Sustained Higher Levels of Plasma hsa-miR-17-5p Expression During Gestational Diabetes Mellitus and Postpartum
Abstract
1. Introduction
2. Results
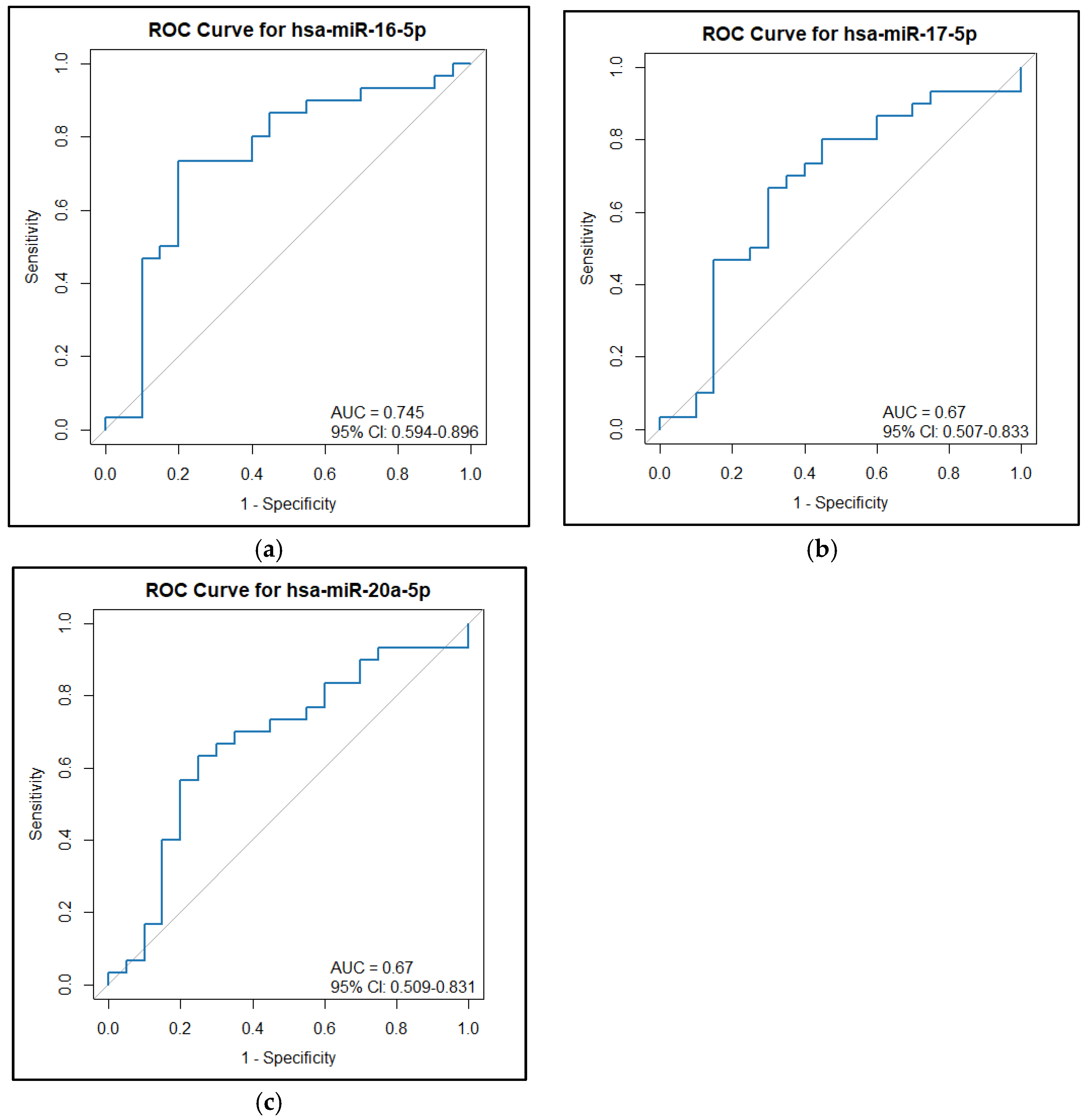
2.1. Clinical Characteristics and miRNA Expression Levels in NGT vs. GDM
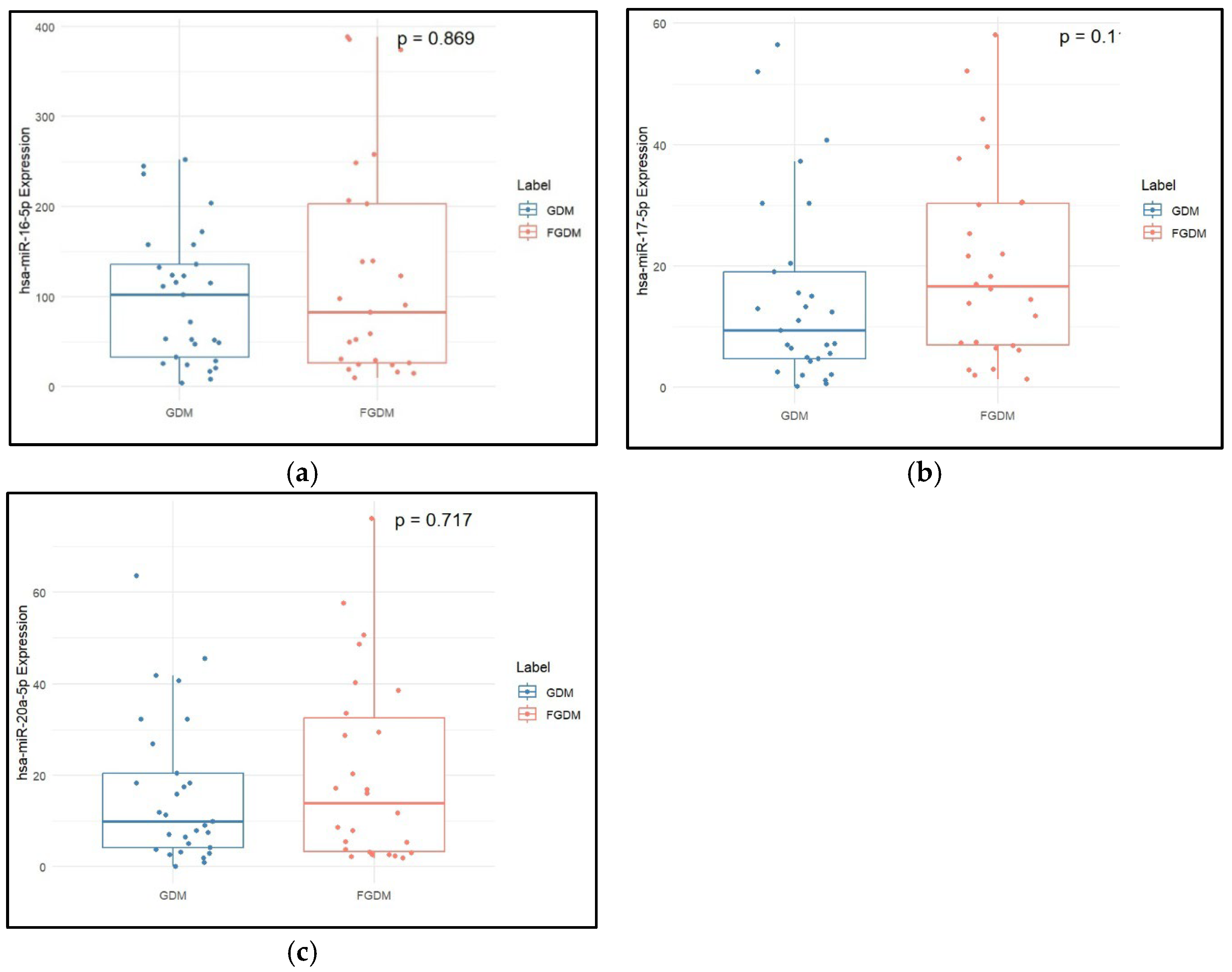
2.2. Clinical Characteristics and miRNA Expression Levels in GDM vs. FGDM
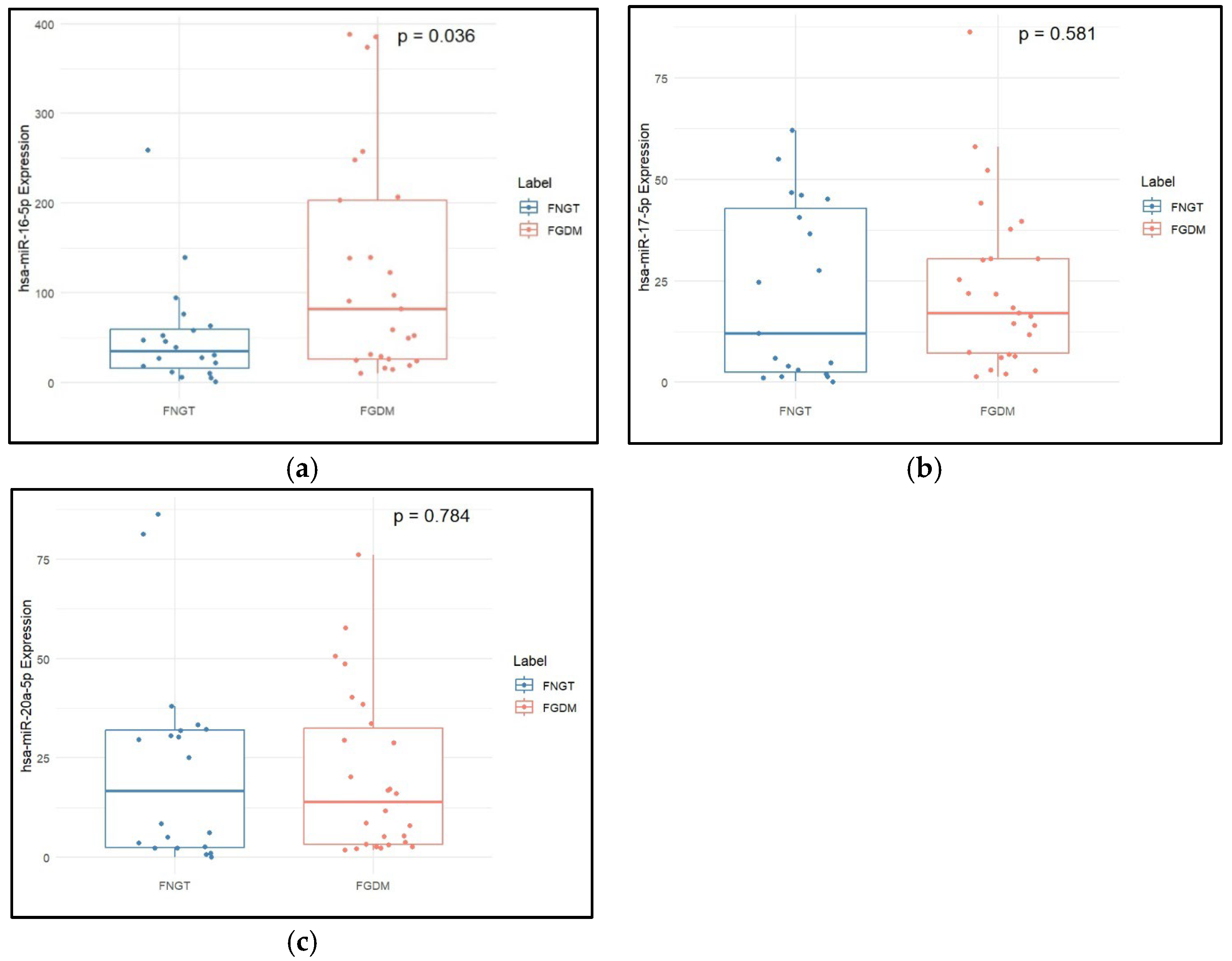
2.3. Clinical Characteristics and miRNA Expression Levels in FNGT vs. FGDM
2.4. Clinical Characteristics and miRNA Expression Levels in NGT vs. FNGT
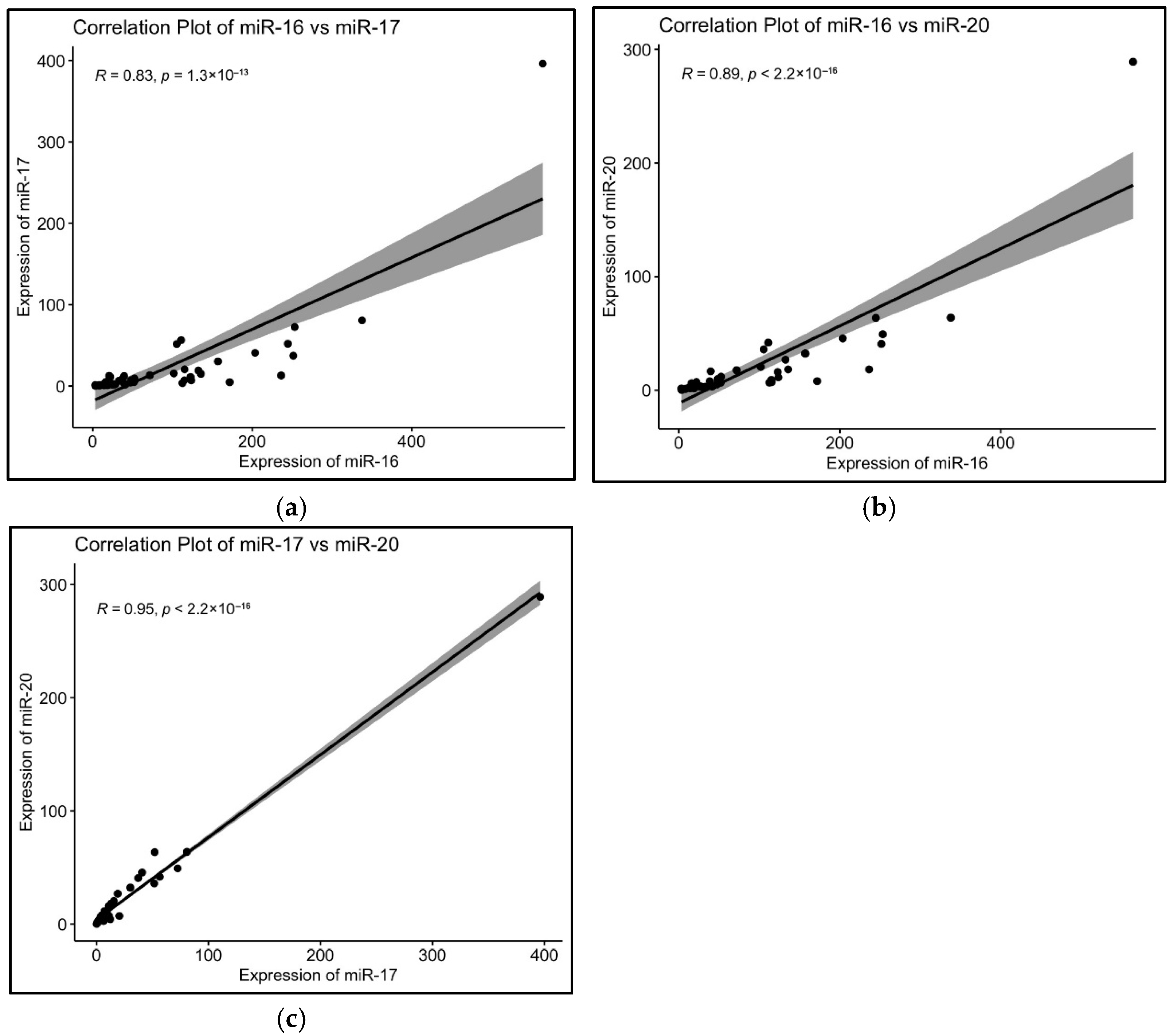
2.5. Correlation Analysis
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Clinical Specimen Collection and Biochemical Parameters
4.3. RT-qPCR for miRNA Expression
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Quintanilla Rodriguez, B.S.; Vadakekut, E.S.; Mahdy, H. Gestational Diabetes. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Choudhury, A.A.; Devi Rajeswari, V. Gestational Diabetes Mellitus—A Metabolic and Reproductive Disorder. Biomed. Pharmacother. Biomed. Pharmacother. 2021, 143, 112183. [Google Scholar] [CrossRef] [PubMed]
- Mogensen, C.S.; Nygaard, M.; Kampmann, U.; Mølgaard, C.; Magkos, F.; Geiker, N.R.W. Maternal Glucose Homeostasis during Pregnancy in Women with Overweight or Obesity and Offspring Metabolic Health. Sci. Rep. 2024, 14, 21398. [Google Scholar] [CrossRef]
- Buchanan, T.A.; Xiang, A.H.; Page, K.A. Gestational Diabetes Mellitus: Risks and Management during and after Pregnancy. Nat. Rev. Endocrinol. 2012, 8, 639–649. [Google Scholar] [CrossRef]
- Ryan, E.A. Diagnosing Gestational Diabetes. Diabetologia 2011, 54, 480–486. [Google Scholar] [CrossRef]
- Bogdanet, D.; O’Shea, P.; Lyons, C.; Shafat, A.; Dunne, F. The Oral Glucose Tolerance Test-Is It Time for a Change?—A Literature Review with an Emphasis on Pregnancy. J. Clin. Med. 2020, 9, 3451. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-N.; Jiang, Y.; Liu, X.-Q.; Yang, M.-M.; Chen, C.; Zhao, B.-H.; Huang, H.-F.; Luo, Q. MiRNAs in Gestational Diabetes Mellitus: Potential Mechanisms and Clinical Applications. J. Diabetes Res. 2021, 2021, 4632745. [Google Scholar] [CrossRef]
- Kitzmiller, J.L.; Dang-Kilduff, L.; Taslimi, M.M. Gestational Diabetes after Delivery. Short-Term Management and Long-Term Risks. Diabetes Care 2007, 30 (Suppl. S2), S225–S235. [Google Scholar] [CrossRef]
- Pallardo, F.; Herranz, L.; Garcia-Ingelmo, T.; Grande, C.; Martin-Vaquero, P.; Jañez, M.; Gonzalez, A. Early Postpartum Metabolic Assessment in Women with Prior Gestational Diabetes. Diabetes Care 1999, 22, 1053–1058. [Google Scholar] [CrossRef]
- Agarwal, M.M.; Punnose, J.; Dhatt, G.S. Gestational Diabetes: Implications of Variation in Post-Partum Follow-up Criteria. Eur. J. Obstet. Gynecol. Reprod. Biol. 2004, 113, 149–153. [Google Scholar] [CrossRef]
- Jang, H.C.; Yim, C.-H.; Han, K.O.; Yoon, H.-K.; Han, I.-K.; Kim, M.-Y.; Yang, J.-H.; Cho, N.H. Gestational Diabetes Mellitus in Korea: Prevalence and Prediction of Glucose Intolerance at Early Postpartum. Diabetes Res. Clin. Pract. 2003, 61, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.-N.; Wang, W.; Huang, X.-M.; Gao, S.-Y. Non-Coding RNAs and Extracellular Vehicles: Their Role in the Pathogenesis of Gestational Diabetes Mellitus. Front. Endocrinol. 2021, 12, 664287. [Google Scholar] [CrossRef] [PubMed]
- Kunysz, M.; Cieśla, M.; Darmochwał-Kolarz, D. Evaluation of miRNA Expression in Patients with Gestational Diabetes Mellitus: Investigating Diagnostic Potential and Clinical Implications. Diabetes Metab. Syndr. Obes. Targets Ther. 2024, 17, 881–891. [Google Scholar] [CrossRef]
- Owen, M.D.; Kennedy, M.G.; Quilang, R.C.; Scott, E.M.; Forbes, K. The Role of microRNAs in Pregnancies Complicated by Maternal Diabetes. Clin. Sci. 2024, 138, 1179–1207. [Google Scholar] [CrossRef]
- Nigi, L.; Grieco, G.E.; Ventriglia, G.; Brusco, N.; Mancarella, F.; Formichi, C.; Dotta, F.; Sebastiani, G. MicroRNAs as Regulators of Insulin Signaling: Research Updates and Potential Therapeutic Perspectives in Type 2 Diabetes. Int. J. Mol. Sci. 2018, 19, 3705. [Google Scholar] [CrossRef]
- Yu, C.-Y.; Yang, C.-Y.; Rui, Z.-L. MicroRNA-125b-5p Improves Pancreatic β-Cell Function through Inhibiting JNK Signaling Pathway by Targeting DACT1 in Mice with Type 2 Diabetes Mellitus. Life Sci. 2019, 224, 67–75. [Google Scholar] [CrossRef]
- da Silva, P.H.C.M.; de Santos, K.F.; da Silva, L.; da Costa, C.C.P.; da Santos, R.S.; da Reis, A.A.S. MicroRNAs Associated with the Pathophysiological Mechanisms of Gestational Diabetes Mellitus: A Systematic Review for Building a Panel of miRNAs. J. Pers. Med. 2023, 13, 1126. [Google Scholar] [CrossRef]
- Elhag, D.A.; Al Khodor, S. Exploring the Potential of microRNA as a Diagnostic Tool for Gestational Diabetes. J. Transl. Med. 2023, 21, 392. [Google Scholar] [CrossRef]
- Trejo-Gonzalez, N.L.; Palomar-Morales, M.; Baiza-Gutman, L.A.; Diaz-Rosas, G.; Ortega-Camarillo, C.; Contreras-Ramos, A. Circulating MicroRNAs Associated with Changes in the Placenta and Their Possible Role in the Fetus During Gestational Diabetes Mellitus: A Review. Metabolites 2025, 15, 367. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Khoshbakht, T.; Hussen, B.M.; Abdullah, S.T.; Taheri, M.; Samadian, M. A Review on the Role of Mir-16-5p in the Carcinogenesis. Cancer Cell Int. 2022, 22, 342. [Google Scholar] [CrossRef] [PubMed]
- Lovis, P.; Gattesco, S.; Regazzi, R. Regulation of the Expression of Components of the Exocytotic Machinery of Insulin-Secreting Cells by microRNAs. Biol. Chem. 2008, 389, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Tang, G.; Özcan, S. Role of MicroRNAs in Diabetes. Biochim. Biophys. Acta 2008, 1779, 697–701. [Google Scholar] [CrossRef]
- Zhao, W.; Gupta, A.; Krawczyk, J.; Gupta, S. The miR-17-92 Cluster: Yin and Yang in Human Cancers. Cancer Treat. Res. Commun. 2022, 33, 100647. [Google Scholar] [CrossRef]
- Tian, L.; Song, Z.; Shao, W.; Du, W.W.; Zhao, L.R.; Zeng, K.; Yang, B.B.; Jin, T. Curcumin Represses Mouse 3T3-L1 Cell Adipogenic Differentiation via Inhibiting miR-17-5p and Stimulating the Wnt Signalling Pathway Effector Tcf7l2. Cell Death Dis. 2017, 8, e2559. [Google Scholar] [CrossRef]
- Zeng, Y.; Wu, Y.; Zhang, Q.; Xiao, X. Non-Coding RNAs: The Link between Maternal Malnutrition and Offspring Metabolism. Front. Nutr. 2022, 9, 1022784. [Google Scholar] [CrossRef]
- Cao, Y.-L.; Jia, Y.-J.; Xing, B.-H.; Shi, D.-D.; Dong, X.-J. Plasma microRNA-16-5p, -17-5p and -20a-5p: Novel Diagnostic Biomarkers for Gestational Diabetes Mellitus. J. Obstet. Gynaecol. Res. 2017, 43, 974–981. [Google Scholar] [CrossRef]
- Gunderson, E.P.; Murtaugh, M.A.; Lewis, C.E.; Quesenberry, C.P.; West, D.S.; Sidney, S. Excess Gains in Weight and Waist Circumference Associated with Childbearing: The Coronary Artery Risk Development in Young Adults Study (CARDIA). Int. J. Obes. 2004, 28, 525–535. [Google Scholar] [CrossRef]
- Olive, V.; Sabio, E.; Bennett, M.J.; De Jong, C.S.; Biton, A.; McGann, J.C.; Greaney, S.K.; Sodir, N.M.; Zhou, A.Y.; Balakrishnan, A.; et al. A Component of the Mir-17-92 Polycistronic Oncomir Promotes Oncogene-Dependent Apoptosis. eLife 2013, 2, e00822. [Google Scholar] [CrossRef]
- Nair, S.; Guanzon, D.; Jayabalan, N.; Lai, A.; Scholz-Romero, K.; Kalita De Croft, P.; Ormazabal, V.; Palma, C.; Diaz, E.; McCarthy, E.A.; et al. Extracellular Vesicle-Associated miRNAs Are an Adaptive Response to Gestational Diabetes Mellitus. J. Transl. Med. 2021, 19, 360. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Van Oostdam, A.S.; Toro-Ortíz, J.C.; López, J.A.; Noyola, D.E.; García-López, D.A.; Durán-Figueroa, N.V.; Martínez-Martínez, E.; Portales-Pérez, D.P.; Salgado-Bustamante, M.; López-Hernández, Y. Placental Exosomes Isolated from Urine of Patients with Gestational Diabetes Exhibit a Differential Profile Expression of microRNAs across Gestation. Int. J. Mol. Med. 2020, 46, 546–560. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Tian, F.; Li, H.; Zhou, Y.; Lu, J.; Ge, Q. Profiling Maternal Plasma microRNA Expression in Early Pregnancy to Predict Gestational Diabetes Mellitus. Int. J. Gynaecol. Obstet. 2015, 130, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Hromadnikova, I.; Kotlabova, K.; Dvorakova, L.; Krofta, L. Diabetes Mellitus and Cardiovascular Risk Assessment in Mothers with a History of Gestational Diabetes Mellitus Based on Postpartal Expression Profile of MicroRNAs Associated with Diabetes Mellitus and Cardiovascular and Cerebrovascular Diseases. Int. J. Mol. Sci. 2020, 21, 2437. [Google Scholar] [CrossRef] [PubMed]
- Dinesen, S.; El-Faitarouni, A.; Dalgaard, L.T. Circulating microRNAs Associated with Gestational Diabetes Mellitus: Useful Biomarkers? J. Endocrinol. 2023, 256, e220170. [Google Scholar] [CrossRef]
- Lewis, K.A.; Chang, L.; Cheung, J.; Aouizerat, B.E.; Jelliffe-Pawlowski, L.L.; McLemore, M.R.; Piening, B.; Rand, L.; Ryckman, K.K.; Flowers, E. Systematic Review of Transcriptome and microRNAome Associations with Gestational Diabetes Mellitus. Front. Endocrinol. 2022, 13, 971354. [Google Scholar] [CrossRef]
- Kaur, J.; Saul, D.; Doolittle, M.L.; Rowsey, J.L.; Vos, S.J.; Farr, J.N.; Khosla, S.; Monroe, D.G. Identification of a Suitable Endogenous Control miRNA in Bone Aging and Senescence. Gene 2022, 835, 146642. [Google Scholar] [CrossRef]
- Xiang, M.; Zeng, Y.; Yang, R.; Xu, H.; Chen, Z.; Zhong, J.; Xie, H.; Xu, Y.; Zeng, X. U6 Is Not a Suitable Endogenous Control for the Quantification of Circulating microRNAs. Biochem. Biophys. Res. Commun. 2014, 454, 210–214. [Google Scholar] [CrossRef]
- Lou, G.; Ma, N.; Xu, Y.; Jiang, L.; Yang, J.; Wang, C.; Jiao, Y.; Gao, X. Differential Distribution of U6 (RNU6-1) Expression in Human Carcinoma Tissues Demonstrates the Requirement for Caution in the Internal Control Gene Selection for microRNA Quantification. Int. J. Mol. Med. 2015, 36, 1400–1408. [Google Scholar] [CrossRef]
- de Santana Silva, Í.T.S.; Fehlberg, H.F.; Ferreira, F.B.; da Silva, M.F.; dos Santos, P.R.; Porto, V.M.; Dias, J.C.T.; Albuquerque, G.R.; Mariano, A.P.M.; Gadelha, S.R.; et al. Identification of SnRNA U6 as an Endogenous Reference Gene for Normalization of MiRNA Expression Data in COVID-19 Patients. Sci. Rep. 2025, 15, 26636. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.-Y.; Cai, G.-Y.; Li, J.-J.; Bu, R.; Wang, N.; Yin, P.; Chen, X.-M. U6 Can Be Used as a Housekeeping Gene for Urinary Sediment miRNA Studies of IgA Nephropathy. Sci. Rep. 2018, 8, 10875. [Google Scholar] [CrossRef] [PubMed]
- Chekka, L.M.S.; Langaee, T.; Johnson, J.A. Comparison of Data Normalization Strategies for Array-Based MicroRNA Profiling Experiments and Identification and Validation of Circulating MicroRNAs as Endogenous Controls in Hypertension. Front. Genet. 2022, 13, 836636. [Google Scholar] [CrossRef]


| Variables | NGT (n= 20) | GDM (n = 30) | p Value |
|---|---|---|---|
| Age (mean ± SD) | 26.30 ± 3.88 | 26.83 ± 4.62 | 0.662 |
| BMI | 22.30 (4.53) | 28.65 (6.76) | 0.004 |
| DBP | 100 (25) | 112 (16) | 0.552 |
| SBP (mean ± SD) | 58.85 ± 10.23 | 65.57 ± 9.85 | 0.026 |
| TGs (mg/dL) | 153.09 (47.04) | 203.20 (95.72) | 0.0004 |
| Total cholesterol (mg/dL) (mean ± SD) | 199.07 ± 46.88 | 181.52 ± 46.34 | 0.200 |
| LDL (mg/dL) (mean ± SD) | 123.24 ± 42.29 | 98.64 ± 41.30 | 0.048 |
| HDL (mg/dL) (mean ± SD) | 43.78 ± 10.10 | 39.11 ± 12.02 | 0.145 |
| FBG (mg/dL) | 81.5 (8.50) | 94.5 (4.75) | 0.0000005 |
| 1 hr BG (mg/dL) (mean ± SD) | 109.95 ± 23.84 | 151.67 ± 37.05 | 0.00001 |
| 2 hr BG(mg/dL) | 86 (20.00) | 128 (73.25) | 0.00001 |
| hsa-miR-16 (2−dCq) | 22.52 (24.42) | 106.44 (115.21) | 0.004 |
| hsa-miR-17 (2−dCq) | 3.35 (8.94) | 10.18 (15.30) | 0.044 |
| Has-miR-20a (2−dCq) | 4.51 (5.29) | 10.56 (20.76) | 0.044 |
| Variables | GDM (n= 30) | FGDM (n = 30) | p Value |
|---|---|---|---|
| TGs (mg/dL) | 203.2 (95.72) | 191.31 (117.50) | 0.0196 |
| Total cholesterol (mg/dL) (mean ± SD) | 181.52 ± 46.34 | 184.57 ± 37.45 | 0.807 |
| LDL (mg/dL) (mean ± SD) | 90.65 (32.98) | 100.20 (27.83) | 0.515 |
| aHDL (mg/dL) (mean ± SD) | 39.11 ± 12.02 | 47.39 ± 11.98 | 0.0254 |
| hsa-miR-16-5p (2−dCq) | 106.44 (115.21) | 110.06 (314.94) | 0.100 |
| hsa-miR-17-5p (2−dCq) | 10.17 (15.29) | 19.97 (31.83) | 0.026 |
| hsa-miR-20a-5p (2−dCq) | 10.56 (20.76) | 17.03 (42.44) | 0.134 |
| Variables | FNGT (n = 20) | FGDM (n = 30) | p Value |
|---|---|---|---|
| TG (mg/dL) | 92.21 (125.04) | 191.32 (117.49) | 0.116 |
| Total cholesterol (mg/dL) (mean ± SD) | 162.84 ± 37.48 | 184.57 ± 37.45 | 0.051 |
| LDL (mg/dL) (mean ± SD) | 94.58 ± 30.28 | 102.49 ± 23.90 | 0.332 |
| HDL (mg/dL) (mean ± SD) | 42.19 ± 11.71 | 47.39 ± 11.98 | 0.134 |
| RBS postpartum | 80.95 (8.49) | 95.54 (17.13) | 0.00078 |
| hsa-miR-16-5p (2−dCq) | 34.86 (42.92) | 110.06 (314.94) | 0.005 |
| hsa-miR-17-5p (2−dCq) | 18.29 (42.56) | 19.97 (31.83) | 0.428 |
| hsa-miR-20a-5p (2−dCq) | 16.76 (29.40) | 17.03 (42.44) | 0.301 |
| Variables | NGT (n = 20) | FNGT (n = 20) | p Value |
|---|---|---|---|
| TG (mg/dL) | 153.09 (47.04) | 92.21 (125.04) | 0.2774 |
| Total cholesterol (mg/dL) (mean ± SD) | 199.07 ± 46.88 | 162.84 ± 37.48 | 0.01718 |
| LDL (mg/dL) (mean ± SD) | 123.24 ± 42.29 | 94.58 ± 30.28 | 0.03459 |
| aHDL (mg/dL) (mean ± SD) | 43.78 ± 10.10 | 42.19 ± 11.71 | 0.6541 |
| hsa-miR-16-5p (2−dCq) | 22.52 (24.42) | 34.86 (42.92) | 0.7012 |
| hsa-miR-17-5p (2−dCq) | 3.35 (8.94) | 18.29 (42.56) | 0.1893 |
| hsa-miR-20a-5p (2−dCq) | 4.51 (5.29) | 16.76 (29.40) | 0.2162 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pillai, A.; Kandi, S.M.; Tripathy, N.; Agarwal, D.; Mukhopadhyay, I.; Mukherjee, B.; Vashum, Y. Sustained Higher Levels of Plasma hsa-miR-17-5p Expression During Gestational Diabetes Mellitus and Postpartum. Epigenomes 2025, 9, 37. https://doi.org/10.3390/epigenomes9040037
Pillai A, Kandi SM, Tripathy N, Agarwal D, Mukhopadhyay I, Mukherjee B, Vashum Y. Sustained Higher Levels of Plasma hsa-miR-17-5p Expression During Gestational Diabetes Mellitus and Postpartum. Epigenomes. 2025; 9(4):37. https://doi.org/10.3390/epigenomes9040037
Chicago/Turabian StylePillai, Arathi, Sibin M Kandi, Nidhi Tripathy, Deeptika Agarwal, Indrani Mukhopadhyay, Bhasker Mukherjee, and Y Vashum. 2025. "Sustained Higher Levels of Plasma hsa-miR-17-5p Expression During Gestational Diabetes Mellitus and Postpartum" Epigenomes 9, no. 4: 37. https://doi.org/10.3390/epigenomes9040037
APA StylePillai, A., Kandi, S. M., Tripathy, N., Agarwal, D., Mukhopadhyay, I., Mukherjee, B., & Vashum, Y. (2025). Sustained Higher Levels of Plasma hsa-miR-17-5p Expression During Gestational Diabetes Mellitus and Postpartum. Epigenomes, 9(4), 37. https://doi.org/10.3390/epigenomes9040037







