Exploration into the MLL4/WRAD Enzyme-Substrate Network: Systematic In Vitro Identification of CFP1 as a Potential Non-Histone Substrate of the MLL4 Lysine Methyltransferase
Abstract
1. Introduction
2. Methods
2.1. Protein Expression and Purification
2.2. SPOT Peptide Array Synthesis and Methylation Assay
2.3. In Vitro Methyltransferase Activity Assay
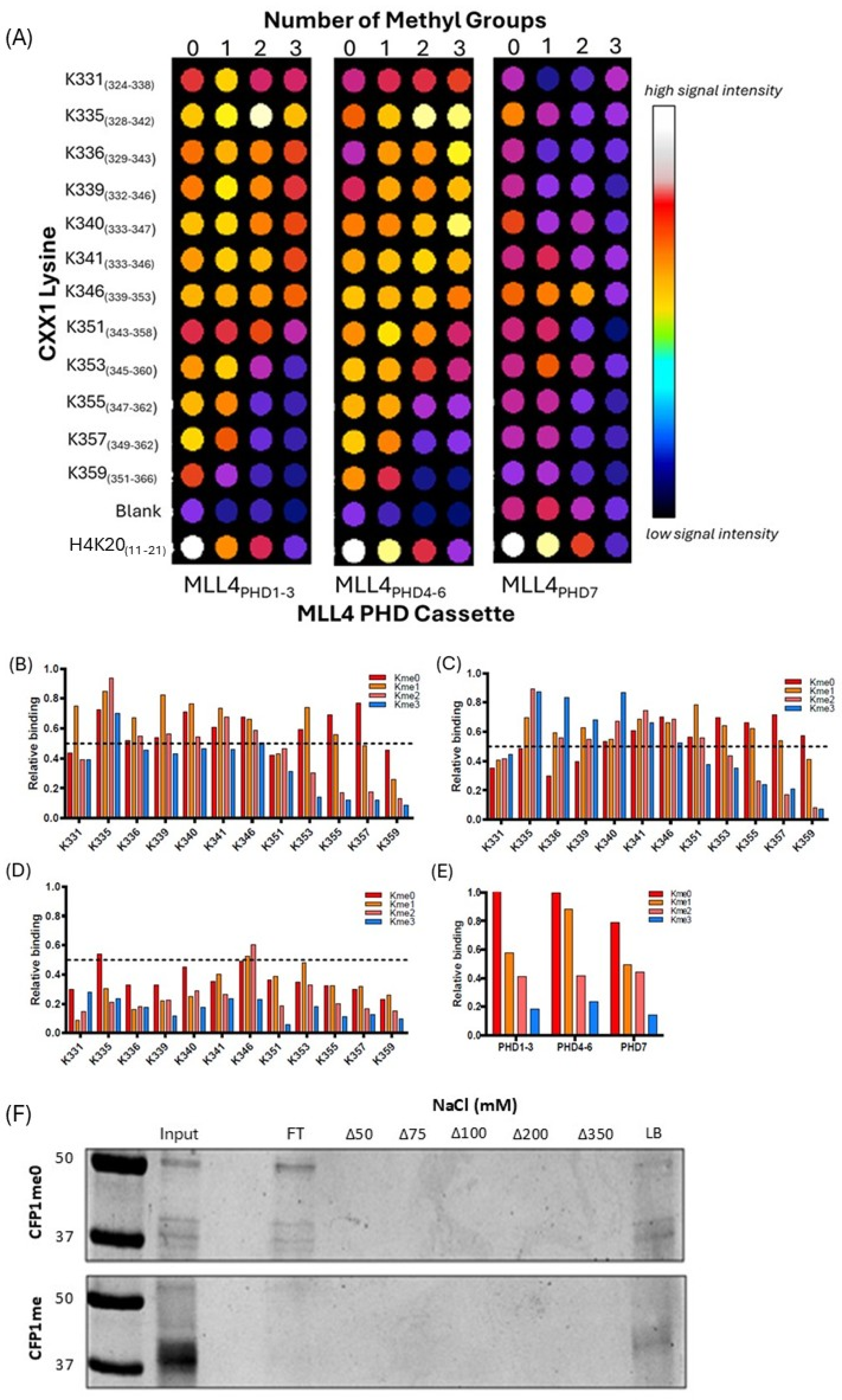
2.4. Peptide Binding Array with MLL4 PHD Domains
2.5. Cloning of Setd1A Fragment
2.6. CFP1 Methylation and Pulldown Assay
3. Results
3.1. A Systematic Approach Uncovers CFP1 K328 as an MLL4 Substrate Candidate
3.2. CFP1 Can Undergo In Vitro Methylation Mediated by MLL4/WRAD at Multiple Sites
3.3. MLL4/WRAD-Mediated CFP1 Methylation Affects MLL4 PHD Cassette Binding
3.4. MLL4/WRAD-Mediated CFP1 Methylation Modulates Setd1A Interaction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Husmann, D.; Gozani, O. Histone Lysine Methyltransferases in Biology and Disease. Nat. Struct. Mol. Biol. 2019, 26, 880–889. [Google Scholar] [CrossRef]
- Greer, E.L.; Shi, Y. Histone Methylation: A Dynamic Mark in Health, Disease and Inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Zhang, Y. The Diverse Functions of Histone Lysine Methylation. Nat. Rev. Mol. Cell. Biol. 2005, 6, 838–849. [Google Scholar] [CrossRef] [PubMed]
- Biggar, K.K.; Li, S.S.C. Non-Histone Protein Methylation as a Regulator of Cellular Signalling and Function. Nat. Rev. Mol. Cell. Biol. 2015, 16, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Falnes, P.Ø. Closing in on Human Methylation—The Versatile Family of Seven-β-Strand (METTL) Methyltransferases. Nucleic Acids Res. 2024, 52, 11423–11441. [Google Scholar] [CrossRef]
- Herz, H.M.; Garruss, A.; Shilatifard, A. SET for Life: Biochemical Activities and Biological Functions of SET Domain-Containing Proteins. Trends Biochem. Sci. 2013, 38, 621–639. [Google Scholar] [CrossRef]
- Shilatifard, A. The COMPASS Family of Histone H3K4 Methylases: Mechanisms of Regulation in Development and Disease Pathogenesis. Annu. Rev. Biochem. 2012, 81, 65–95. [Google Scholar] [CrossRef]
- Eissenberg, J.C.; Shilatifard, A. Histone H3 Lysine 4 (H3K4) Methylation in Development and Differentiation. Dev. Biol. 2010, 339, 240–249. [Google Scholar] [CrossRef]
- Mohan, M.; Herz, H.-M.; Smith, E.R.; Zhang, Y.; Jackson, J.; Washburn, M.P.; Florens, L.; Eissenberg, J.C.; Shilatifard, A. The COMPASS Family of H3K4 Methylases in Drosophila. Mol. Cell. Biol. 2011, 31, 4310–4318. [Google Scholar] [CrossRef]
- Ardehali, M.B.; Mei, A.; Zobeck, K.L.; Caron, M.; Lis, J.T.; Kusch, T. Drosophila Set1 Is the Major Histone H3 Lysine 4 Trimethyltransferase with Role in Transcription. EMBO J. 2011, 30, 2817–2828. [Google Scholar] [CrossRef]
- Schuettengruber, B.; Bourbon, H.M.; Di Croce, L.; Cavalli, G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 2017, 171, 34–57. [Google Scholar] [CrossRef] [PubMed]
- Schuettengruber, B.; Martinez, A.M.; Iovino, N.; Cavalli, G. Trithorax Group Proteins: Switching Genes on and Keeping Them Active. Nat. Rev. Mol. Cell. Biol. 2011, 12, 799–814. [Google Scholar] [CrossRef] [PubMed]
- Bledau, A.S.; Schmidt, K.; Neumann, K.; Hill, U.; Ciotta, G.; Gupta, A.; Torres, D.C.; Fu, J.; Kranz, A.; Stewart, A.F.; et al. The H3K4 Methyltransferase Setd1a Is First Required at the Epiblast Stage, Whereas Setd1b Becomes Essential after Gastrulation. Development 2014, 141, 1022–1035. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Wang, P.F.; Lee, J.S.; Martin-Brown, S.; Florens, L.; Washburn, M.; Shilatifard, A. Molecular Regulation of H3K4 Trimethylation by Wdr82, a Component of Human Set1/COMPASS. Mol. Cell. Biol. 2008, 28, 7337–7344. [Google Scholar] [CrossRef]
- Lee, J.-H.; Skalnik, D.G. Wdr82 Is a C-Terminal Domain-Binding Protein That Recruits the Setd1A Histone H3-Lys4 Methyltransferase Complex to Transcription Start Sites of Transcribed Human Genes. Mol. Cell. Biol. 2008, 28, 609–618. [Google Scholar] [CrossRef]
- Clouaire, T.; Webb, S.; Skene, P.; Illingworth, R.; Kerr, A.; Andrews, R.; Lee, J.H.; Skalnik, D.; Bird, A. Cfp1 Integrates Both CpG Content and Gene Activity for Accurate H3K4me3 Deposition in Embryonic Stem Cells. Genes Dev. 2012, 26, 1714–1728. [Google Scholar] [CrossRef]
- Li, Y.; Han, J.; Zhang, Y.; Cao, F.; Liu, Z.; Li, S.; Wu, J.; Hu, C.; Wang, Y.; Shuai, J.; et al. Structural Basis for Activity Regulation of MLL Family Methyltransferases. Nature 2016, 530, 447–452. [Google Scholar] [CrossRef]
- Haddad, J.F.; Yang, Y.; Takahashi, Y.H.; Joshi, M.; Chaudhary, N.; Woodfin, A.R.; Benyoucef, A.; Yeung, S.; Brunzelle, J.S.; Skiniotis, G.; et al. Structural Analysis of the Ash2L/Dpy-30 Complex Reveals a Heterogeneity in H3K4 Methylation. Structure 2018, 26, 1594–1603.e4. [Google Scholar] [CrossRef]
- Zhang, P.; Chaturvedi, C.P.; Tremblay, V.; Cramet, M.; Brunzelle, J.S.; Skiniotis, G.; Brand, M.; Shilatifard, A.; Couture, J.F. A Phosphorylation Switch on RbBP5 Regulates Histone H3 Lys4 Methylation. Genes Dev. 2015, 29, 123–128. [Google Scholar] [CrossRef]
- Milne, T.A.; Briggs, S.D.; Brock, H.W.; Martin, M.E.; Gibbs, D.; Allis, C.D.; Hess, J.L. MLL Targets SET Domain Methyltransferase Activity to Hox Gene Promoters. Mol. Cell 2002, 10, 1107–1117. [Google Scholar] [CrossRef]
- Slany, R.K. The Molecular Biology of Mixed Lineage Leukemia. Haematologica 2009, 94. [Google Scholar] [CrossRef]
- Rao, R.C.; Dou, Y. Hijacked in Cancer: The KMT2 (MLL) Family of Methyltransferases. Nat. Rev. Cancer 2015, 15, 334–346. [Google Scholar] [CrossRef] [PubMed]
- Froimchuk, E.; Jang, Y.; Ge, K. Histone H3 Lysine 4 Methyltransferase KMT2D. Gene 2017, 627, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Huang, Y.; Mencius, J.; Li, Y.; Zhao, L.; Luo, W.; Chen, Y.; Quan, S. Distinct Kinetic Mechanisms of H3K4 Methylation Catalyzed by MLL3 and MLL4 Core Complexes. J. Biol. Chem. 2021, 296, 100635. [Google Scholar] [CrossRef] [PubMed]
- Jain, K.; Fraser, C.S.; Marunde, M.R.; Parker, M.M.; Sagum, C.; Burg, J.M.; Hall, N.; Popova, I.K.; Rodriguez, K.L.; Vaidya, A.; et al. Characterization of the Plant Homeodomain (PHD) Reader Family for Their Histone Tail Interactions. Epigenetics Chromatin 2020, 13, 3. [Google Scholar] [CrossRef]
- Lavery, W.J.; Barski, A.; Wiley, S.; Schorry, E.K.; Lindsley, A.W. KMT2C/D COMPASS Complex-Associated Diseases [KCDCOM-ADs]: An Emerging Class of Congenital Regulopathies. Clin. Epigenetics 2020, 12, 10. [Google Scholar] [CrossRef]
- Ng, S.B.; Bigham, A.W.; Buckingham, K.J.; Hannibal, M.C.; McMillin, M.J.; Gildersleeve, H.I.; Beck, A.E.; Tabor, H.K.; Cooper, G.M.; Mefford, H.C.; et al. Exome Sequencing Identifies MLL2 Mutations as a Cause of Kabuki Syndrome. Nat. Genet. 2010, 42, 790–793. [Google Scholar] [CrossRef]
- Zaidi, S.; Choi, M.; Wakimoto, H.; Ma, L.; Jiang, J.; Overton, J.D.; Romano-Adesman, A.; Bjornson, R.D.; Breitbart, R.E.; Brown, K.K.; et al. De Novo Mutations in Histone-Modifying Genes in Congenital Heart Disease. Nature 2013, 498, 220–223. [Google Scholar] [CrossRef]
- Cenik, B.K.; Shilatifard, A. COMPASS and SWI/SNF Complexes in Development and Disease. Nat. Rev. Genet. 2021, 22, 38–58. [Google Scholar] [CrossRef]
- Egolf, S.; Zou, J.; Anderson, A.; Simpson, C.L.; Aubert, Y.; Prouty, S.; Ge, K.; Seykora, J.T.; Capell, B.C. MLL4 Mediates Differentiation and Tumor Suppression through Ferroptosis. Sci. Adv. 2021, 7. [Google Scholar] [CrossRef]
- Schäfer, F.; Seip, N.; Maertens, B.; Block, H.; Kubicek, J. Purification of GST-Tagged Proteins. Methods Enzymol. 2015, 559, 127–139. [Google Scholar]
- Block, H.; Maertens, B.; Spriestersbach, A.; Brinker, N.; Kubicek, J.; Fabis, R.; Labahn, J.; Schäfer, F. Chapter 27 Immobilized-Metal Affinity Chromatography (IMAC). A Review. Methods Enzymol. 2009, 463, 439–473. [Google Scholar] [PubMed]
- Rowe, E.M.; Biggar, K.K. An Optimized Method Using Peptide Arrays for the Identification of in Vitro Substrates of Lysine Methyltransferase Enzymes. MethodsX 2018, 5, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, K.; Zegzouti, H.; Goueli, S.A. Methyltransferase-Glo: A Universal, Bioluminescent and Homogenous Assay for Monitoring All Classes of Methyltransferases. Epigenomics 2016, 8, 321–339. [Google Scholar] [CrossRef] [PubMed]
- Dhar, S.S.; Lee, S.H.; Kan, P.Y.; Voigt, P.; Ma, L.; Shi, X.; Reinberg, D.; Lee, M.G. Trans-Tail Regulation of MLL4-Catalyzed H3K4 Methylation by H4R3 Symmetric Dimethylation Is Mediated by a Tandem PHD of MLL4. Genes Dev. 2012, 26, 2749–2762. [Google Scholar] [CrossRef]
- Topcu, E.; Biggar, K.K. PeSA: A Software Tool for Peptide Specificity Analysis. Comput. Biol. Chem. 2019, 83, 107145. [Google Scholar] [CrossRef]
- Zhang, Y.; Mittal, A.; Reid, J.; Reich, S.; Gamblin, S.J.; Wilson, J.R. Evolving Catalytic Properties of the MLL Family SET Domain. Structure 2015, 23, 1921–1933. [Google Scholar] [CrossRef]
- Lanouette, S.; Davey, J.A.; Elisma, F.; Ning, Z.; Figeys, D.; Chica, R.A.; Couture, J.F. Discovery of Substrates for a SET Domain Lysine Methyltransferase Predicted by Multistate Computational Protein Design. Structure 2015, 23, 206–215. [Google Scholar] [CrossRef]
- Butler, J.S.; Lee, J.H.; Skalnik, D.G. CFP1 Interacts with DNMT1 Independently of Association with the Setd1 Histone H3K4 Methyltransferase Complexes. DNA Cell Biol. 2008, 2, 533–543. [Google Scholar] [CrossRef]
- Shin Voo, K.; Carlone, D.L.; Jacobsen, B.M.; Flodin, A.; Skalnik, D.G. Cloning of a Mammalian Transcriptional Activator That Binds Unmethylated CpG Motifs and Shares a CXXC Domain with DNA Methyltransferase, Human Trithorax, and Methyl-CpG Binding Domain Protein 1. Mol. Cell. Biol. 2000, 20, 2108–2121. [Google Scholar] [CrossRef]
- Beurton, F.; Stempor, P.; Caron, M.; Appert, A.; Dong, Y.; Chen, R.A.J.; Cluet, D.; Couté, Y.; Herbette, M.; Huang, N.; et al. Physical and Functional Interaction between SET1/COMPASS Complex Component CFP-1 and a Sin3S HDAC Complex in C. elegans. Nucleic Acids Res. 2019, 47, 11164–11180. [Google Scholar] [CrossRef] [PubMed]
- van Nuland, R.; Smits, A.H.; Pallaki, P.; Jansen, P.W.T.C.; Vermeulen, M.; Timmers, H.T.M. Quantitative Dissection and Stoichiometry Determination of the Human SET1/MLL Histone Methyltransferase Complexes. Mol. Cell. Biol. 2013, 33, 2067–2077. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Stamatoyannopoulos, J.A.; Bailey, T.L.; Noble, W.S. Quantifying Similarity between Motifs. Genome Biol. 2007, 8, R24. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Wang, C.; Xu, S.; Cho, Y.W.; Wang, L.; Feng, X.; Baldridge, A.; Sartorelli, V.; Zhuang, L.; Peng, W.; et al. H3K4 Mono- and Di-Methyltransferase MLL4 Is Required for Enhancer Activation during Cell Differentiation. eLife 2013. [Google Scholar] [CrossRef]
- Wu, Z.; Cheng, Z.; Sun, M.; Wan, X.; Liu, P.; He, T.; Tan, M.; Zhao, Y. A Chemical Proteomics Approach for Global Analysis of Lysine Monomethylome Profiling. Mol. Cell. Proteom. 2015, 14, 329–339. [Google Scholar] [CrossRef]
- Gaurav, N.; Kutateladze, T.G. Non-Histone Binding Functions of PHD Fingers. Trends Biochem. Sci. 2023, 48, 610–617. [Google Scholar] [CrossRef]
- Li, Y.; Li, H. Many Keys to Push: Diversifying the “readership” of Plant Homeodomain Fingers. Acta Biochim. Biophys. Sin. 2012, 44, 28–39. [Google Scholar] [CrossRef]
- Becht, D.C.; Mohid, S.A.; Lee, J.E.; Zandian, M.; Benz, C.; Biswas, S.; Sinha, V.K.; Ivarsson, Y.; Ge, K.; Zhang, Y.; et al. MLL4 Binds TET3. Structure 2024, 32, 706–714. [Google Scholar] [CrossRef]
- Lee, J.H.; Skalnik, D.G. CpG-Binding Protein Is a Nuclear Matrix- and Euchromatin-Associated Protein Localized to Nuclear Speckles Containing Human Trithorax: Identification of Nuclear Matrix Targeting Signals. J. Biol. Chem. 2002, 277, 42259–42267. [Google Scholar] [CrossRef]
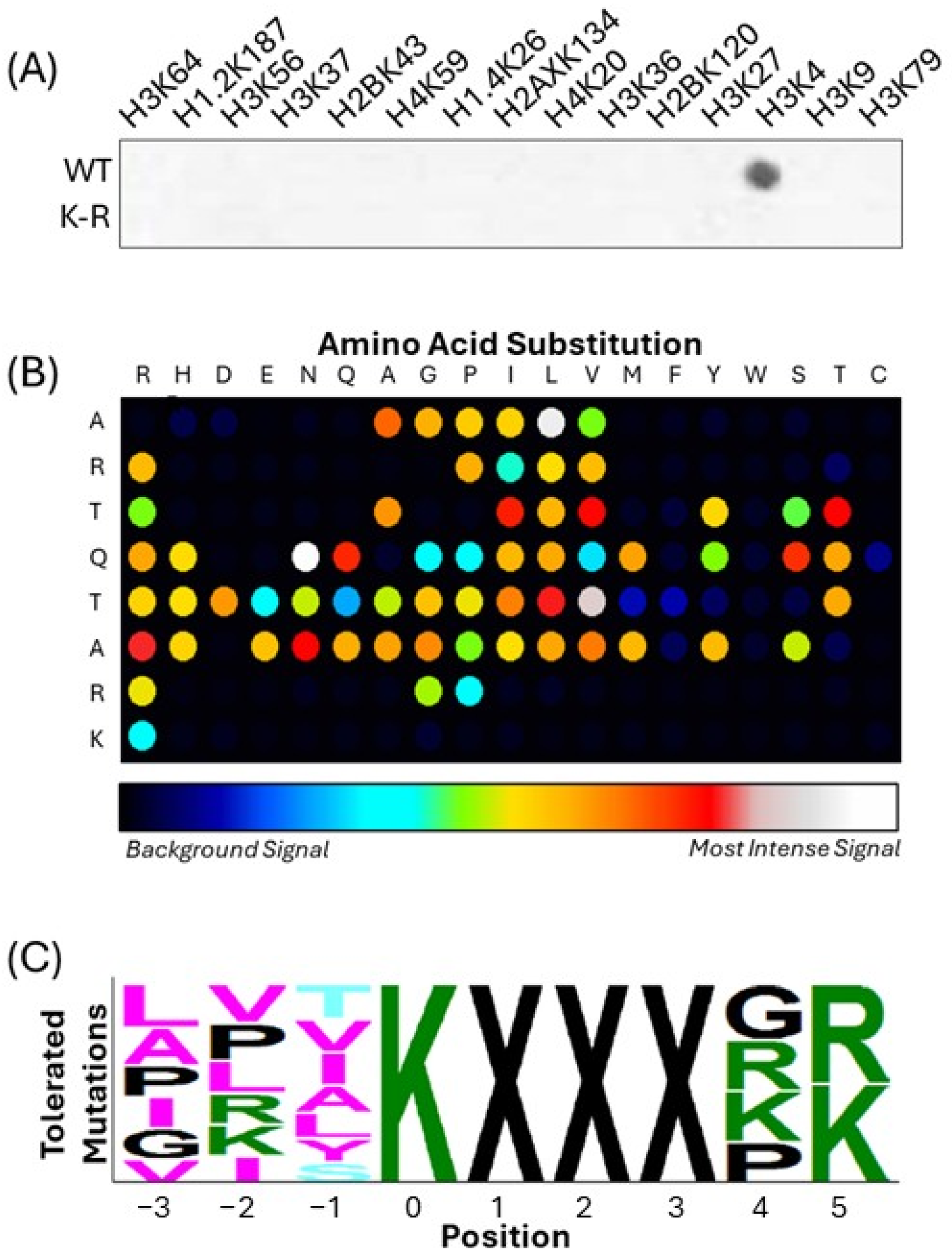
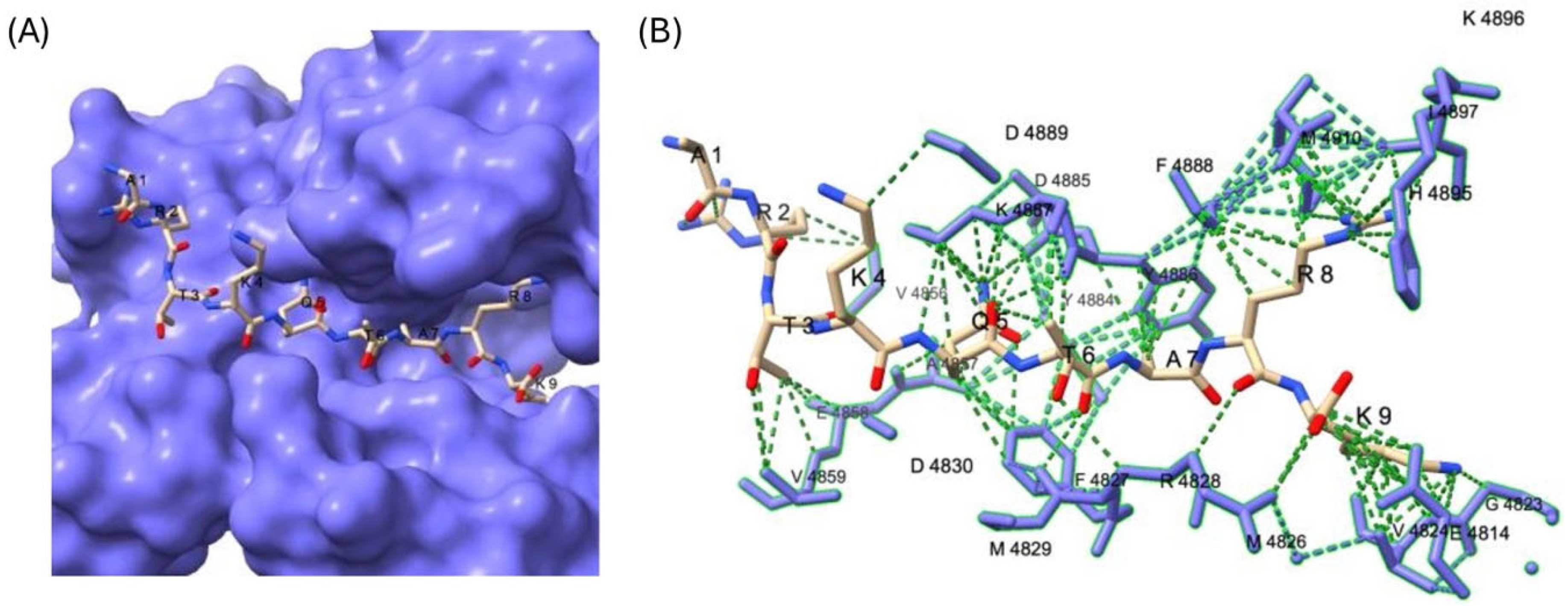
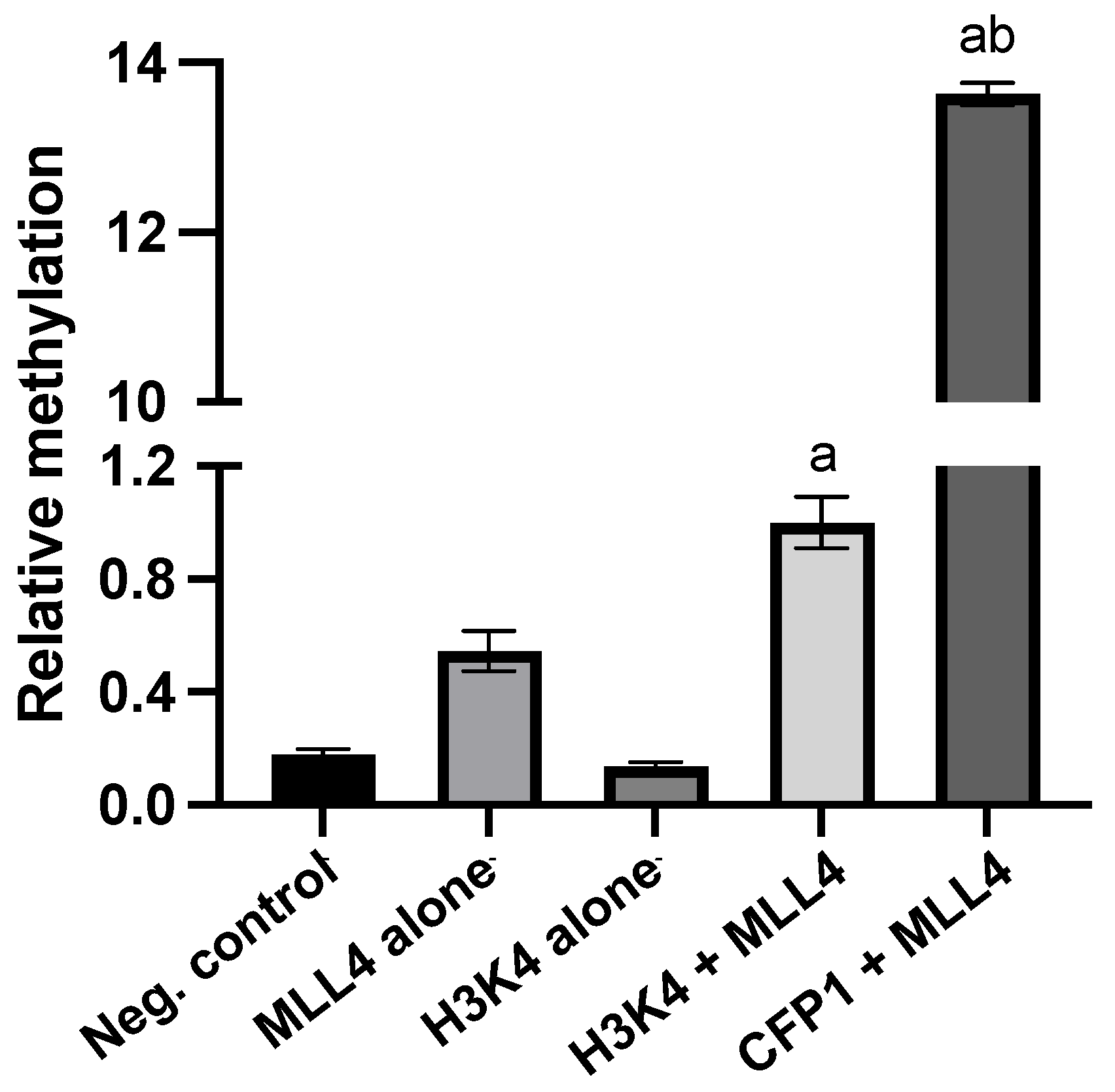
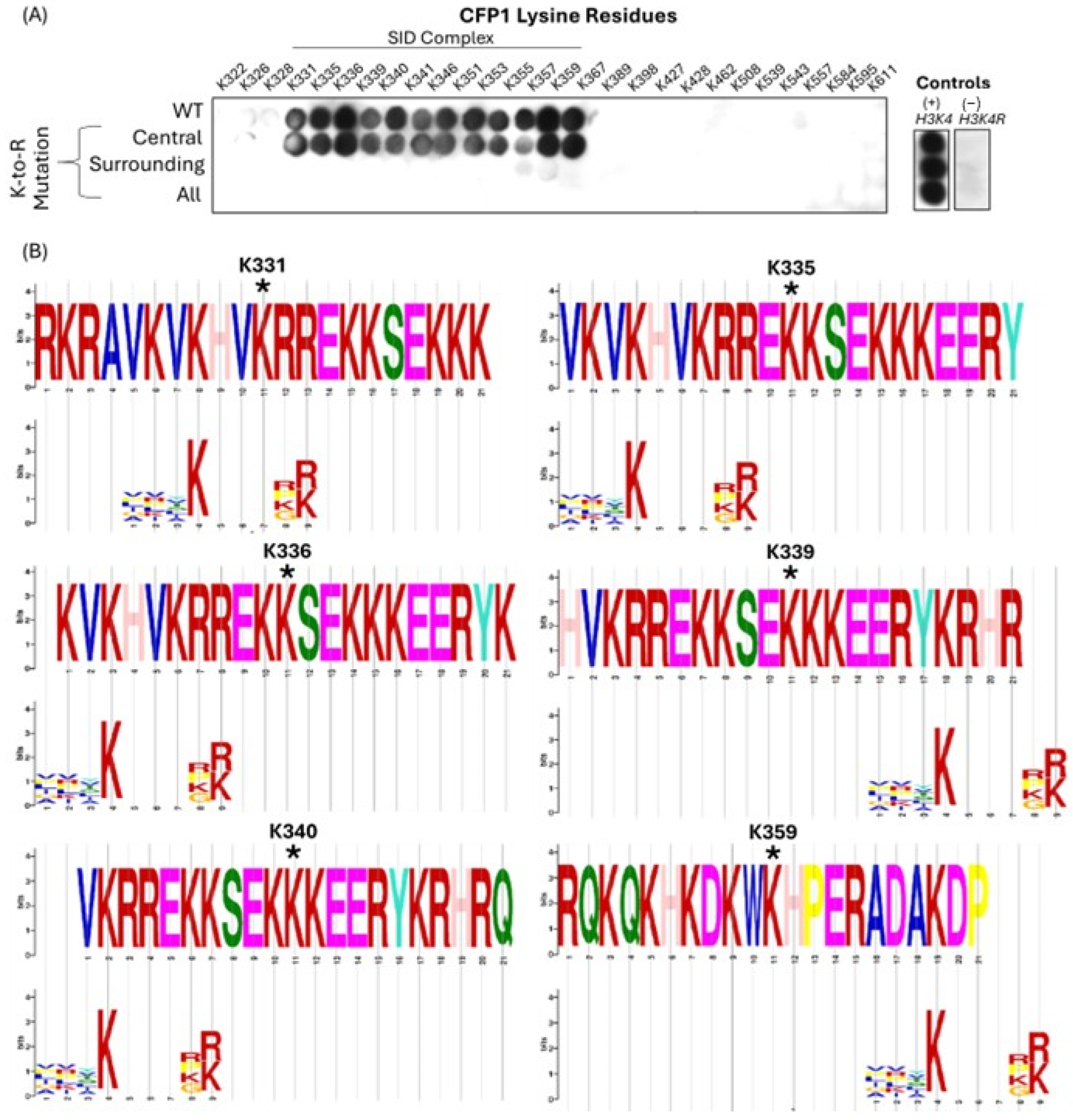
| Uniprot ID | Gene Name | Residue | Sequence |
|---|---|---|---|
| P11388 | TOP2A_HUMAN | 1286 | AFKPIKKGKKR |
| P68431 | H31_HUMAN | 4 | ARTKQTARK |
| Q92831 | KAT2B_HUMAN | 672 | KEIIKKLIERK |
| Q9H2G4 | TSPYL2_HUMAN | 407 | RIKRKKQEMKK |
| Q9P0U4 | CXXC1_HUMAN | 328 | RAVKVKHVKRR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boulter, M.; Collins, R.; Biggar, K.K. Exploration into the MLL4/WRAD Enzyme-Substrate Network: Systematic In Vitro Identification of CFP1 as a Potential Non-Histone Substrate of the MLL4 Lysine Methyltransferase. Epigenomes 2025, 9, 41. https://doi.org/10.3390/epigenomes9040041
Boulter M, Collins R, Biggar KK. Exploration into the MLL4/WRAD Enzyme-Substrate Network: Systematic In Vitro Identification of CFP1 as a Potential Non-Histone Substrate of the MLL4 Lysine Methyltransferase. Epigenomes. 2025; 9(4):41. https://doi.org/10.3390/epigenomes9040041
Chicago/Turabian StyleBoulter, Mullen, Ryan Collins, and Kyle K. Biggar. 2025. "Exploration into the MLL4/WRAD Enzyme-Substrate Network: Systematic In Vitro Identification of CFP1 as a Potential Non-Histone Substrate of the MLL4 Lysine Methyltransferase" Epigenomes 9, no. 4: 41. https://doi.org/10.3390/epigenomes9040041
APA StyleBoulter, M., Collins, R., & Biggar, K. K. (2025). Exploration into the MLL4/WRAD Enzyme-Substrate Network: Systematic In Vitro Identification of CFP1 as a Potential Non-Histone Substrate of the MLL4 Lysine Methyltransferase. Epigenomes, 9(4), 41. https://doi.org/10.3390/epigenomes9040041





