmental Challenges; Springer: Berlin/Heidelberg, Germany, 2017; pp. 359–413. [Google Scholar]
- Annacondia, M.L.; Magerøy, M.H.; Martinez, G. Stress Response Regulation by Epigenetic Mechanisms: Changing of the Guards. Physiol. Plant. 2018, 162, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Begcy, K.; Dresselhaus, T. Epigenetic Responses to Abiotic Stresses during Reproductive Development in Cereals. Plant Reprod. 2018, 31, 343–355. [Google Scholar] [CrossRef]
- Subhanullah, M.; Hassan, N.; Ali, S.; Saleh, I.A.; Ilyas, M.; Rawan, B.; Ullah, W.; Iqbal, B.; Okla, M.K.; Alaraidh, I.A.; et al. The Detrimental Effects of Heavy Metals on Tributaries Exert Pressure on Water Quality, Crossocheilus Aplocheilus, and the Well-Being of Human Health. Sci. Rep. 2024, 14, 2868. [Google Scholar] [CrossRef]
- Zhao, H.; Wu, Y.; Lan, X.; Yang, Y.; Wu, X.; Du, L. Comprehensive Assessment of Harmful Heavy Metals in Contaminated Soil in Order to Score Pollution Level. Sci. Rep. 2022, 12, 3552. [Google Scholar] [CrossRef]
- López-González, D.; Bruno, L.; Díaz-Tielas, C.; Lupini, A.; Aci, M.M.; Talarico, E.; Madeo, M.L.; Muto, A.; Sánchez-Moreiras, A.M.; Araniti, F. Short-Term Effects of Trans-Cinnamic Acid on the Metabolism of Zea mays L. Roots. Plants 2023, 12, 189. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, J.; Zhao, L.; Yang, S.; Song, Y. Impact of heavy metal stresses on the growth and auxin homeostasis of Arabidopsis seedlings. Biometals 2015, 28, 123–132. [Google Scholar] [CrossRef]
- Luo, Q.; Sun, L.; Hu, X.; Zhou, R. The variation of root exudates from the hyperaccumulator Sedum alfredii under cadmium stress: Metabonomics analysis. PLoS ONE 2014, 9, e115581. [Google Scholar] [CrossRef]
- Shafiq, S.; Zeb, Q.; Ali, A.; Sajjad, Y.; Nazir, R.; Widemann, E.; Liu, L. Lead, Cadmium and Zinc Phytotoxicity Alter DNA Methylation Levels to Confer Heavy Metal Tolerance in Wheat. Int. J. Mol. Sci. 2019, 20, 4676. [Google Scholar] [CrossRef]
- He, J.-Y.; Ren, Y.-F.; Zhu, C.; Jiang, D.-A. Effects of cadmium stress on seed germination, seedling growth and seed amylase activities in rice (Oryza sativa). Rice Sci. 2008, 15, 319–325. [Google Scholar] [CrossRef]
- Fan, S.K.; Ye, J.Y.; Zhang, L.L.; Chen, H.S.; Zhang, H.H.; Zhu, Y.X.; Liu, X.X.; Jin, C.W. Inhibition of DNA Demethylation Enhances Plant Tolerance to Cadmium Toxicity by Improving Iron Nutrition. Plant Cell Environ. 2020, 43, 275–291. [Google Scholar] [CrossRef]
- Ankush; Ritambhara; Lamba, S.; Deepika; Prakash, R. Cadmium in Environment-An Overview. In Cadmium Toxicity in Water: Challenges and Solutions; Jha, A.K., Kumar, N., Eds.; Springer Nature: Cham, Switzerland, 2024; pp. 3–20. [Google Scholar]
- Mourato, M.; Pinto, F.; Moreira, I.; Sales, J.; Leitão, I.; Martins, L.L. The Effect of Cd Stress in Mineral Nutrient Uptake in Plants. In Cadmium Toxicity and Tolerance in Plants; Elsevier: Amsterdam, The Netherlands, 2019; pp. 327–348. [Google Scholar]
- Brunetti, P.; Zanella, L.; De Paolis, A.; Di Litta, D.; Cecchetti, V.; Falasca, G.; Barbieri, M.; Altamura, M.M.; Costantino, P.; Cardarelli, M. Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis. J. Exp. Bot. 2015, 66, 3815–3829. [Google Scholar] [CrossRef]
- Guan, M.Y.; Zhang, H.H.; Pan, W.; Jin, C.W.; Lin, X.Y. Sulfide Alleviates Cadmium Toxicity in Arabidopsis Plants by Altering the Chemical Form and the Subcellular Distribution of Cadmium. Sci. Total Environ. 2018, 627, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, A.; Prasad, M. Exogenous Citrate and Malate Alleviate Cadmium Stress in Oryza sativa L.: Probing Role of Cadmium Localization and Iron Nutrition. Ecotoxicol. Environ. Saf. 2018, 166, 215–222. [Google Scholar] [CrossRef]
- Mathur, J.; Chauhan, P. Mechanism of Toxic Metal Uptake and Transport in Plants. In Sustainable Solutions for Elemental Deficiency and Excess in Crop Plants; Springer: Berlin/Heidelberg, Germany, 2020; pp. 335–349. [Google Scholar]
- Metwally, A.; Safronova, V.I.; Belimov, A.A.; Dietz, K.-J. Genotypic variation of the response to cadmium toxicity in Pisum sativum L. J. Exp. Bot. 2005, 56, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Serrano, M.; Romero-Puertas, M.C.; Zabalza, A.; Corpas, F.J.; Gómez, M.; Del Río, L.A.; Sandalio, L.M. Cadmium effect on oxidative metabolism of pea (Pisum sativum L.) roots. Imaging of reactive oxygen species and nitric oxide accumulation in vivo. Plant Cell Environ. 2006, 29, 1532–1544. [Google Scholar] [CrossRef] [PubMed]
- Hačkuličová, D.; Labancová, E.; Vivodová, Z.; Danchenko, M.; Holeková, K.; Bajus, M.; Kučerová, D.; Baráth, P.; Kollárová, K. Modification of peroxidase activity and proteome in maize exposed to cadmium in the presence of galactoglucomannan oligosaccharides. Ecotoxicol. Environ. Saf. 2025, 290, 117732. [Google Scholar] [CrossRef]
- Khan, N.M.; Ali, A.; Wan, Y.; Zhou, G. Genome-Wide Identification of Heavy-Metal ATPases Genes in Areca catechu: Investigating Their Functionality under Heavy Metal Exposure. BMC Plant Biol. 2024, 24, 484. [Google Scholar] [CrossRef]
- Bruno, L.; Araniti, F.; Talarico, E.; Greco, E.; Muto, A.; Pacenza, M.; Chiappetta, A.; Bitonti, M.B. Transcriptomic and metabolomic analysis of the main stress-related pathways in the DNA methylation-defective ddc mutant of Arabidopsis thaliana exposed to cadmium. Plant Biosyst. 2025. [Google Scholar] [CrossRef]
- Pacenza, M.; Muto, A.; Chiappetta, A.; Mariotti, L.; Talarico, E.; Picciarelli, P.; Picardi, E.; Bruno, L.; Bitonti, M.B. In Arabidopsis thaliana Cd differentially impacts on hormone genetic pathways in the methylation defective ddc mutant compared to wild type. Sci. Rep. 2021, 11, 10965. [Google Scholar] [CrossRef] [PubMed]
- Galviz, Y.C.; Ribeiro, R.V.; Souza, G.M. Yes, plants do have memory. Theor. Exp. Plant Physiol. 2020, 32, 195–202. [Google Scholar] [CrossRef]
- Chandra, R.; Kumar, V. Phytoextraction of heavy metals by potential native plants and their microscopic observation of root growing on stabilised distillery sludge as a prospective tool for in situ phytoremediation of industrial waste. Environ. Sci. Pollut. Res. Int. 2017, 24, 2605–2619. [Google Scholar] [CrossRef]
- Makarova, A.; Nikulina, E.; Avdeenkova, T.; Pishaeva, K. The Improved Phytoextraction of Heavy Metals and the Growth of Trifolium repens L.: The Role of K2HEDP and Plant Growth Regulators Alone and in Combination. Sustainability. 2021, 13, 2432. [Google Scholar] [CrossRef]
- Abid, H.; Mahroof, S.; Ahmad, S.M.; Sadia, S.; Iqbal, U.; Mehmood, A.; Shehzad, M.A.; Basit, A.; Tahir, M.M.; Awan, U.A.; et al. Harnessing native plants for sustainable heavy metal phytoremediation in crushing industry soils of Muzaffarabad. Environ. Technol. Innov. 2025, 38, 104141. [Google Scholar] [CrossRef]
- Roca Paixão, J.F.; Gillet, F.X.; Ribeiro, T.P.; Bournaud, C.; Lourenço-Tessutti, I.T.; Noriega, D.D.; Melo, B.P.; de Almeida-Engler, J.; Grossi-de-Sa, M.F. Improved drought stress tolerance in Arabidopsis by CRISPR/dCas9 fusion with a histone AcetylTransferase. Sci. Rep. 2019, 9, 8080. [Google Scholar] [CrossRef] [PubMed]
- Gallo-Franco, J.J.; Sosa, C.C.; Ghneim-Herrera, T.; Quimbaya, M. Epigenetic Control of Plant Response to Heavy Metal Stress: A New View on Aluminum Tolerance. Front. Plant Sci. 2020, 11, 602625. [Google Scholar] [CrossRef]
- Cheng, M.; Zhou, Q.; Wang, L.; Jiao, Y.; Liu, Y.; Tan, L.; Zhu, H.; Nagawa, S.; Wei, H.; Yang, Z.; et al. A New Mechanism by Which Environmental Hazardous Substances Enhance Their Toxicities to Plants. J. Hazard. Mater. 2022, 421, 126802. [Google Scholar] [CrossRef] [PubMed]
- Talarico, E.; Zambelli, A.; Araniti, F.; Greco, E.; Chiappetta, A.; Bruno, L. Unravelling the Epigenetic Code: DNA Methylation in Plants and Its Role in Stress Response. Epigenomes 2024, 8, 30. [Google Scholar] [CrossRef]
- Feng, S.J.; Liu, X.S.; Ma, L.Y.; Rono, J.K.; Yang, Z.M. Identification of Epigenetic Mechanisms in Paddy Crop Associated with Lowering Environmentally Related Cadmium Risks to Food Safety. Environ. Pollut. 2020, 256, 113464. [Google Scholar] [CrossRef]
- Mitra, S.; Chakraborty, A.J.; Tareq, A.M.; Emran, T.B.; Nainu, F.; Khusro, A.; Idris, A.M.; Khandaker, M.U.; Osman, H.; Alhumaydhi, F.A.; et al. Impact of Heavy Metals on the Environment and Human Health: Novel Therapeutic Insights to Counter the Toxicity. J. King Saud Univ.-Sci. 2022, 34, 101865. [Google Scholar] [CrossRef]
- Remy, E.; Duque, P. Assessing Tolerance to Heavy-Metal Stress in Arabidopsis thaliana Seedlings. In Environmental Responses in Plants: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2016; pp. 197–208. [Google Scholar]
- Huihui, Z.; Xin, L.; Zisong, X.; Yue, W.; Zhiyuan, T.; Meijun, A.; Yuehui, Z.; Wenxu, Z.; Nan, X.; Guangyu, S. Toxic effects of heavy metals Pb and Cd on mulberry (Morus alba L.) seedling leaves: Photosynthetic function and reactive oxygen species (ROS) metabolism responses. Ecotoxicol. Environ. Saf. 2020, 1, 195–110469. [Google Scholar] [CrossRef]
- Pan, C.; Lu, H.; Yang, C.; Wang, L.; Chen, J.; Yan, C. Comparative Transcriptome Analysis Reveals Different Functions of Kandelia obovata Superoxide Dismutases in Regulation of Cadmium Translocation. Sci. Total Environ. 2021, 771, 144922. [Google Scholar] [CrossRef]
- Bruno, L.; Talarico, E.; Cabeiras-Freijanes, L.; Madeo, M.L.; Muto, A.; Minervino, M.; Lucini, L.; Miras-Moreno, B.; Sofo, A.; Araniti, F. Coumarin Interferes with Polar Auxin Transport Altering Microtubule Cortical Array Organization in Arabidopsis thaliana (L.) Heynh. Root Apical Meristem. Int. J. Mol. Sci. 2021, 22, 7305. [Google Scholar] [CrossRef]
- Li, Z.; Yuan, Y.; Xiang, L.; Su, Q.; Liu, Z.; Wu, W.; Huang, Y.; Tu, S. Silicon-Rich Materials Detoxify Multiple Heavy Metals in Wheat by Regulating Oxidative Stress and Heavy Metal Subcellular Distribution. Sustainability 2022, 14, 16417. [Google Scholar] [CrossRef]
- Gichner, T.; Žnidar, I.; Száková, J. Evaluation of DNA damage and mutagenicity induced by lead in tobacco plants. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2008, 652, 186–190. [Google Scholar] [CrossRef]
- Han, M.; Chen, Z.; Sun, G.; Feng, Y.; Guo, Y.; Bai, S.; Yan, X. Nano-Fe3O4: Enhancing the tolerance of Elymus nutans to Cd stress through regulating programmed cell death. Environ. Pollut. 2024, 360, 124711. [Google Scholar] [CrossRef]
- Liu, J.G.; Liang, J.S.; Li, K.Q.; Zhang, Z.J.; Yu, B.Y.; Lu, X.L.; Yang, J.C.; Zhu, Q.S. Correlations between cadmium and mineral nutrients in absorption and accumulation in various genotypes of rice under cadmium stress. Chemosphere 2003, 52, 1467–1473. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, V.; Nogueira, G.; Monteiro, G.; Júnior, W.; de Freitas, J.M.N.; Neto, C. Influence of Heavy Metals on the Nitrogen Metabolism in Plants. In Nitrogen in Agriculture: Physiological, Agricultural and Ecological Aspects; Books on Demand: Singapore, 2021. [Google Scholar]
- Sharma, P.; Dubey, R.S. Involvement of oxidative stress and role of antioxidative defense system in growing rice seedlings exposed to toxic concentrations of aluminium. Plant Cell Rep. 2007, 26, 2027–2038. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, A.; AbdElgawad, H.; Fidalgo, F.; Teixeira, J.; Matos, M.; Tamagnini, P.; Fernandes, R.; Figueiredo, F.; Azenha, M.; Teles, L.O.; et al. Subcellular compartmentalization of aluminium reduced its hazardous impact on rye photosynthesis. Environ. Pollut. 2022, 315, 120313. [Google Scholar] [CrossRef]
- Chao, D.Y.; Chen, Y.; Chen, J.; Shi, S.; Chen, Z.; Wang, C.; Danku, J.M.; Zhao, F.J.; Salt, D.E. Salt Genome-wide association mapping identifies a new arsenate reductase enzyme critical for limiting arsenic accumulation in plants. PLoS Biol. 2024, 12, e1002009. [Google Scholar]
- Tyagi, P.; Singh, A.; Ranjan, R. Hazardous Elements in Plants: Sources, Effect and Management. In Hazardous and Trace Materials in Soil and Plants; Elsevier: Amsterdam, The Netherlands, 2022; pp. 113–128. [Google Scholar]
- Singh, S.; Parihar, P.; Singh, R.; Singh, V.P.; Prasad, S.M. Heavy Metal Tolerance in Plants: Role of Transcriptomics, Proteomics, Metabolomics, and Ionomics. Front. Plant Sci. 2016, 6, 1143. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, R.; Ferreira, C.S.; Viela, F.L. Determination of Soluble/Exchangeable Metals in Peri-Urban Farmland (Ribeira Dos Covões) of Central Portugal. In Proceedings of the 4th World Congress on New Technologies (NewTech’18), Madrid, Spain, 19–21 August 2018. [Google Scholar]
- Sharma, J.K.; Kumar, N.; Singh, N.P.; Santal, A.R. Phytoremediation Technologies and Their Mechanism for Removal of Heavy Metal from Contaminated Soil: An Approach for a Sustainable Environment. Front. Plant Sci. 2023, 14, 1076876. [Google Scholar] [CrossRef] [PubMed]
- Priya, A.K.; Muruganandam, M.; Ali, S.S.; Kornaros, M. Clean-Up of Heavy Metals from Contaminated Soil by Phytoremediation: A Multidisciplinary and Eco-Friendly Approach. Toxics 2023, 11, 422. [Google Scholar] [CrossRef]
- Shah, F.U.R.; Ahmad, N.; Masood, K.R.; Peralta-Videa, J.R.; Ahmad, F.u.D. Heavy Metal Toxicity in Plants. In Plant Adaptation and Phytoremediation; Springer: Dordrecht, The Netherlands, 2010; pp. 71–97. [Google Scholar]
- Mahmud, J.A.; Hasanuzzaman, M.; Nahar, K.; Bhuyan, M.H.M.B.; Fujita, M. Insights into citric acid-induced cadmium tolerance and phytoremediation in Brassica juncea L.: Coordinated functions of metal chelation, antioxidant defense and glyoxalase systems. Ecotoxicol. Environ. Saf. 2018, 147, 990–1001. [Google Scholar] [CrossRef]
- Liu, X.; Chen, J.; Wang, G.H.; Wang, W.H.; Shen, Z.J.; Luo, M.R.; Gao, G.F.; Simon, M.; Ghoto, K.; Zheng, H.L. Hydrogen sulfide alleviates zinc toxicity by reducing zinc uptake and regulating genes expression of antioxidative enzymes and metallothioneins in roots of the cadmium/zinc hyperaccumulator Solanum nigrum L. Plant Soil. 2016, 400, 177–192. [Google Scholar] [CrossRef]
- Yang, X.-E.; Jin, X.-F.; Feng, Y.; Islam, E. Molecular Mechanisms and Genetic Basis of Heavy Metal Tolerance/Hyperaccumulation in Plants. J. Integr. Plant Biol. 2005, 47, 1025–1035. [Google Scholar] [CrossRef]
- Fernandez, L.R.; Vandenbussche, G.; Roosens, N.; Govaerts, C.; Goormaghtigh, E.; Verbruggen, N. Metal binding properties and structure of a type III metallothionein from the metal hyperaccumulator plant Noccaea caerulescens. Biochim. Biophys. Acta 2012, 1824, 1016–1023. [Google Scholar] [CrossRef]
- Bhat, J.A.; Shivaraj, S.; Singh, P.; Navadagi, D.B.; Tripathi, D.K.; Dash, P.K.; Solanke, A.U.; Sonah, H.; Deshmukh, R. Role of Silicon in Mitigation of Heavy Metal Stresses in Crop Plants. Plants 2019, 8, 71. [Google Scholar] [CrossRef]
- Rai, G.K.; Bhat, B.A.; Mushtaq, M.; Tariq, L.; Rai, P.K.; Basu, U.; Dar, A.A.; Islam, S.T.; Dar, T.U.; Bhat, J.A. Insights into Decontamination of Soils by Phytoremediation: A Detailed Account on Heavy Metal Toxicity and Mitigation Strategies. Physiol. Plant. 2021, 173, 287–304. [Google Scholar] [CrossRef]
- Tiwari, S.; Lata, C. Heavy Metal Stress, Signaling, and Tolerance Due to Plant-Associated Microbes: An Over-1369 view. Front. Plant Sci. 2018, 9, 452. [Google Scholar] [CrossRef]
- Briffa, J.; Sinagra, E.; Blundell, R. Heavy Metal Pollution in the Environment and Their Toxicological Effects on Humans. Heliyon 2020, 6, e04691. [Google Scholar] [CrossRef]
- Ahmad, Z.; Khan, S.M.; Page, S.E.; Balzter, H.; Ullah, A.; Ali, S.; Jehangir, S.; Ejaz, U.; Afza, R.; Razzaq, A.; et al. Environmental Sustainability and Resilience in a Polluted Ecosystem via Phytoremediation of Heavy Metals and Plant Physiological Adaptations. J. Clean. Prod. 2023, 385, 135733. [Google Scholar] [CrossRef]
- Gyamfi, E.; Appiah-Adjei, E.K.; Adjei, K.A. Potential Heavy Metal Pollution of Soil and Water Resources from Artisanal Mining in Kokoteasua, Ghana. Groundw. Sustain. Dev. 2019, 8, 450–456. [Google Scholar] [CrossRef]
- Akoto, O.; Yakubu, S.; Ofori, L.A.; Bortey-Sam, N.; Boadi, N.O.; Horgah, J.; Sackey, L.N. Multivariate Studies and Heavy Metal Pollution in Soil from Gold Mining Area. Heliyon 2023, 9, e12661. [Google Scholar] [CrossRef]
- Akinbile, C.O.; Erazua, A.E.; Babalola, T.E.; Ajibade, F.O. Environmental Implications of Animal Wastes Pollution on Agricultural Soil and Water Quality. Soil Water Res. 2016, 11, 172–180. [Google Scholar] [CrossRef]
- Tiwari, R.M.; Liu, J.; Xie, Y.; Yao, S.; Liu, S.; Wu, S.; Liu, J.; Qian, H.; Lei, Z.; Zhang, H.; et al. Decoupling the Impact of Biodiversity and Environmental Factors on the Biomass and Biomass Growth of Trees in Subtropical Forests. J. Plant Ecol. 2023, 16, rtac040. [Google Scholar] [CrossRef]
- Ismael, M.A.; Elyamine, A.M.; Moussa, M.G.; Cai, M.; Zhao, X.; Hu, C. Cadmium in plants: Uptake, toxicity, and its interactions with selenium fertilizers. Metallomics 2019, 11, 255–277. [Google Scholar] [CrossRef]
- Ranieri, E.; Moustakas, K.; Barbafieri, M.; Ranieri, A.C.; Herrera-Melián, J.A.; Petrella, A.; Tommasi, F. Phytoextraction technologies for mercury-and chromium-contaminated soil: A review. J. Chem. Technol. Biotech. 2020, 95, 317–327. [Google Scholar] [CrossRef]
- Abbas, G.; Murtaza, B.; Bibi, I.; Shahid, M.; Niazi, N.K.; Khan, M.I.; Amjad, M.; Hussain, M.; Natasha. Arsenic Uptake, Toxicity, Detoxification, and Speciation in Plants: Physiological, Biochemical, and Molecular Aspects. Int. J. Environ. Res. Public Health 2018, 15, 59. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Y.; Ippolito, J.A.; Xing, W.; Qiu, K.; Yang, H. Lead smelting effects heavy metal concentrations in soils, wheat, and potentially humans. Environ. Pollut. 2020, 257, 113641. [Google Scholar] [CrossRef] [PubMed]
- Ackova, D.G. Heavy metals and their general toxicity on plants. Plant Sci. Today. 2018, 5, 15–19. [Google Scholar]
- Zhang, B.; Li, Z.; Feng, Y.; Qaharaduqin, S.; Liu, W.; Yan, Y. Impact of Cd and Pb on the photosynthetic and antioxidant systems of Hemerocallis citrina Baroni as revealed by physiological and transcriptomic analyses. Plant Cell Rep. 2024, 43, 226. [Google Scholar] [CrossRef]
- Nazar, R.; Iqbal, N.; Masood, A.; Syeed, S.; Khan, N.A. Cadmium Toxicity in Plants and Role of Mineral Nutrients in Its Alleviation. Am. J. Plant Sci. 2012, 3, 1476–1489. [Google Scholar] [CrossRef]
- Molas, J. Changes of chloroplast ultrastructure and total chlorophyll concentration in cabbage leaves caused by excess of organic Ni (II) complexes. Environ. Exp. Bot. 2002, 47, 115–126. [Google Scholar] [CrossRef]
- Sofo, A.; Khan, N.A.; D’ Ippolito, I.; Reyes, F. Subtoxic levels of some heavy metals cause differential root-shoot structure, morphology and auxins levels in Arabidopsis thaliana. Plant Physiol. Biochem. 2022, 173, 68–75. [Google Scholar] [CrossRef]
- Bruno, L.; Pacenza, M.; Forgione, I.; Lamerton, L.R.; Greco, M.; Chiappetta, A.; Bitonti, M.B. In Arabidopsis thaliana Cadmium Impact on the Growth of Primary Root by Altering SCR Expression and Auxin-Cytokinin Cross-Talk. Front. Plant Sci. 2017, 8, 1323. [Google Scholar] [CrossRef]
- Araniti, F.; Talarico, E.; Madeo, M.L.; Greco, E.; Minervino, M.; Álvarez-Rodríguez, S.; Muto, A.; Ferrari, M.; Chiappetta, A.; Bruno, L. Short-Term Exposition to Acute Cadmium Toxicity Induces the Loss of Root Gravitropic Stimuli Perception through PIN2-Mediated Auxin Redistribution in Arabidopsis thaliana (L.) Heynh. Plant Sci. 2023, 332, 111726. [Google Scholar] [CrossRef]
- Bruno, L.; Talarico, E.; Madeo, M.L.; Muto, A.; Minervino, M.; Araniti, F.; Bitonti, M.B.; Chiappetta, A. Cadmium Affects Cell Niches Maintenance in Arabidopsis thaliana Post-Embryonic Shoot and Root Apical Meristem by Altering the Expression of WUS/WOX Homolog Genes and Cytokinin Accumulation. Plant Physiol. Biochem. 2021, 167, 785–794. [Google Scholar]
- Tavarez, M.; Grusak, M.A.; Sankaran, R.P. Effects of Zinc Fertilization on Grain Cadmium Accumulation, Gene Expression, and Essential Mineral Partitioning in Rice. Agronomy 2022, 12, 2182. [Google Scholar] [CrossRef]
- Hédiji, H.; Djebali, W.; Belkadhi, A.; Cabasson, C.; Moing, A.; Rolin, D.; Brouquisse, R.; Gallusci, P.; Chaïbi, W. Impact of long-term cadmium exposure on mineral content of Solanum lycopersicum plants: Consequences on fruit production. S. Afr. J. Bot. 2015, 97, 176–181. [Google Scholar] [CrossRef]
- Tricker, P.J. Transgenerational inheritance or resetting of stress-induced epigenetic modifications: Two sides of the same coin. Front. Plant Sci. 2015, 6, 699. [Google Scholar] [CrossRef]
- Boyko, A.; Blevins, T.; Yao, Y.; Golubov, A.; Bilichak, A.; Ilnytskyy, Y.; Hollunder, J.; Meins, F., Jr.; Kovalchuk, I. Transgenerational Adaptation of Arabidopsis to Stress Requires DNA Methylation and the Function of Dicer-Like Proteins. PLoS ONE 2010, 5, e9514. [Google Scholar] [CrossRef]
- Bilichak, A.; Ilnytskyy, Y.; Wóycicki, R.; Kepeshchuk, N.; Fogen, D.; Kovalchuk, I. The elucidation of stress memory inheritance in Brassica rapa plants. Front. Plant Sci. 2015, 6, 5. [Google Scholar] [CrossRef]
- Srivastava, S.; Suprasanna, P. MicroRNAs: Tiny, Powerful Players of Metal Stress Responses in Plants. Plant Physiol. Biochem. 2021, 166, 928–938. [Google Scholar] [CrossRef]
- Dumanović, J.; Nepovimova, E.; Natić, M.; Kuča, K.; Jaćević, V. The Significance of Reactive Oxygen Species and Antioxidant Defense System in Plants: A Concise Overview. Front. Plant Sci. 2021, 11, 552969. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Wang, R.; Sun, X.; Liu, L.; Liu, P.; Tang, J.; Zhang, C.; Liu, H. Heavy Metal Stress in Plants: Ways to Alleviate with Exogenous Substances. Sci. Total Environ. 2023, 897, 165397. [Google Scholar] [CrossRef]
- Sharma, S.S.; Dietz, K.-J.; Mimura, T. Vacuolar Compartmentalization as Indispensable Component of Heavy Metal Detoxification in Plants. Plant Cell Environ. 2016, 39, 1112–1126. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Martinoia, E.; Lee, Y. Vacuolar Transporters for Cadmium and Arsenic in Plants and Their Applications in Phytoremediation and Crop Development. Plant Cell Physiol. 2018, 59, 1317–1325. [Google Scholar] [CrossRef]
- Yan, A.; Wang, Y.; Tan, S.N.; Mohd Yusof, M.L.; Ghosh, S.; Chen, Z. Phytoremediation: A Promising Approach for Revegetation of Heavy Metal-Polluted Land. Front. Plant Sci. 2020, 11, 359. [Google Scholar] [CrossRef] [PubMed]
- Rao, K.P.; Vani, G.; Kumar, K.; Wankhede, D.P.; Misra, M.; Gupta, M.; Sinha, A.K. Arsenic stress activates MAP kinase in rice roots and leaves. Arch. Biochem. Biophys. 2011, 506, 73–82. [Google Scholar] [CrossRef]
- Yeh, C.-M.; Hung, W.-C.; Huang, H.-J. Copper treatment activates mitogen-activated protein kinase signalling in rice. Physiol. Plant. 2003, 119, 392–399. [Google Scholar] [CrossRef]
- Dutta, S.; Mitra, M.; Agarwal, P.; Mahapatra, K.; De, S.; Sett, U.; Roy, S. Oxidative and Genotoxic Damages in Plants in Response to Heavy Metal Stress and Maintenance of Genome Stability. Plant Signal. Behav. 2018, 13, e1460048. [Google Scholar] [CrossRef]
- Phillips, T. The Role of Methylation in Gene Expression. Nat. Educ. 2008, 1, 116. [Google Scholar]
- Panchin, A.Y.; Makeev, V.J.; Medvedeva, Y.A. Preservation of Methylated CpG Dinucleotides in Human CpG Islands. Biol. Direct 2016, 11, 1–15. [Google Scholar] [CrossRef]
- Forgione, I.; Wołoszyńska, M.; Pacenza, M.; Chiappetta, A.; Greco, M.; Araniti, F.; Abenavoli, M.R.; Van Lijsebettens, M.; Bitonti, M.B.; Bruno, L. Hypomethylated drm1 drm2 cmt3 Mutant Phenotype of Arabidopsis thaliana Is Related to Auxin Pathway Impairment. Plant Sci. 2019, 280, 383–396. [Google Scholar] [CrossRef]
- Forgione, I.; Muto, A.; Woloszynska, M.; Chiappetta, A.A.; Ferrari, M.; Van Lijsebettens, M.; Bitonti, M.B.; Bruno, L. Epigenetic Mechanisms Affect the Curled Leaf Phenotype in the Hypomethylated ddc Mutant of Arabidopsis thaliana. Plant Sci. 2022, 319, 111254. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Narayana Chevala, V.; Shankar, R.; Jain, M. Divergent DNA Methylation Patterns Associated with Gene Expression in Rice Cultivars with Contrasting Drought and Salinity Stress Response. Sci. Rep. 2015, 5, 14922. [Google Scholar] [CrossRef]
- Akhter, Z.; Bi, Z.; Ali, K.; Sun, C.; Fiaz, S.; Haider, F.U.; Bai, J. In Response to Abiotic Stress, DNA Methylation Confers Epigenetic Changes in Plants. Plants 2021, 10, 1096. [Google Scholar] [CrossRef]
- Woloszynska, M.; Le Gall, S.; Neyt, P.; Boccardi, T.M.; Grasser, M.; Längst, G.; Aesaert, S.; Coussens, G.; Dhondt, S.; Van De Slijke, E.; et al. Histone 2B monoubiquitination complex integrates transcript elongation with RNA processing at circadian clock and flowering regulators. Proc. Natl. Acad. Sci. USA 2019, 116, 8060–8069. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, X.; Yang, Y.; Zhang, H.; Zhu, W.; Nie, W.-F. The Histone Variant Sl_H2A. Z Regulates Carotenoid Biosynthesis and Gene Expression during Tomato Fruit Ripening. Hortic. Res. 2021, 8, 85. [Google Scholar] [CrossRef] [PubMed]
- Weiner, A.; Hsieh, T.-H.S.; Appleboim, A.; Chen, H.V.; Rahat, A.; Amit, I.; Rando, O.J.; Friedman, N. High-Resolution Chromatin Dynamics during a Yeast Stress Response. Mol. Cell 2015, 58, 371–386. [Google Scholar] [CrossRef]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of Post-Transcriptional Regulation by microRNAs: Are the Answers in Sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.K. RNA-directed DNA methylation and demethylation in plants. Sci. China C. Life Sci. 2009, 52, 331–343. [Google Scholar] [CrossRef] [PubMed]
- Imaduwage, I.; Hewadikaram, M. Predicted Roles of Long Non-Coding RNAs in Abiotic Stress Tolerance Responses of Plants. Mol. Hortic. 2024, 4, 20. [Google Scholar] [CrossRef]
- Ou, X.; Zhang, Y.; Xu, C.; Lin, X.; Zang, Q.; Zhuang, T.; Jiang, L.; Von Wettstein, D.; Liu, B. Transgenerational Inheritance of Modified DNA Methylation Patterns and Enhanced Tolerance Induced by Heavy Metal Stress in Rice (Oryza sativa L.). PLoS ONE 2012, 7, e41143. [Google Scholar] [CrossRef]
- Akimoto, K.; Katakami, H.; Kim, H.J.; Ogawa, E.; Sano, C.M.; Wada, Y.; Sano, H. Epigenetic inheritance in rice plants. Ann. Bot. 2007, 100, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Lukić, N.; Schurr, F.M.; Trifković, T.; Kukavica, B.; Walter, J. Transgenerational stress memory in plants is mediated by upregulation of the antioxidative system. Environ. Exp. Bot. 2023, 205, 105129. [Google Scholar] [CrossRef]
- Ding, Y.; Ding, L.; Xia, Y.; Wang, F.; Zhu, C. Emerging Roles of microRNAs in Plant Heavy Metal Tolerance and Homeostasis. J. Agric. Food Chem. 2020, 68, 1958–1965. [Google Scholar] [CrossRef]
- Nilsson, E.E.; Skinner, M.K. Environmentally Induced Epigenetic Transgenerational Inheritance of Disease Susceptibility. Transl. Res. 2015, 165, 12–17. [Google Scholar] [CrossRef]
- Skinner, M.K. Environmental Stress and Epigenetic Transgenerational Inheritance. BMC Med. 2014, 12, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Skinner, M.K.; Haque, C.G.-B.M.; Nilsson, E.; Bhandari, R.; McCarrey, J.R. Correction: Environmentally Induced Transgenerational Epigenetic Reprogramming of Primordial Germ Cells and the Subsequent Germ Line. PLoS ONE 2013, 8, 10–1371. [Google Scholar] [CrossRef]
- Beck, D.; Nilsson, E.E.; Ben Maamar, M.; Skinner, M.K. Environmental Induced Transgenerational Inheritance Impacts Systems Epigenetics in Disease Etiology. Sci. Rep. 2022, 12, 5452. [Google Scholar] [CrossRef]
- Rendina González, A.P.; Preite, V.; Verhoeven, K.J.F.; Latzel, V. Transgenerational Effects and Epigenetic Memory in the Clonal Plant Trifolium repens. Front. Plant Sci. 2018, 9, 1677. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-N.; Zhu, C.; Jiang, J.; Zhang, H.; Zhu, J.-K.; Duan, C.-G. Epigenetic Regulation in Plant Abiotic Stress Responses. J. Integr. Plant Biol. 2020, 62, 563–580. [Google Scholar] [CrossRef] [PubMed]
- Jogam, P.; Sandhya, D.; Alok, A.; Peddaboina, V.; Allini, V.R.; Zhang, B. A Review on CRISPR/Cas-Based Epigenetic Regulation in Plants. Int. J. Biol. Macromol. 2022, 219, 1261–1271. [Google Scholar] [CrossRef]
- Abdulraheem, M.I.; Xiong, Y.; Moshood, A.Y.; Cadenas-Pliego, G.; Zhang, H.; Hu, J. Mechanisms of Plant Epigenetic Regulation in Response to Plant Stress: Recent Discoveries and Implications. Plants 2024, 13, 163. [Google Scholar] [CrossRef]
- Tonosaki, K.; Fujimoto, R.; Dennis, E.S.; Raboy, V.; Osabe, K. Will Epigenetics Be a Key Player in Crop Breeding? Front. Plant Sci. 2022, 13, 958350. [Google Scholar] [CrossRef]
- Mozgova, I.; Wildhaber, T.; Liu, Q.; Abou-Mansour, E.; L’Haridon, F.; Métraux, J.-P.; Gruissem, W.; Hofius, D.; Hennig, L. Chromatin Assembly Factor CAF-1 Represses Priming of Plant Defence Response Genes. Nat. Plants 2015, 1, 1–8. [Google Scholar] [CrossRef]
- Hossain, Z.; Huq, F. Studies on the interaction between Cd (2+) ions and DNA. J. Inorg. Biochem. 2002, 90, 85–96. [Google Scholar] [CrossRef]
- Shafiq, S.; Ali, A.; Sajjad, Y.; Zeb, Q.; Shahzad, M.; Khan, A.R.; Nazir, R.; Widemann, E. The Interplay between Toxic and Essential Metals for Their Uptake and Translocation Is Likely Governed by DNA Methylation and Histone Deacetylation in Maize. Int. J. Mol. Sci. 2020, 21, 6959. [Google Scholar] [CrossRef]
- Niekerk, L.-A.; Carelse, M.F.; Bakare, O.O.; Mavumengwana, V.; Keyster, M.; Gokul, A. The Relationship between Cadmium Toxicity and the Modulation of Epigenetic Traits in Plants. Int. J. Mol. Sci. 2021, 22, 7046. [Google Scholar] [CrossRef]
- Żabka, A.; Winnicki, K.; Polit, J.T.; Wróblewski, M.; Maszewski, J. Cadmium (II)-Induced Oxidative Stress Results in Replication Stress and Epigenetic Modifications in Root Meristem Cell Nuclei of Vicia Faba. Cells 2021, 10, 640. [Google Scholar] [CrossRef]
- Zhang, H.; Lang, Z.; Zhu, J.-K. Dynamics and Function of DNA Methylation in Plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef] [PubMed]
- Bräutigam, K.; Cronk, Q. DNA Methylation and the Evolution of Developmental Complexity in Plants. Front. Plant Sci. 2018, 9, 1447. [Google Scholar] [CrossRef] [PubMed]
- Greco, M.; Chiappetta, A.; Bruno, L.; Bitonti, M.B. In Posidonia oceanica Cadmium Induces Changes in DNA Methylation and Chromatin Patterning. J. Exp. Bot. 2012, 63, 695–709. [Google Scholar] [CrossRef]
- Bartels, A.; Han, Q.; Nair, P.; Stacey, L.; Gaynier, H.; Mosley, M.; Huang, Q.Q.; Pearson, J.K.; Hsieh, T.-F.; An, Y.-Q.C.; et al. Dynamic DNA Methylation in Plant Growth and Development. Int. J. Mol. Sci. 2018, 19, 2144. [Google Scholar] [CrossRef]
- Park, J.; Song, W.; Ko, D.; Eom, Y.; Hansen, T.H.; Schiller, M.; Lee, T.G.; Martinoia, E.; Lee, Y. The Phytochelatin Transporters AtABCC1 and AtABCC2 Mediate Tolerance to Cadmium and Mercury. Plant J. 2012, 69, 278–288. [Google Scholar] [CrossRef]
- Jiang, J.; Ding, A.B.; Liu, F.; Zhong, X. Linking Signaling Pathways to Histone Acetylation Dynamics in Plants. J. Exp. Bot. 2020, 71, 5179–5190. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Li, L.; Han, X.; Liu, T.; Jin, P.; Cai, L.; Xu, M.; Zhang, T.; Zhang, F.; Chen, J.; et al. Genome-Wide Identification of the Histone Acetyltransferase Gene Family in Triticum aestivum. BMC Genom. 2021, 22, 49. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Ye, Y.; Jiang, Z.; Wang, Y.; Zhu, C. MicroRNA390 Is Involved in Cadmium Tolerance and Accumulation in Rice. Front. Plant Sci. 2016, 7, 235. [Google Scholar] [CrossRef]
- Carbone, F.; Bruno, L.; Perrotta, G.; Bitonti, M.B.; Muzzalupo, I.; Chiappetta, A. Identification of miRNAs Involved in Fruit Ripening by Deep Sequencing of Olea europaea L. Transcriptome. PLoS ONE 2019, 14, e0221460. [Google Scholar] [CrossRef]
- Zhou, Z.S.; Song, J.B.; Yang, Z.M. Genome-Wide Identification of Brassica napus microRNAs and Their Targets in Response to Cadmium. J. Exp. Bot. 2012, 63, 4597–4613. [Google Scholar] [CrossRef]
- Gielen, H.; Remans, T.; Vangronsveld, J.; Cuypers, A. MicroRNAs in Metal Stress: Specific Roles or Secondary Responses? Int. J. Mol. Sci. 2012, 13, 15826–15847. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Zheng, W.; Cao, F.; Wu, F. Identification and Comparative Analysis of the microRNA Transcriptome in Roots of Two Contrasting Tobacco Genotypes in Response to Cadmium Stress. Sci. Rep. 2016, 6, 32805. [Google Scholar] [CrossRef]
- Qiu, Z.; Hai, B.; Guo, J.; Li, Y.; Zhang, L. Characterization of Wheat miRNAs and Their Target Genes Responsive to Cadmium Stress. Plant Physiol. Biochem. 2016, 101, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Serrano, M.; Romero-Puertas, M.C.; Pazmiño, D.M.; Testillano, P.S.; Risueño, M.C.; Del Río, L.A.; Sandalio, L.M. Cellular response of pea plants to cadmium toxicity: Cross talk between reactive oxygen species, nitric oxide, and calcium. Plant Physiol. 2009, 150, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Romero-Puertas, M.C.; Corpas, F.J.; Rodríguez-Serrano, M.; Gómez, M.; Del Río, L.A.; Sandalio, L.M. Differential expression and regulation of antioxidative enzymes by cadmium in pea plants. J. Plant Physiol. 2007, 164, 1346–1357. [Google Scholar] [CrossRef]
- Zhou, M.; Zheng, S.; Liu, R.; Lu, L.; Zhang, C.; Zhang, L.; Yant, L.; Wu, Y. The Genome-Wide Impact of Cadmium on microRNA and mRNA Expression in Contrasting Cd Responsive Wheat Genotypes. BMC Genom. 2019, 20, 615. [Google Scholar] [CrossRef]
- Shiv, A.; Krishna, H.; Sinha, N.; Priyadarshini, P.; Sahu, S.; Jain, N.; Singh, P.K.; Prabhu, K.V. Leaf Rust Responsive miRNA Mediated Regulation of Puccinia triticina Genes during Host Pathogen Interaction in Wheat. Indian J. Genet. Plant Breed. 2023, 83, 185–194. [Google Scholar]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Lu, X.; Chen, X.; Mu, M.; Wang, J.; Wang, X.; Wang, D.; Yin, Z.; Fan, W.; Wang, S.; Guo, L.; et al. Genome-Wide Analysis of Long Noncoding RNAs and Their Responses to Drought Stress in Cotton (Gossypium hirsutum L.). PLoS ONE 2016, 11, e0156723. [Google Scholar] [CrossRef] [PubMed]
- Gui, Q.; Yang, Z.; Chen, C.; Yang, F.; Wang, S.; Dong, R. Identification and Characterization of Long Noncoding RNAs Involved in the Aluminum Stress Response in Medicago truncatula via Genome-Wide Analysis. Front. Plant Sci. 2022, 13, 1017869. [Google Scholar] [CrossRef]
- Luo, C.; He, B.; Shi, P.; Xi, J.; Gui, H.; Pang, B.; Cheng, J.; Hu, F.; Chen, X.; Lv, Y. Transcriptome Dynamics Uncovers Long Non-Coding RNAs Response to Salinity Stress in Chenopodium quinoa. Front. Plant Sci. 2022, 13, 988845. [Google Scholar] [CrossRef]
- Zhou, X.; Cui, J.; Meng, J.; Luan, Y. Interactions and Links among the Noncoding RNAs in Plants under Stresses. Theor. Appl. Genet. 2020, 133, 3235–3248. [Google Scholar] [CrossRef]
- Shuai, P.; Liang, D.; Tang, S.; Zhang, Z.; Ye, C.-Y.; Su, Y.; Xia, X.; Yin, W. Genome-Wide Identification and Functional Prediction of Novel and Drought-Responsive lincRNAs in Populus trichocarpa. J. Exp. Bot. 2014, 65, 4975–4983. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ma, R.; Cui, D.; Cao, Q.; Shan, Z.; Jiao, Z. Physio-Biochemical and Molecular Mechanism Underlying the Enhanced Heavy Metal Tolerance in Highland Barley Seedlings Pre-Treated with Low-Dose Gamma Irradiation. Sci. Rep. 2017, 7, 14233. [Google Scholar] [CrossRef]
- Feng, S.J.; Zhang, X.D.; Liu, X.S.; Tan, S.K.; Chu, S.S.; Meng, J.G.; Zhao, K.X.; Zheng, J.F.; Yang, Z.M. Characterization of Long Non-Coding RNAs Involved in Cadmium Toxic Response in Brassica napus. RSC Adv. 2016, 6, 82157–82173. [Google Scholar] [CrossRef]
- Bukhari, S.A.H.; Shang, S.; Zhang, M.; Zheng, W.; Zhang, G.; Wang, T.-Z.; Shamsi, I.H.; Wu, F. Genome-Wide Identification of Chromium Stress-Responsive Micro RNAs and Their Target Genes in Tobacco (Nicotiana tabacum) Roots. Environ. Toxicol. Chem. 2015, 34, 2573–2582. [Google Scholar] [CrossRef]
- An, Y.; Su, H.; Niu, Q.; Yin, S. Integrated Analysis of Coding and Non-Coding RNAs Reveals the Molecular Mechanism Underlying Salt Stress Response in Medicago truncatula. Front. Plant Sci. 2022, 13, 891361. [Google Scholar] [CrossRef]
- Feng, S.J.; Liu, X.S.; Tao, H.; Tan, S.K.; Chu, S.S.; Oono, Y.; Zhang, X.D.; Chen, J.; Yang, Z.M. Variation of DNA Methylation Patterns Associated with Gene Expression in Rice (Oryza sativa) Exposed to Cadmium. Plant Cell Environ. 2016, 39, 2629–2649. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Wang, Y.; Zhang, S.; Guo, Q.; Jin, Y.; Chen, J.; Gao, Y.; Ma, H. Transcriptomic and Physiological Analyses of Medicago sativa L. Roots in Response to Lead Stress. PLoS ONE 2017, 12, e0175307. [Google Scholar] [CrossRef] [PubMed]
- Basharat, S.; Shaheen, I.; Waseem, M.; Liu, P. The Comprehensive Identification of lncRNA Landscape Associated with Cadmium and Arsenic Stress Responses in Huanghuazhan Rice. Genet. Resour. Crop Evol. 2025, 72, 5997–6011. [Google Scholar] [CrossRef]
- Feng, X.; Chen, X.; Meng, Q.; Song, Z.; Zeng, J.; He, X.; Wu, F.; Ma, W.; Liu, W. Comparative Long Non-Coding Transcriptome Analysis of Three Contrasting Barley Varieties in Response to Aluminum Stress. Int. J. Mol. Sci. 2024, 25, 9181. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Ding, Y.; Tan, Z.; Wang, J.; Zhang, D.; Wang, Y. Identification and Characterization of Cadmium Stress-Related LncRNAs from Betula platyphylla. Plant Sci. 2020, 299, 110601. [Google Scholar] [CrossRef]
- Zhou, X.-Y.; Wang, N.-H.; Qiu, C.-W.; Luo, L.; Zhang, M.; Zhang, S.; Gao, Z.-F.; Ahmed, I.M.; Wu, F. Transcriptome Profiling Uncovers the lncRNA-Mediated Regulatory Networks Associated with Tolerance to Cadmium Stress in Barley. Environ. Exp. Bot. 2023, 206, 105156. [Google Scholar] [CrossRef]
- Foysal, M.R.A.; Qiu, C.-W.; Wu, F. Comparative Transcriptome Analysis Reveals Key Long Noncoding RNAs for Cadmium Tolerance in Tibetan Hull-Less Barley. Front. Plant Sci. 2025, 16, 1572490. [Google Scholar] [CrossRef]
- Dong, B.; Meng, D.; Song, Z.; Cao, H.; Du, T.; Qi, M.; Wang, S.; Xue, J.; Yang, Q.; Fu, Y. CcNFYB3-CcMATE35 and LncRNA CcLTCS-CcCS Modules Jointly Regulate the Efflux and Synthesis of Citrate to Enhance Aluminium Tolerance in Pigeon pea. Plant Biotechnol. J. 2024, 22, 181–199. [Google Scholar] [CrossRef]
- Qiu, C.-W.; Richmond, M.; Ma, Y.; Zhang, S.; Liu, W.; Feng, X.; Ahmed, I.M.; Wu, F. Melatonin Enhances Cadmium Tolerance in Rice via Long Non-Coding RNA-Mediated Modulation of Cell Wall and Photosynthesis. J. Hazard. Mater. 2024, 465, 133251. [Google Scholar] [CrossRef]
- Zhu, M.; Liu, Y.; Bai, H.; Zhang, W.; Liu, H.; Qiu, Z. Integrated Physio-Biochemical and RNA Sequencing Analysis Revealed Mechanisms of Long Non-Coding RNA-Mediated Response to Cadmium Toxicity in Wheat. Plant Physiol. Biochem. 2023, 203, 108028. [Google Scholar] [CrossRef]
- Zhao, S.; Bai, H.; Fan, Z.; Zhu, M.; Qiu, Z. A Long Non-Coding RNA lncRNA18313 Regulates Resistance against Cadmium Stress in Wheat. Front. Plant Sci. 2025, 16, 1583758. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Yi, Z.; Lv, S.; Zhang, B.; Guo, Z.; Li, Y. Uncovering the Key lncRNAs in Regulating Cadmium Accumulation and Translocation in Sweet sorghum. Planta 2025, 261, 1–15. [Google Scholar] [CrossRef]
- Duarte-Aké, F.; Us-Camas, R.; De-la-Peña, C. Epigenetic Regulation in Heterosis and Environmental Stress: The Challenge of Producing Hybrid Epigenomes to Face Climate Change. Epigenomes 2023, 7, 14. [Google Scholar] [CrossRef]
- Shen, L.; Yu, H. Epitranscriptome engineering in crop improvement. Mol. Plant. 2021, 14, 1418–1420. [Google Scholar] [CrossRef]
- Gajardo, H.A.; Gómez-Espinoza, O.; Boscariol Ferreira, P.; Carrer, H.; Bravo, L.A. The Potential of CRISPR/Cas Technology to Enhance Crop Performance on Adverse Soil Conditions. Plants 2023, 12, 1892. [Google Scholar] [CrossRef]
- Luo, F.; Zhu, D.; Zou, R.; Duan, W.; Yan, Y. Wheat (Triticum aestivum L.) Selenium-Binding Protein-A Enhances Cadmium Tolerance via Interaction between CXXC Motif and Cadmium and Detoxification. Authorea Prepr. 2020. [Google Scholar] [CrossRef]
- Meng, L.; Zhang, J.; Clarke, N. A Critical Review of Recent Advances in Maize Stress Molecular Biology. Int. J. Mol. Sci. 2024, 25, 12383. [Google Scholar] [CrossRef]
- Qiao, K.; Tian, Y.; Hu, Z.; Chai, T. Wheat Cell Number Regulator CNR10 Enhances the Tolerance, Translocation, and Accumulation of Heavy Metals in Plants. Environ. Sci. Technol. 2018, 53, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.; Wan, X. Epigenome and Epitranscriptome: Potential Resources for Crop Improvement. Int. J. Mol. Sci. 2021, 22, 12912. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Singh, A.K.; Mohapatra, T. Epigenetics: History, Present Status and Future Perspective. Indian J. Genet. Plant Breed. 2017, 77, 445–463. [Google Scholar] [CrossRef]
- Kakoulidou, I.; Avramidou, E.V.; Baránek, M.; Brunel-Muguet, S.; Farrona, S.; Johannes, F.; Kaiserli, E.; Lieberman-Lazarovich, M.; Martinelli, F.; Mladenov, V.; et al. Epigenetics for Crop Improvement in Times of Global Change. Biology 2021, 10, 766. [Google Scholar] [CrossRef]
- Haroon, M.; Zafar, M.M.; Farooq, M.A.; Afzal, R.; Batool, M.; Idrees, F.; Babar, U.; Khan, A.S.; Mo, H.; Li, L.; et al. Conventional Breeding, Molecular Breeding and Speed Breeding; Brave Approaches to Revamp the Production of Cereal Crops. Preprints 2020. [Google Scholar] [CrossRef]
- Lv, Y.; Zhu, X.; Zhang, M.; Liu, X.; Wang, J. In-Situ Bioremediation of Multiple Heavy Metals Contaminated Farmland Soil by Sulfate-Reducing Bacteria. Pol. J. Environ. Stud. 2022, 31, 1747–1755. [Google Scholar] [CrossRef]
- Guarino, F.; Miranda, A.; Castiglione, S.; Cicatelli, A. Arsenic phytovolatilization and epigenetic modifications in Arundo donax L. assisted by a PGPR consortium. Chemosphere 2020, 251, 126310. [Google Scholar] [CrossRef]
- Ikram, M.; Ali, N.; Jan, G.; Jan, F.G.; Rahman, I.U.; Iqbal, A.; Hamayun, M. IAA Producing Fungal Endophyte Penicillium roqueforti thom., Enhances Stress Tolerance and Nutrients Uptake in Wheat Plants Grown on Heavy Metal Contaminated Soils. PLoS ONE 2018, 13, e0208150. [Google Scholar] [CrossRef] [PubMed]
- Cong, W.; Miao, Y.; Xu, L.; Zhang, Y.; Yuan, C.; Wang, J.; Zhuang, T.; Lin, X.; Jiang, L.; Wang, N.; et al. Transgenerational Memory of Gene Expression Changes Induced by Heavy Metal Stress in Rice (Oryza sativa L.). BMC Plant Biol. 2019, 19, 282. [Google Scholar] [CrossRef]
- Rahavi, M.R.; Migicovsky, Z.; Titov, V.; Kovalchuk, I. Transgenerational Adaptation to Heavy Metal Salts in Arabidopsis. Front. Plant Sci. 2011, 2, 91. [Google Scholar] [CrossRef]
- Xue, T.; Li, N.; He, F.; Liu, J. Effects of Exogennous Ebr on Physiological And Biochemical Characteristics of Soybean Under Cadmium Stress. Bangladesh J. Bot. 2024, 53, 327–335. [Google Scholar] [CrossRef]
- Shah, N.; Qadir, M.; Irshad, M.; Hussain, A.; Hamayun, M.; Murad, W.; Khan, A.; Al-Harrasi, A. Enhancement of Cadmium Phytoremediation Potential of Helianthus annuus L. with Application of EDTA and IAA. Metabolites 2022, 12, 1049. [Google Scholar] [CrossRef]
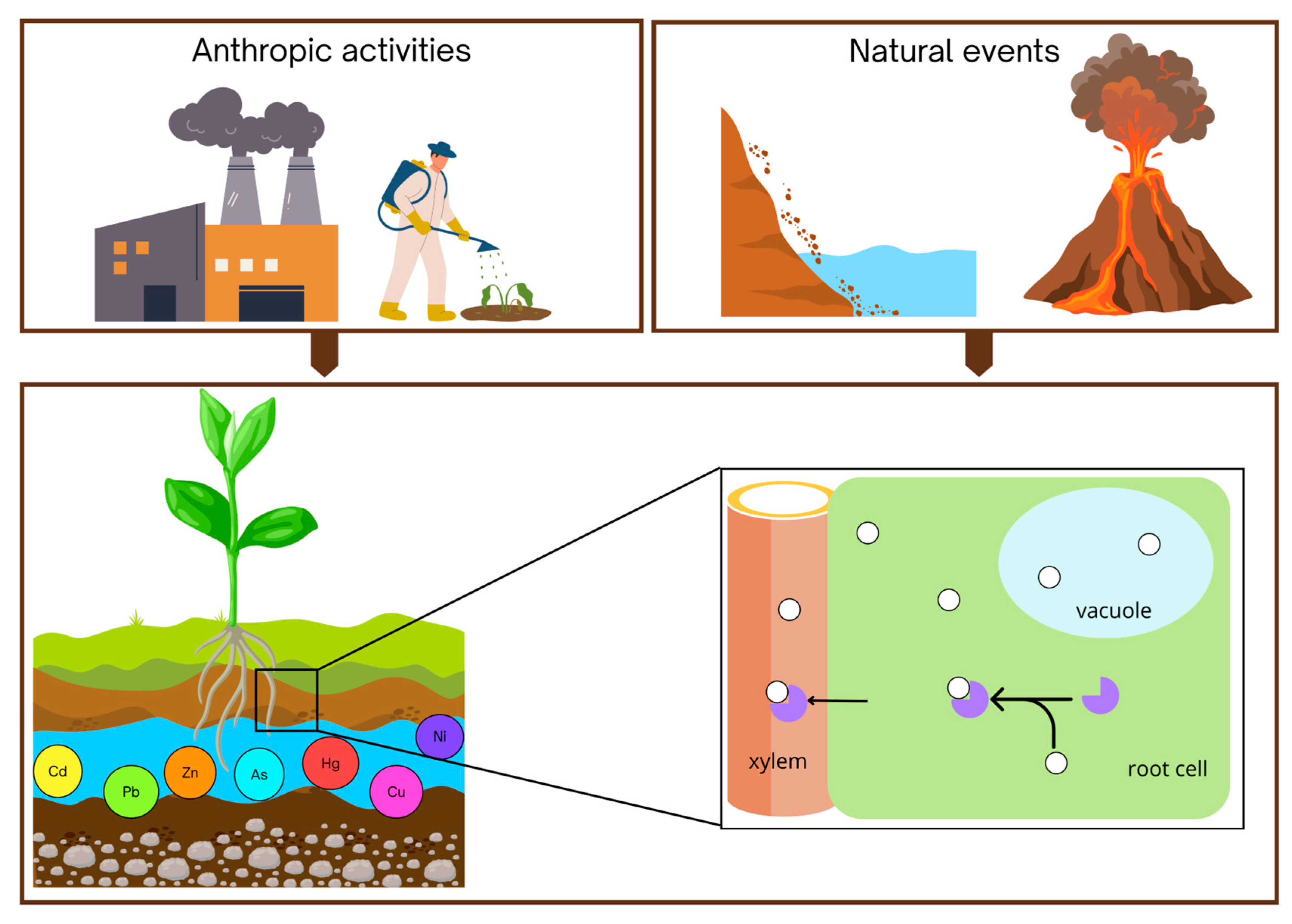

| Metal | Sources | Citations |
|---|---|---|
| Cadmium | Sedimentary rocks and marine phosphates, production of alloys, pigments, batteries, mining operations, industrial manufacturing, phosphate-based fertilizers | [60,66] |
| Mercury | Coal combustion, industrial processes (production of caustic soda, nuclear reactors, antifungal agents, solvent for reactive and precious metal) electrical industry (switches, thermostats, batteries), cement production | [60,67] |
| Arsenicum | Volcanic eruptions and soil erosion, mining runoff, agricultural pesticides, dyestuffs | [60,68] |
| Lead | Industrial emissions, lead-based paints, batteries, fossil fuels burning, mining, manufacturing | [60,69] |
| Copper | Industrial discharges, farming activities, corrosion of plumbing systems, alloys, electric circuit boards, electromagnets | [60,70] |
| Zinc | Industrial waste, mining by-products, sewage sludge, Ni-Zn batteries, manufacture of plastics, paints | [60,70] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Greco, E.; Talarico, E.; Guarasci, F.; Camoli, M.; Palermo, A.M.; Zambelli, A.; Chiappetta, A.; Araniti, F.; Bruno, L. Epigenetic Mechanisms of Plant Adaptation to Cadmium and Heavy Metal Stress. Epigenomes 2025, 9, 43. https://doi.org/10.3390/epigenomes9040043
Greco E, Talarico E, Guarasci F, Camoli M, Palermo AM, Zambelli A, Chiappetta A, Araniti F, Bruno L. Epigenetic Mechanisms of Plant Adaptation to Cadmium and Heavy Metal Stress. Epigenomes. 2025; 9(4):43. https://doi.org/10.3390/epigenomes9040043
Chicago/Turabian StyleGreco, Eleonora, Emanuela Talarico, Francesco Guarasci, Marina Camoli, Anna Maria Palermo, Alice Zambelli, Adriana Chiappetta, Fabrizio Araniti, and Leonardo Bruno. 2025. "Epigenetic Mechanisms of Plant Adaptation to Cadmium and Heavy Metal Stress" Epigenomes 9, no. 4: 43. https://doi.org/10.3390/epigenomes9040043
APA StyleGreco, E., Talarico, E., Guarasci, F., Camoli, M., Palermo, A. M., Zambelli, A., Chiappetta, A., Araniti, F., & Bruno, L. (2025). Epigenetic Mechanisms of Plant Adaptation to Cadmium and Heavy Metal Stress. Epigenomes, 9(4), 43. https://doi.org/10.3390/epigenomes9040043








