A Pilot Epigenome-Wide Study of Posttraumatic Growth: Identifying Novel Candidates for Future Research
Abstract
1. Introduction
2. Results
2.1. Candidate Gene Analysis
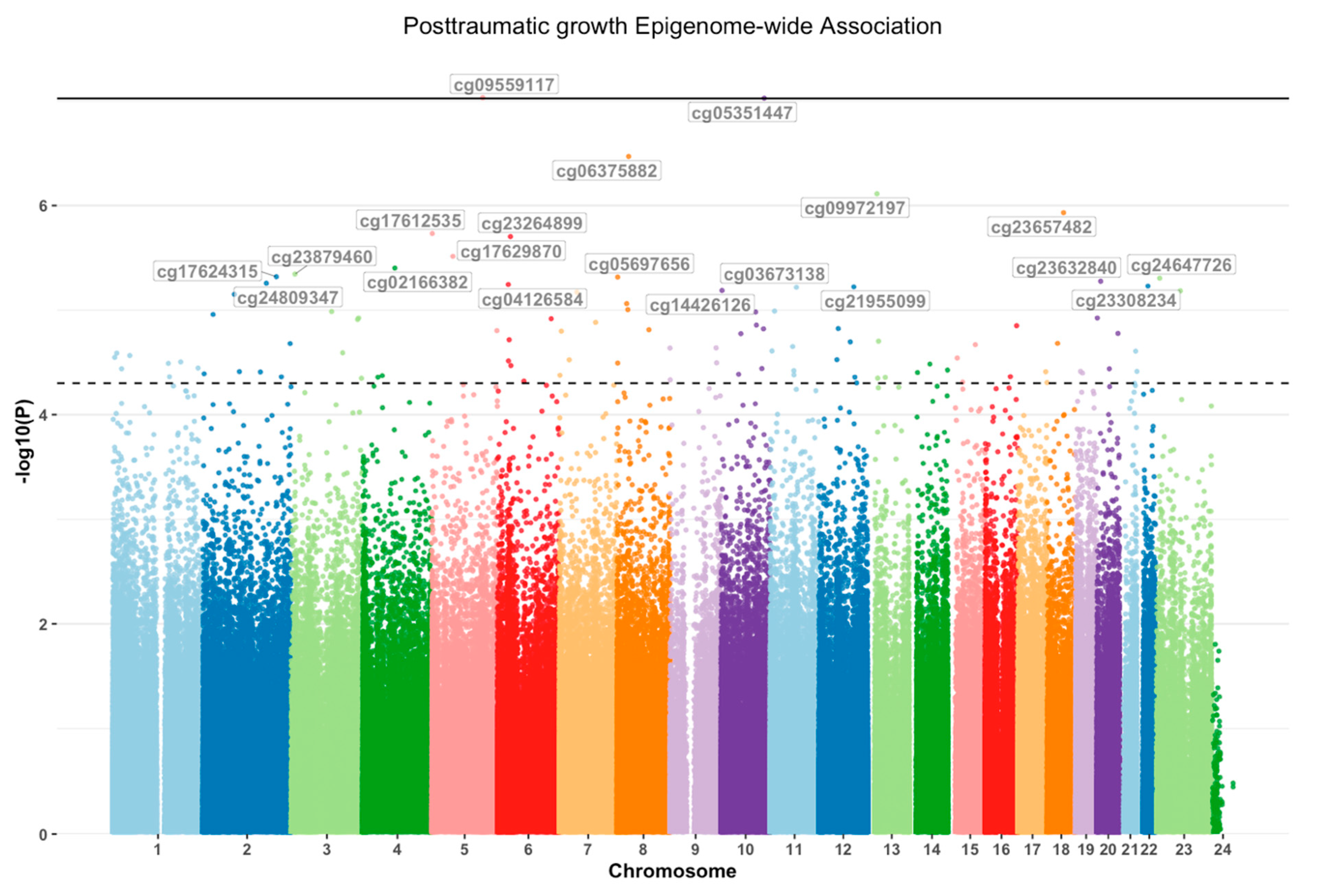
2.2. EWAS of PTG
2.3. Overlap Between PTG and PTSD
3. Discussion
4. Methodology
4.1. Participants
4.2. Assessments
4.3. Posttraumatic Growth Inventory X
4.4. Posttraumatic Stress Disorder Checklist for DSM-V
4.5. Experiments
4.6. Statistical Analysis and Power Calculations
4.7. Replication Cohort
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tedeschi, R.G.; Calhoun, L.G. Posttraumatic growth: Conceptual foundations and empirical evidence. Psychol. Inq. 2004, 15, 1–18. [Google Scholar] [CrossRef]
- Tominaga, Y.; Goto, T.; Shelby, J.; Oshio, A.; Nishi, D.; Takahashi, S. Secondary trauma and posttraumatic growth among mental health clinicians involved in disaster relief activities following the 2011 Tohoku earthquake and tsunami in Japan. Couns. Psychol. Q. 2020, 33, 427–447. [Google Scholar] [CrossRef]
- Feder, A.; Southwick, S.M.; Goetz, R.R.; Wang, Y.; Alonso, A.; Smith, B.W.; Buchholz, K.R.; Waldeck, T.; Ameli, R.; Moore, J. Posttraumatic growth in former Vietnam prisoners of war. Psychiatry 2008, 71, 359–370. [Google Scholar] [CrossRef]
- Sattler, D.N.; Boyd, B.; Kirsch, J. Trauma-exposed firefighters: Relationships among posttraumatic growth, posttraumatic stress, resource availability, coping and critical incident stress debriefing experience. Stress Health 2014, 30, 356–365. [Google Scholar] [CrossRef]
- Calhoun, L.G.; Tedeschi, R.G. The foundations of posttraumatic growth: An expanded framework. In Handbook of Posttraumatic Growth; Routledge: London, UK, 2014; pp. 3–23. [Google Scholar]
- American Psychiatric Association. Posttraumatic Stress Disorder. In Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychiatric Association: Arlington, VA, USA, 2013. [Google Scholar] [CrossRef]
- Bernhard, A.; Martinelli, A.; Ackermann, K.; Saure, D.; Freitag, C.M. Association of trauma, posttraumatic stress disorder and conduct disorder: A systematic review and meta-analysis. Neurosci. Biobehav. Rev. 2018, 91, 153–169. [Google Scholar] [CrossRef]
- Breslau, N. The epidemiology of trauma, PTSD, and other posttrauma disorders. Trauma Violence Abus. 2009, 10, 198–210. [Google Scholar] [CrossRef]
- Mehta, D.; Miller, O.; Bruenig, D.; David, G.; Shakespeare-Finch, J. A systematic review of DNA methylation and gene expression studies in posttraumatic stress disorder, posttraumatic growth, and resilience. J. Trauma. Stress 2020, 33, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Laufer, A.; Solomon, Z. Posttraumatic symptoms and posttraumatic growth among Israeli youth exposed to terror incidents. J. Soc. Clin. Psychol. 2006, 25, 429. [Google Scholar] [CrossRef]
- Powell, S.; Rosner, R.; Butollo, W.; Tedeschi, R.G.; Calhoun, L.G. Posttraumatic growth after war: A study with former refugees and displaced people in Sarajevo. J. Clin. Psychol. 2003, 59, 71–83. [Google Scholar] [CrossRef]
- Taku, K.; Calhoun, L.G.; Cann, A.; Tedeschi, R.G. The role of rumination in the coexistence of distress and posttraumatic growth among bereaved Japanese university students. Death Stud. 2008, 32, 428–444. [Google Scholar] [CrossRef] [PubMed]
- Kılıç, C.; Ulusoy, M. Psychological effects of the November 1999 earthquake in Turkey: An epidemiological study. Acta Psychiatr. Scand. 2003, 108, 232–238. [Google Scholar] [CrossRef]
- Ho, S.M.; Kwong-Lo, R.S.; Mak, C.W.; Wong, J.S. Fear of severe acute respiratory syndrome (SARS) among health care workers. J. Consult. Clin. Psychol. 2005, 73, 344. [Google Scholar] [CrossRef]
- Shakespeare-Finch, J.; Lurie-Beck, J. A meta-analytic clarification of the relationship between posttraumatic growth and symptoms of posttraumatic distress disorder. J. Anxiety Disord. 2014, 28, 223–229. [Google Scholar] [CrossRef]
- Broekman, B.F.; Olff, M.; Boer, F. The genetic background to PTSD. Neurosci. Biobehav. Rev. 2007, 31, 348–362. [Google Scholar] [CrossRef] [PubMed]
- Nievergelt, C.M.; Maihofer, A.X.; Klengel, T.; Atkinson, E.G.; Chen, C.-Y.; Choi, K.W.; Coleman, J.R.; Dalvie, S.; Duncan, L.E.; Gelernter, J. International meta-analysis of PTSD genome-wide association studies identifies sex-and ancestry-specific genetic risk loci. Nat. Commun. 2019, 10, 4558. [Google Scholar] [CrossRef]
- Dell’Osso, L.; Carpita, B.; Nardi, B.; Bonelli, C.; Calvaruso, M.; Cremone, I.M. Biological correlates of post-traumatic growth (PTG): A literature review. Brain Sci. 2023, 13, 305. [Google Scholar] [CrossRef] [PubMed]
- Dunn, E.C.; Solovieff, N.; Lowe, S.R.; Gallagher, P.J.; Chaponis, J.; Rosand, J.; Koenen, K.C.; Waters, M.C.; Rhodes, J.E.; Smoller, J.W. Interaction between genetic variants and exposure to Hurricane Katrina on post-traumatic stress and post-traumatic growth: A prospective analysis of low income adults. J. Affect. Disord. 2014, 152–154, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Amstadter, A.B.; Koenen, K.C.; Ruggiero, K.J.; Acierno, R.; Galea, S.; Kilpatrick, D.G.; Gelernter, J. Variant in RGS2 moderates posttraumatic stress symptoms following potentially traumatic event exposure. J. Anxiety Disord. 2009, 23, 369–373. [Google Scholar] [CrossRef][Green Version]
- Kimple, A.J.; Soundararajan, M.; Hutsell, S.Q.; Roos, A.K.; Urban, D.J.; Setola, V.; Temple, B.R.; Roth, B.L.; Knapp, S.; Willard, F.S.; et al. Structural determinants of G-protein alpha subunit selectivity by regulator of G-protein signaling 2 (RGS2). J. Biol. Chem. 2009, 284, 19402–19411. [Google Scholar] [CrossRef]
- Jjingo, D.; Conley, A.B.; Soojin, V.Y.; Lunyak, V.V.; Jordan, I.K. On the presence and role of human gene-body DNA methylation. Oncotarget 2012, 3, 462. [Google Scholar] [CrossRef]
- Smith, A.K.; Conneely, K.N.; Kilaru, V.; Mercer, K.B.; Weiss, T.E.; Bradley, B.; Tang, Y.; Gillespie, C.F.; Cubells, J.F.; Ressler, K.J. Differential immune system DNA methylation and cytokine regulation in post-traumatic stress disorder. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2011, 156, 700–708. [Google Scholar] [CrossRef]
- Bick, J.; Naumova, O.; Hunter, S.; Barbot, B.; Lee, M.; Luthar, S.S.; Raefski, A.; Grigorenko, E.L. Childhood adversity and DNA methylation of genes involved in the hypothalamus–pituitary–adrenal axis and immune system: Whole-genome and candidate-gene associations. Dev. Psychopathol. 2012, 24, 1417–1425. [Google Scholar] [CrossRef]
- Cimino, P.J.; Yang, Y.; Li, X.; Hemingway, J.F.; Cherne, M.K.; Khademi, S.B.; Fukui, Y.; Montine, K.S.; Montine, T.J.; Keene, C.D. Ablation of the microglial protein DOCK2 reduces amyloid burden in a mouse model of Alzheimer’s disease. Exp. Mol. Pathol. 2013, 94, 366–371. [Google Scholar] [CrossRef]
- Mehta, D.; Bruenig, D.; Carrillo-Roa, T.; Lawford, B.; Harvey, W.; Morris, C.P.; Smith, A.K.; Binder, E.B.; Young, R.M.; Voisey, J. Genomewide DNA methylation analysis in combat veterans reveals a novel locus for PTSD. Acta Psychiatr. Scand. 2017, 136, 493–505. [Google Scholar] [CrossRef] [PubMed]
- Snijders, C.; Maihofer, A.X.; Ratanatharathorn, A.; Baker, D.G.; Boks, M.P.; Geuze, E.; Jain, S.; Kessler, R.C.; Pishva, E.; Risbrough, V.B. Longitudinal epigenome-wide association studies of three male military cohorts reveal multiple CpG sites associated with post-traumatic stress disorder. Clin. Epigenetics 2020, 12, 11. [Google Scholar] [CrossRef] [PubMed]
- Miller, O.; Shakespeare-Finch, J.; Bruenig, D.; Mehta, D. DNA methylation of NR3C1 and FKBP5 is associated with posttraumatic stress disorder, posttraumatic growth, and resilience. Psychol. Trauma Theory Res. Pract. Policy 2020, 12, 750. [Google Scholar] [CrossRef]
- Carlson, E.B.; Palmieri, P.A.; Field, N.P.; Dalenberg, C.J.; Macia, K.S.; Spain, D.A. Contributions of risk and protective factors to prediction of psychological symptoms after traumatic experiences. Compr. Psychiatry 2016, 69, 106–115. [Google Scholar] [CrossRef]
- Mansell, G.; Gorrie-Stone, T.J.; Bao, Y.; Kumari, M.; Schalkwyk, L.S.; Mill, J.; Hannon, E. Guidance for DNA methylation studies: Statistical insights from the Illumina EPIC array. BMC Genom. 2019, 20, 366. [Google Scholar] [CrossRef]
- Kandaswamy, R.; Hannon, E.; Arseneault, L.; Mansell, G.; Sugden, K.; Williams, B.; Burrage, J.; Staley, J.R.; Pishva, E.; Dahir, A.; et al. DNA methylation signatures of adolescent victimization: Analysis of a longitudinal monozygotic twin sample. Epigenetics 2021, 16, 1169–1186. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Wu, Q.; Maniatis, T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell 1999, 97, 779–790. [Google Scholar] [CrossRef]
- Suderman, M.; McGowan, P.O.; Sasaki, A.; Huang, T.C.T.; Hallett, M.T.; Meaney, M.J.; Turecki, G.; Szyf, M. Conserved epidenetic sensitivity to early life experiences in the rat and human hippocampus. Proc. Natl. Acad. Sci. USA 2012, 109, 17266–17272. [Google Scholar] [CrossRef]
- Lachman, H.M.; Petruolo, O.A.; Pedrosa, E.; Novak, T.; Nolan, K.; Stopkova, P. Analysis of protocadherin alpha gene deletion variant in bipolar disorder and schizophrenia. Psychiatr. Genet. 2008, 18, 110–115. [Google Scholar] [CrossRef]
- Shao, Z.; Noh, H.; Kim, W.B.; Ni, P.; Nguyen, C.; Cote, S.; Noyes, E.; Zhao, J.; Parsons, T.; Park, J.; et al. Dysregulated protocadherin-pathway activity as an intrinsic defect in induced pluripotent stem cell–derived cortical interneurons from subjects with schizophrenia. Nat. Neurosci. 2019, 22, 229–242. [Google Scholar] [CrossRef]
- Paban, V.; Ogier, M.; Chambon, C.; Fernandez, N.; Davidsson, J.; Risling, M.; Alescio-Lautier, B. Molecular gene expression following blunt and rotational models of traumatic brain injury parallel injuries associated with stroke and depression. J. Transl. Sci. 2016, 2, 330–339. [Google Scholar] [CrossRef]
- Bharadwaj, R.A.; Jaffe, A.E.; Chen, Q.; Deep-Soboslay, A.; Goldman, A.L.; Mighdoll, M.I.; Cotoia, J.A.; Brandtjen, A.C.; Shin, J.; Hyde, T.M. Genetic risk mechanisms of posttraumatic stress disorder in the human brain. J. Neurosci. Res. 2018, 96, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Mellon, S.H.; Gautam, A.; Hammamieh, R.; Jett, M.; Wolkowitz, O.M. Metabolism, metabolomics, and inflammation in posttraumatic stress disorder. Biol. Psychiatry 2018, 83, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Chen, T.; Bosco, D.B.; Xie, M.; Zheng, J.; Dheer, A.; Ying, Y.; Wu, Q.; Lennon, V.A.; Wu, L.J. The complement C3-C3aR pathway mediates microglia–astrocyte interaction following status epilepticus. Glia 2021, 69, 1155–1169. [Google Scholar] [CrossRef]
- Iqbal, Z.; Vandeweyer, G.; van der Voet, M.; Waryah, A.M.; Zahoor, M.Y.; Besseling, J.A.; Roca, L.T.; Vulto-van Silfhout, A.T.; Nijhof, B.; Kramer, J.M.; et al. Homozygous and heterozygous disruptions of ANK3: At the crossroads of neurodevelopmental and psychiatric disorders. Hum. Mol. Genet. 2013, 22, 1960–1970. [Google Scholar] [CrossRef]
- Klein, S.; Lee, H.; Ghahremani, S.; Kempert, P.; Ischander, M.; Teitell, M.A.; Nelson, S.F.; Martinez-Agosto, J.A. Expanding the phenotype of mutations in DICER1: Mosaic missense mutations in the RNase IIIb domain of DICER1 cause GLOW syndrome. J. Med. Genet. 2014, 51, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Koenen, K.C.; Moffitt, T.E.; Poulton, R.; Martin, J.; Caspi, A. Early childhood factors associated with the development of post-traumatic stress disorder: Results from a longitudinal birth cohort. Psychol. Med. 2007, 37, 181–192. [Google Scholar] [CrossRef]
- Hijazi, A.M.; Keith, J.A.; O’Brien, C. Predictors of posttraumatic growth in a multiwar sample of US Combat veterans. Peace Confl. J. Peace Psychol. 2015, 21, 395. [Google Scholar] [CrossRef]
- Logue, M.W.; Solovieff, N.; Leussis, M.P.; Wolf, E.J.; Melista, E.; Baldwin, C.; Koenen, K.C.; Petryshen, T.L.; Miller, M.W. The ankyrin-3 gene is associated with posttraumatic stress disorder and externalizing comorbidity. Psychoneuroendocrinology 2013, 38, 2249–2257. [Google Scholar] [CrossRef]
- Ferreira, M.A.; O’Donovan, M.C.; Meng, Y.A.; Jones, I.R.; Ruderfer, D.M.; Jones, L.; Fan, J.; Kirov, G.; Perlis, R.H.; Green, E.K. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat. Genet. 2008, 40, 1056–1058. [Google Scholar] [CrossRef]
- Wingo, A.P.; Almli, L.M.; Stevens, J.S.; Klengel, T.; Uddin, M.; Li, Y.; Bustamante, A.C.; Lori, A.; Koen, N.; Stein, D.J. DICER1 and microRNA regulation in post-traumatic stress disorder with comorbid depression. Nat. Commun. 2015, 6, 10106. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Yamasue, H.; Tochigi, M.; Takei, K.; Suga, M.; Abe, O.; Yamada, H.; Rogers, M.A.; Aoki, S.; Sasaki, T.; et al. Effect of tryptophan hydroxylase-2 gene variants on amygdalar and hippocampal volumes. Brain Res. 2010, 1331, 51–57. [Google Scholar] [CrossRef]
- Neves, I.; Dinis-Oliveira, R.J.; Magalhães, T. Epigenomic mediation after adverse childhood experiences: A systematic review and meta-analysis. Forensic Sci. Res. 2021, 6, 103–114. [Google Scholar] [CrossRef]
- Nolan, K.A.; Volavka, J.; Lachman, H.M.; Saito, T. An association between a polymorphism of the tryptophan hydroxylase gene and aggression in schizophrenia and schizoaffective disorder. Psychiatr. Genet. 2000, 10, 109–115. [Google Scholar] [CrossRef]
- Mohandas, N.; Bass-Stringer, S.; Maksimovic, J.; Crompton, K.; Loke, Y.J.; Walstab, J.; Reid, S.M.; Amor, D.J.; Reddihough, D.; Craig, J.M. Epigenome-wide analysis in newborn blood spots from monozygotic twins discordant for cerebral palsy reveals consistent regional differences in DNA methylation. Clin. Epigenetics 2018, 10, 25. [Google Scholar] [CrossRef]
- Tedeschi, R.G.; Cann, A.; Taku, K.; Senol-Durak, E.; Calhoun, L.G. The posttraumatic growth inventory: A revision integrating existential and spiritual change. J. Trauma. Stress 2017, 30, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Weathers, F.W.; Litz, B.T.; Keane, T.M.; Palmieri, P.A.; Marx, B.P.; Schnurr, P.P. The PTSD Checklist for DSM-5 (pcl-5). Scale Available from the National Center for PTSD. 2013. Available online: https://www.ptsd.va.gov/ (accessed on 25 March 2020).
- Blevins, C.A.; Weathers, F.W.; Davis, M.T.; Witte, T.K.; Domino, J.L. The posttraumatic stress disorder checklist for DSM-5 (PCL-5): Development and initial psychometric evaluation. J. Trauma. Stress 2015, 28, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Wockner, L.F.; Noble, E.P.; Lawford, B.R.; Young, R.M.; Morris, C.P.; Whitehall, V.L.; Voisey, J. Genome-wide DNA methylation analysis of human brain tissue from schizophrenia patients. Transl. Psychiatry 2014, 4, e339. [Google Scholar] [CrossRef] [PubMed]
- Barfield, R.T.; Almli, L.M.; Kilaru, V.; Smith, A.K.; Mercer, K.B.; Duncan, R.; Klengel, T.; Mehta, D.; Binder, E.B.; Epstein, M.P.; et al. Accounting for population stratification in DNA methylation studies. Genet. Epidemiolgy 2014, 38, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Mehta, D.; Gonik, M.; Klengel, T.; Rex-Haffner, M.; Menke, A.; Rubel, J.; Mercer, K.B.; Putz, B.; Bradley, B.; Holsboer, F.; et al. Using polymorphisms in FKBP5 to define biologically distinct subtypes of posttraumatic stress disorder: Evidence from endocrine and gene expression studies. Arch. Gen. Psychiatry 2011, 68, 901–910. [Google Scholar] [CrossRef]
- Mehta, D.; Klengel, T.; Conneely, K.N.; Smith, A.K.; Altmann, A.; Pace, T.W.; Rex-Haffner, M.; Loeschner, A.; Gonik, M.; Mercer, K.B.; et al. Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proc. Natl. Acad. Sci. USA 2013, 110, 8302–8307. [Google Scholar] [CrossRef]
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef]
- Middleton, L.Y.M.; Dou, J.; Fisher, J.; Heiss, J.A.; Nguyen, V.K.; Just, A.C.; Faul, J.; Ware, E.B.; Mitchell, C.; Colacino, J.A.; et al. Saliva cell type DNA methylation reference panel for epidemiological studies in children. Epigenetics 2021, 17, 161–177. [Google Scholar] [CrossRef]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 48. [Google Scholar] [CrossRef]
- Tsai, P.-C.; Bell, J.T. Power and sample size estimation for epigenome-wide association scans to detect differential DNA methylation. Int. J. Epidemiol. 2015, 44, 1429–1441. [Google Scholar] [CrossRef] [PubMed]
| Demographics/Traits | Minimum | Maximum | Mean [SE]/N [%] |
|---|---|---|---|
| Overall Sample | |||
| Age (in years) | 17 | 43 | 23.44 [1.080] |
| Sex—Male | 15 [38.5%] | ||
| - Female | 24 [61.5%] | ||
| Ethnicity | |||
| - Caucasian | 35 [89.7%] | ||
| - Asian | 2 [5.1%] | ||
| - African American | 1 [2.6%] | ||
| - Aboriginal/Torres Strait Islander | 1 [2.6%] | ||
| Body Mass index/BMI | 17.1 | 36.2 | 24.88 [0.75] |
| Current alcohol use | 28 [71.8%] | ||
| Current medication | 11 [28.2%] | ||
| Current smoking | 5 [12.8%] | ||
| Current drugs | 1 [2.6%] | ||
| Baseline—at start of paramedicine course | |||
| Posttraumatic growth Inventory Score | 6 | 120 | 72.05 [4.74] |
| Appreciation of Life | 0 | 5 | 3.48 [0.19] |
| Personal Strength | 0 | 5 | 3.36 [0.19] |
| New Possibilities | 0 | 5 | 2.80 [0.24] |
| Relating to Others. | 0.43 | 4.86 | 2.82 [0.20] |
| Spiritual and existential change | 0 | 4.83 | 2.33 [0.21] |
| PTSD Symptoms Score (PCL) | 0 | 50 | 16.82 [2.28] |
| PCL cluster B score | 0 | 18 | 3.56 [0.67] |
| PCL cluster C score | 0 | 8 | 1.95 [0.36] |
| PCL cluster D score | 0 | 21 | 6.26 [0.92] |
| PCL cluster E score | 0 | 12 | 5.05 [0.66] |
| Posttraumatic growth Inventory Score | 6 | 120 | 72.05 [4.74] |
| Follow-up—post trauma exposure | |||
| Posttraumatic growth Inventory Score | 10 | 114 | 64.14 [3.95] |
| Appreciation of Life | 0.33 | 4.67 | 2.99 [0.17] |
| Personal Strength | 0.25 | 4.75 | 3.12 [0.18] |
| New Possibilities | 0 | 4.8 | 2.36 [0.20] |
| Relating to Others. | 0.57 | 4.71 | 2.75 [0.17] |
| Spiritual and existential change | 0.17 | 4.2 | 1.86 [0.19] |
| PTSD Symptoms Score (PCL) | 0 | 50 | 12.83 [2.27] |
| PCL cluster B score | 0 | 13 | 3 [0.59] |
| PCL cluster C score | 0 | 8 | 1.37 [0.35] |
| PCL cluster D score | 0 | 20 | 3.97 [0.85] |
| PCL cluster E score | 0 | 15 | 4.49 [0.70] |
| Candidate Genes | No. of CpGs Tested | No of CpGs with p ≤ 0.05 | Survive Bonferroni (N) |
|---|---|---|---|
| HDAC4 | 503 | 6 | NO |
| CACNA1C | 298 | 23 | NO |
| RORA | 237 | 13 | NO |
| ANK3 | 160 | 13 | YES (1) |
| DOCK2 | 106 | 6 | NO |
| NOS1AP | 94 | 12 | NO |
| NR3C1 | 89 | 6 | NO |
| NLGN1 | 86 | 7 | NO |
| BDNF | 84 | 5 | NO |
| SLC6A3 | 81 | 8 | NO |
| WWC1 | 77 | 5 | NO |
| CRHR1 | 69 | 5 | NO |
| ANKRD55 | 58 | 3 | NO |
| NR3C2 | 53 | 7 | NO |
| COMT | 47 | 5 | NO |
| DICER1 | 57 | 7 | YES (1) |
| FKBP5 | 45 | 4 | NO |
| HEXDC | 44 | 1 | NO |
| CAMKMT | 44 | 1 | NO |
| CRHR2 | 41 | 5 | NO |
| DRD2 | 41 | 3 | NO |
| ADCYAP1 | 40 | 2 | NO |
| PDE1A | 40 | 3 | NO |
| MAN2C1 | 37 | 1 | NO |
| ADCYAP1R1 | 36 | 4 | NO |
| OXTR | 36 | 1 | NO |
| CNR1 | 35 | 5 | NO |
| PRTFDC1 | 35 | 4 | NO |
| LY9 | 34 | 4 | NO |
| TPH2 | 33 | 2 | NO |
| SLC6A4 | 31 | 3 | NO |
| FOS | 26 | 3 | NO |
| GABRA2 | 26 | 4 | NO |
| SLC18A2 | 26 | 1 | NO |
| ALOX12 | 24 | 3 | NO |
| NPY | 22 | 1 | NO |
| HTR1A | 21 | 3 | NO |
| SKA2 | 21 | 1 | YES (1) |
| IL12B | 19 | 2 | YES (1) |
| RGS2 | 18 | 2 | NO |
| DBH | 18 | 1 | NO |
| AIM2 | 16 | 1 | NO |
| OPRL1 | 40 | 3 | NO |
| ZNF626 | 14 | 2 | NO |
| GBP1 | 13 | 1 | NO |
| PRR11 | 13 | 2 | NO |
| TPH1 | 11 | 2 | YES (1) |
| Total | 2999 | 236 | 5 |
| cpg | p-Value PTG | Chromosome | Basepair | Gene Symbol |
|---|---|---|---|---|
| cg09559117 | 9.28 × 10−8 | 5 | 140173855 | PCDHA2;PCDHA1 |
| cg05351447 | 9.39 × 10−8 | 10 | 119120604 | PDZD8 |
| cg06375882 | 3.39 × 10−7 | 8 | 32113523 | NRG1 |
| cg09972197 | 7.70 × 10−7 | 13 | 26301550 | ATP8A2 |
| cg23657482 | 1.17 × 10−6 | 18 | 45102036 | |
| cg17612535 | 1.85 × 10−6 | 5 | 932900 | |
| cg23264899 | 1.98 × 10−6 | 6 | 35765259 | CLPS |
| cg17629870 | 3.06 × 10−6 | 5 | 57756980 | PLK2 |
| cg02166382 | 3.96 × 10−6 | 4 | 88496363 | |
| cg23879460 | 4.52 × 10−6 | 3 | 10806569 | LOC285370 |
| cg17624315 | 4.79 × 10−6 | 2 | 202289200 | TRAK2 |
| cg05697656 | 4.83 × 10−6 | 8 | 1897697 | ARHGEF10 |
| cg24647726 | 4.95 × 10−6 | X | 11128608 | HCCS |
| cg23632840 | 5.29 × 10−6 | 20 | 10414722 | C20orf94;MKKS |
| cg24809347 | 5.52 × 10−6 | 2 | 174723194 | |
| cg04126584 | 5.69 × 10−6 | 6 | 29920309 | |
| cg23308234 | 5.89 × 10−6 | 22 | 29965207 | NIPSNAP1 |
| cg21955099 | 5.99 × 10−6 | 12 | 96005661 | |
| cg03673138 | 6.04 × 10−6 | 11 | 72385963 | PDE2A |
| cg14426126 | 6.49 × 10−6 | 10 | 2394012 | |
| cg17733714 | 6.55 × 10−6 | X | 68114285 | |
| cg10626169 | 6.73 × 10−6 | 7 | 48319696 | ABCA13 |
| cg18825430 | 7.06 × 10−6 | 2 | 86422958 | IMMT |
| cg07572251 | 8.66 × 10−6 | 8 | 26688088 | ADRA1A |
| cg00739259 | 9.89 × 10−6 | 8 | 29858411 | |
| cg13332953 | 1.02 × 10−5 | 11 | 12003759 | DKK3 |
| cg07479253 | 1.03 × 10−5 | 3 | 111904892 | SLC9A10 |
| cg06789550 | 1.04 × 10−5 | 10 | 95462915 | C10orf4 |
| cg16745960 | 1.10 × 10−5 | 2 | 27549918 | GTF3C2 |
| cg02754380 | 1.19 × 10−5 | 3 | 186369639 | FETUB |
| cg01858394 | 1.19 × 10−5 | 20 | 1277043 | SNPH |
| cg14673315 | 1.21 × 10−5 | 6 | 148336294 | |
| cg00018767 | 1.23 × 10−5 | 3 | 183693809 | ABCC5 |
| cg10714329 | 1.31 × 10−5 | 7 | 100027122 | MEPCE;ZCWPW1 |
| cg14192396 | 1.39 × 10−5 | 10 | 97416393 | ALDH18A1 |
| cg26384474 | 1.41 × 10−5 | 16 | 86702325 | |
| cg12831349 | 1.50 × 10−5 | 12 | 52935087 | |
| cg01399353 | 1.51 × 10−5 | 10 | 117114665 | ATRNL1 |
| cg12533940 | 1.54 × 10−5 | 8 | 88056685 | CNBD1 |
| cg13810079 | 1.57 × 10−5 | 5 | 179484006 | RNF130 |
| cg00730549 | 1.59 × 10−5 | 7 | 5430660 | TNRC18 |
| cg09887207 | 1.67 × 10−5 | 20 | 58249281 | PHACTR3 |
| cg19492498 | 1.68 × 10−5 | 10 | 54531460 | MBL2 |
| cg24478695 | 1.92 × 10−5 | 6 | 32363167 | BTNL2 |
| cg03929569 | 1.98 × 10−5 | 13 | 30689009 | |
| cg06740227 | 2.01 × 10−5 | 12 | 86229804 | RASSF9 |
| cg14263702 | 2.08 × 10−5 | 18 | 28651637 | DSC2;DSC2 |
| cg01804434 | 2.09 × 10−5 | 2 | 240456931 | |
| cg08343397 | 2.14 × 10−5 | 15 | 75340982 | PPCDC |
| cg24078577 | 2.23 × 10−5 | 11 | 62160859 | ASRGL1 |
| cg09039879 | 2.30 × 10−5 | 9 | 127230734 | |
| cg13793478 | 2.31 × 10−5 | 9 | 109039 | |
| cg17748470 | 2.46 × 10−5 | 11 | 4969161 | OR51A4 |
| cg23107033 | 2.47 × 10−5 | 21 | 44166176 | PDE9A |
| cg27218767 | 2.56 × 10−5 | 3 | 142442934 | TRPC1 |
| cg13085232 | 2.58 × 10−5 | 1 | 10802080 | CASZ1 |
| cg26563242 | 2.72 × 10−5 | 1 | 46797699 | |
| cg27170935 | 2.83 × 10−5 | 1 | 5221521 | |
| cg03858387 | 2.87 × 10−5 | 15 | 25199164 | SNRPN;SNURF |
| cg05435504 | 2.98 × 10−5 | 12 | 49251596 | RND1 |
| cg11908057 | 2.99 × 10−5 | 7 | 27171154 | HOXA4 |
| cg01316659 | 3.06 × 10−5 | 6 | 30418115 | |
| cg27045794 | 3.13 × 10−5 | 1 | 187412747 | |
| cg08727313 | 3.19 × 10−5 | 9 | 128734485 | |
| cg06879681 | 3.21 × 10−5 | 8 | 1900524 | ARHGEF10 |
| cg10228283 | 3.23 × 10−5 | 1 | 153234387 | LOR |
| cg03492327 | 3.28 × 10−5 | 14 | 57273276 | OTX2 |
| cg00733115 | 3.39 × 10−5 | 6 | 37105406 | |
| cg04501323 | 3.58 × 10−5 | 1 | 235267609 | |
| cg17619701 | 3.62 × 10−5 | 10 | 112610100 | |
| cg21765224 | 3.64 × 10−5 | 20 | 34359771 | PHF20 |
| cg21528040 | 3.64 × 10−5 | 1 | 24195227 | FUCA1 |
| cg15895505 | 3.74 × 10−5 | 14 | 105903354 | MTA1 |
| cg14669919 | 3.79 × 10−5 | 11 | 65340482 | FAM89B |
| cg04664999 | 3.85 × 10−5 | 19 | 14185985 | |
| cg00499599 | 3.85 × 10−5 | 21 | 47706392 | C21orf57;MCM3AP |
| cg17362661 | 3.87 × 10−5 | 2 | 100210490 | AFF3 |
| cg12010144 | 3.88 × 10−5 | 17 | 76733624 | CYTH1 |
| cg07568040 | 3.90 × 10−5 | 2 | 158454401 | ACVR1C |
| cg18887769 | 3.95 × 10−5 | 14 | 22945181 | |
| cg14251798 | 4.00 × 10−5 | 19 | 19545333 | MIR640;GATAD2A |
| cg06836148 | 4.07 × 10−5 | 2 | 2957515 | LINC01250 |
| cg08920628 | 4.11 × 10−5 | 10 | 48354911 | ZNF488 |
| cg20827116 | 4.17 × 10−5 | 11 | 65627404 | MUS81 |
| cg11980004 | 4.20 × 10−5 | 7 | 1571105 | MAFK |
| cg24383710 | 4.23 × 10−5 | 4 | 53916546 | SCFD2 |
| cg02645135 | 4.33 × 10−5 | 16 | 69516238 | |
| cg13056505 | 4.34 × 10−5 | 1 | 156378014 | C1orf61 |
| cg22202891 | 4.35 × 10−5 | 2 | 216001968 | ABCA12 |
| cg09468051 | 4.38 × 10−5 | 4 | 41879262 | |
| cg23053746 | 4.38 × 10−5 | 12 | 98811404 | |
| cg13290331 | 4.40 × 10−5 | 13 | 49068807 | RCBTB2 |
| cg01660473 | 4.48 × 10−5 | 13 | 28395757 | |
| cg01243529 | 4.49 × 10−5 | 3 | 194223220 | |
| cg01183384 | 4.65 × 10−5 | 9 | 716332 | KANK1 |
| cg02281539 | 4.78 × 10−5 | 6 | 73273769 | |
| cg20259534 | 4.88 × 10−5 | 15 | 40453036 | BUB1B |
| cg18456621 | 4.93 × 10−5 | 17 | 80297270 | |
| cg19359858 | 4.97 × 10−5 | 12 | 103667687 | C12orf42 |
| Pathways (p < 5 × 10−5 CpGs genes) | Number of genes | p-value | FDR p-value |
| ABC transporters | 3 | 1.22 × 10−4 | 2.76 × 10−2 |
| Pathways (p < 0.001 CpGs genes) | Number of genes | p-value | FDR p-value |
| Phospholipase D signaling pathway | 21 | 2.44 × 10−5 | 6.14 × 10−3 |
| Axon guidance | 23 | 4.42 × 10−5 | 6.14 × 10−3 |
| EGFR tyrosine kinase inhibitor resistance | 14 | 5.65 × 10−5 | 6.14 × 10−3 |
| Morphine addiction | 14 | 2.71 × 10−4 | 2.17 × 10−2 |
| Dopaminergic synapse | 17 | 5.05 × 10−4 | 2.17 × 10−2 |
| Ras signaling pathway | 25 | 5.16 × 10−4 | 2.17 × 10−2 |
| AMPK signaling pathway | 16 | 5.42 × 10−4 | 2.17 × 10−2 |
| Inflammatory mediator regulation of TRP channels | 14 | 6.57 × 10−4 | 2.17 × 10−2 |
| Choline metabolism in cancer | 14 | 6.57 × 10−4 | 2.17 × 10−2 |
| GABAergic synapse | 13 | 6.66 × 10−4 | 2.17 × 10−2 |
| MAPK signaling pathway | 29 | 8.63 × 10−4 | 2.51 × 10−2 |
| Glutamatergic synapse | 15 | 9.22 × 10−4 | 2.51 × 10−2 |
| Autophagy | 16 | 1.11 × 10−3 | 2.58 × 10−2 |
| Thyroid hormone signaling pathway | 15 | 1.11 × 10−3 | 2.58 × 10−2 |
| Relaxin signaling pathway | 16 | 1.31 × 10−3 | 2.83 × 10−2 |
| Longevity regulating pathway | 10 | 1.39 × 10−3 | 2.83 × 10−2 |
| ErbB signaling pathway | 12 | 1.59 × 10−3 | 3.06 × 10−2 |
| Endocrine resistance | 13 | 1.85 × 10−3 | 3.34 × 10−2 |
| Proteoglycans in cancer | 21 | 2.09 × 10−3 | 3.59 × 10−2 |
| Endocytosis | 24 | 2.35 × 10−3 | 3.83 × 10−2 |
| Fc epsilon RI signaling pathway | 10 | 2.83 × 10−3 | 4.13 × 10−2 |
| Serotonergic synapse | 14 | 2.85 × 10−3 | 4.13 × 10−2 |
| Endocrine and other factor-regulated calcium reabsorption | 8 | 2.91 × 10−3 | 4.13 × 10−2 |
| Sphingolipid signaling pathway | 14 | 3.62 × 10−3 | 4.91 × 10−2 |
| Cell adhesion molecules (CAMs) | 16 | 3.76 × 10−3 | 4.91 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rubens, M.; Pinto, P.R.; Sathyanarayanan, A.; Miller, O.; Mullens, A.B.; Bruenig, D.; Obst, P.; Shakespeare-Finch, J.; Mehta, D. A Pilot Epigenome-Wide Study of Posttraumatic Growth: Identifying Novel Candidates for Future Research. Epigenomes 2025, 9, 39. https://doi.org/10.3390/epigenomes9040039
Rubens M, Pinto PR, Sathyanarayanan A, Miller O, Mullens AB, Bruenig D, Obst P, Shakespeare-Finch J, Mehta D. A Pilot Epigenome-Wide Study of Posttraumatic Growth: Identifying Novel Candidates for Future Research. Epigenomes. 2025; 9(4):39. https://doi.org/10.3390/epigenomes9040039
Chicago/Turabian StyleRubens, Mackenzie, Paul Ruiz Pinto, Anita Sathyanarayanan, Olivia Miller, Amy B. Mullens, Dagmar Bruenig, Patricia Obst, Jane Shakespeare-Finch, and Divya Mehta. 2025. "A Pilot Epigenome-Wide Study of Posttraumatic Growth: Identifying Novel Candidates for Future Research" Epigenomes 9, no. 4: 39. https://doi.org/10.3390/epigenomes9040039
APA StyleRubens, M., Pinto, P. R., Sathyanarayanan, A., Miller, O., Mullens, A. B., Bruenig, D., Obst, P., Shakespeare-Finch, J., & Mehta, D. (2025). A Pilot Epigenome-Wide Study of Posttraumatic Growth: Identifying Novel Candidates for Future Research. Epigenomes, 9(4), 39. https://doi.org/10.3390/epigenomes9040039







