Innate Immune Surveillance and Recognition of Epigenetic Marks
Abstract
1. Introduction
2. Epigenetic Alterations as Danger Signals: Conceptual Framework
3. Molecular Mechanisms of Epigenetic Alteration Recognition by PRRs
3.1. Sensing Unmethylated DNA Patterns
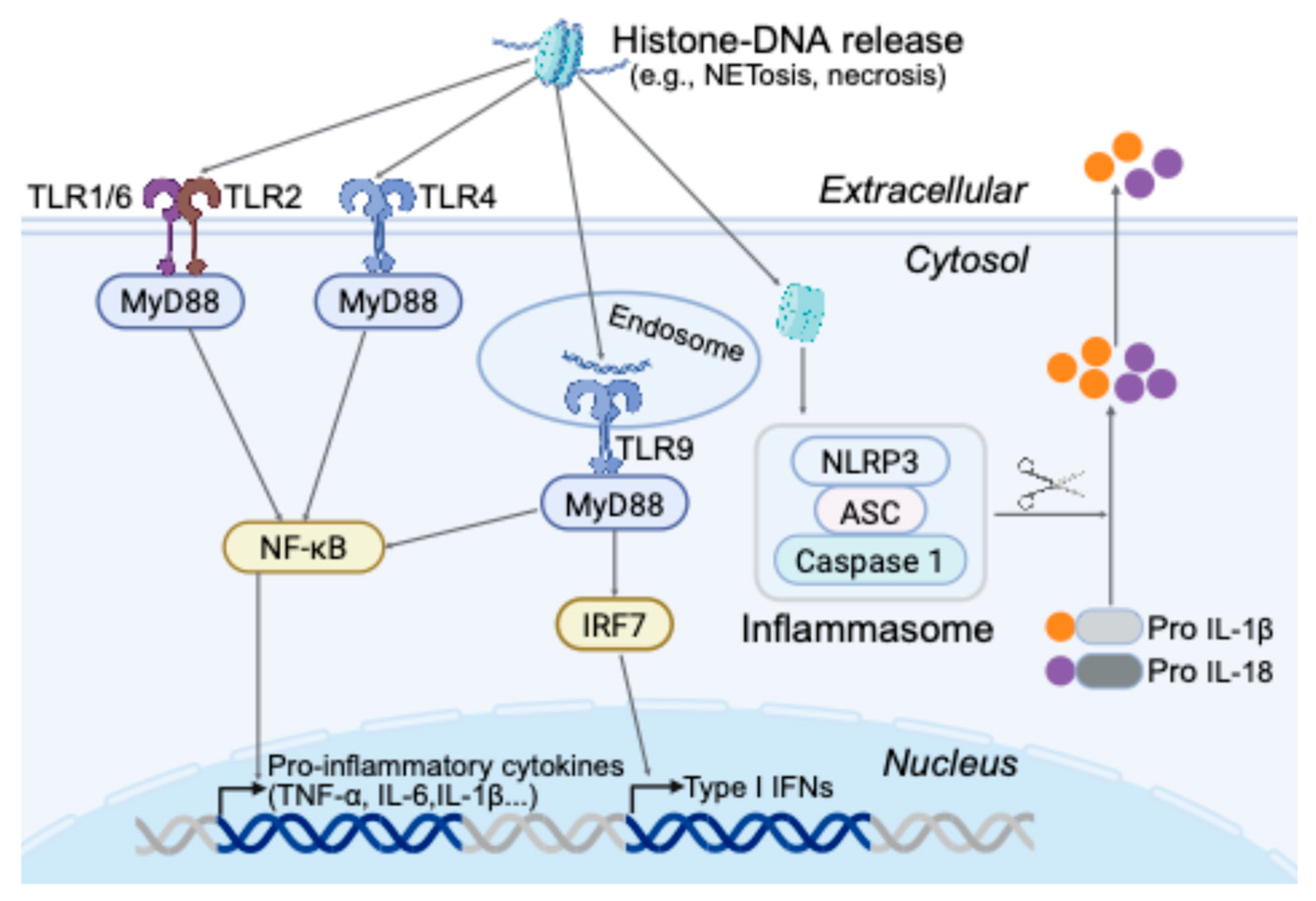
3.2. Extracellular Chromatin and Histones as DAMPs
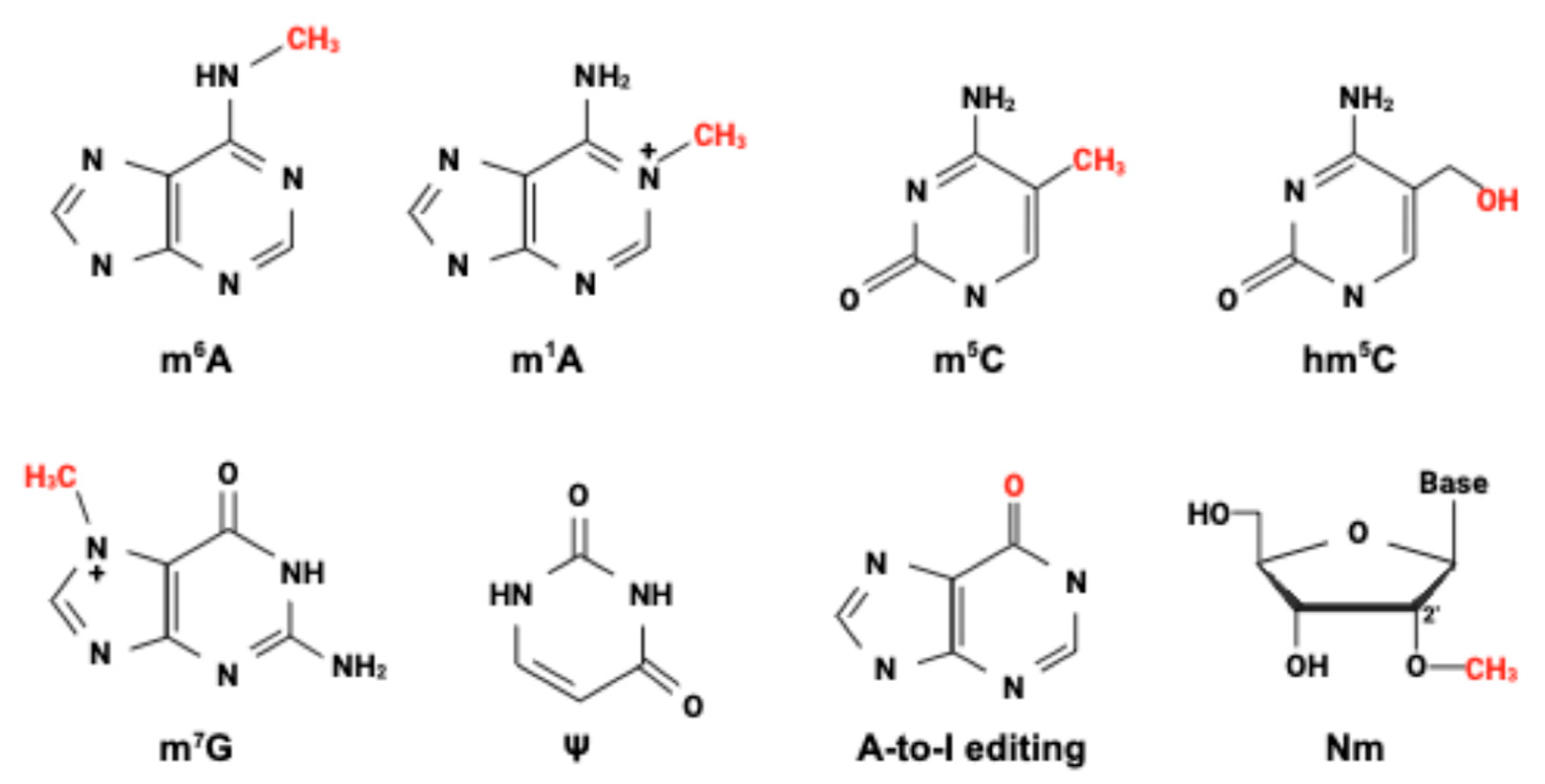
3.3. Epitranscriptomic Modifications and RNA Sensing
4. Evidence from Cancer Models and Disease Contexts
4.1. Viral Mimicry in Tumors: Endogenous Retroviruses and Interferon Activation
4.2. DNA Damage, Micronuclei, and cGAS-STING in Cancer
4.3. Immunogenic Cell Death and Epigenetic Modulation
5. Can Epigenetic Readers Function as Pattern Recognition Receptors?
6. Conclusions and Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Janeway, C.A., Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 1989, 54 Pt 1, 1–13. [Google Scholar] [CrossRef]
- Janeway, C.A., Jr.; Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 2002, 20, 197–216. [Google Scholar] [CrossRef]
- Medzhitov, R.; Janeway, C., Jr. Innate immune recognition: Mechanisms and pathways. Immunol. Rev. 2000, 173, 89–97. [Google Scholar] [CrossRef]
- Matzinger, P. Tolerance, danger, and the extended family. Annu. Rev. Immunol. 1994, 12, 991–1045. [Google Scholar] [CrossRef]
- Matzinger, P. The danger model: A renewed sense of self. Science 2002, 296, 301–305. [Google Scholar] [CrossRef]
- Seong, S.Y.; Matzinger, P. Hydrophobicity: An ancient damage-associated molecular pattern that initiates innate immune responses. Nat. Rev. Immunol. 2004, 4, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Jiang, W.; Zhou, R. DAMPs and DAMP-sensing receptors in inflammation and diseases. Immunity 2024, 57, 752–771. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Mattei, A.L.; Bailly, N.; Meissner, A. DNA methylation: A historical perspective. Trends Genet. 2022, 38, 676–707. [Google Scholar] [CrossRef]
- Li, X.; Ye, Y.; Peng, K.; Zeng, Z.; Chen, L.; Zeng, Y. Histones: The critical players in innate immunity. Front. Immunol. 2022, 13, 1030610. [Google Scholar] [CrossRef] [PubMed]
- Silk, E.; Zhao, H.; Weng, H.; Ma, D. The role of extracellular histone in organ injury. Cell Death Dis. 2017, 8, e2812. [Google Scholar] [CrossRef]
- Yang, T.; Peng, J.; Zhang, Z.; Chen, Y.; Liu, Z.; Jiang, L.; Jin, L.; Han, M.; Su, B.; Li, Y. Emerging therapeutic strategies targeting extracellular histones for critical and inflammatory diseases: An updated narrative review. Front. Immunol. 2024, 15, 1438984. [Google Scholar] [CrossRef] [PubMed]
- Latz, E.; Schoenemeyer, A.; Visintin, A.; Fitzgerald, K.A.; Monks, B.G.; Knetter, C.F.; Lien, E.; Nilsen, N.J.; Espevik, T.; Golenbock, D.T. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 2004, 5, 190–198. [Google Scholar] [CrossRef]
- Hemmi, H.; Takeuchi, O.; Kawai, T.; Kaisho, T.; Sato, S.; Sanjo, H.; Matsumoto, M.; Hoshino, K.; Wagner, H.; Takeda, K.; et al. A Toll-like receptor recognizes bacterial DNA. Nature 2000, 408, 740–745. [Google Scholar] [CrossRef] [PubMed]
- West, A.P.; Shadel, G.S. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat. Rev. Immunol. 2017, 17, 363–375. [Google Scholar] [CrossRef]
- Beltran-Garcia, J.; Osca-Verdegal, R.; Perez-Cremades, D.; Novella, S.; Hermenegildo, C.; Pallardo, F.V.; Garcia-Gimenez, J.L. Extracellular Histones Activate Endothelial NLRP3 Inflammasome and are Associated with a Severe Sepsis Phenotype. J. Inflamm. Res. 2022, 15, 4217–4238. [Google Scholar] [CrossRef]
- Chen, G.Y.; Nunez, G. Sterile inflammation: Sensing and reacting to damage. Nat. Rev. Immunol. 2010, 10, 826–837. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, X.; Monestier, M.; Esmon, N.L.; Esmon, C.T. Extracellular histones are mediators of death through TLR2 and TLR4 in mouse fatal liver injury. J. Immunol. 2011, 187, 2626–2631. [Google Scholar] [CrossRef]
- Chen, R.; Ishak, C.A.; De Carvalho, D.D. Endogenous Retroelements and the Viral Mimicry Response in Cancer Therapy and Cellular Homeostasis. Cancer Discov. 2021, 11, 2707–2725. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M.; Dawson, M.A.; Kadoch, C.; Rassool, F.V.; Jones, P.A.; Baylin, S.B. The Epigenetic Hallmarks of Cancer. Cancer Discov. 2024, 14, 1783–1809. [Google Scholar] [CrossRef]
- Nishiyama, A.; Nakanishi, M. Navigating the DNA methylation landscape of cancer. Trends Genet. 2021, 37, 1012–1027. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, M.; Wang, Y. The roles of histone modifications in tumorigenesis and associated inhibitors in cancer therapy. J. Natl. Cancer Cent. 2022, 2, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Locatelli, M.; Faure-Dupuy, S. Virus hijacking of host epigenetic machinery to impair immune response. J. Virol. 2023, 97, e0065823. [Google Scholar] [CrossRef]
- Roh, J.S.; Sohn, D.H. Damage-Associated Molecular Patterns in Inflammatory Diseases. Immune Netw. 2018, 18, e27. [Google Scholar] [CrossRef]
- Miller, S.I.; Ernst, R.K.; Bader, M.W. LPS, TLR4 and infectious disease diversity. Nat. Rev. Microbiol. 2005, 3, 36–46. [Google Scholar] [CrossRef]
- Smith, K.D.; Andersen-Nissen, E.; Hayashi, F.; Strobe, K.; Bergman, M.A.; Barrett, S.L.; Cookson, B.T.; Aderem, A. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat. Immunol. 2003, 4, 1247–1253. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, H.; Wang, J.; Chen, J.; Guo, Z.; Liu, Y.; Hua, H. Exploiting RIG-I-like receptor pathway for cancer immunotherapy. J. Hematol. Oncol. 2023, 16, 8. [Google Scholar] [CrossRef]
- Zhong, H.; Li, X.; Zhou, S.; Jiang, P.; Liu, X.; Ouyang, M.; Nie, Y.; Chen, X.; Zhang, L.; Liu, Y.; et al. Interplay between RAGE and TLR4 Regulates HMGB1-Induced Inflammation by Promoting Cell Surface Expression of RAGE and TLR4. J. Immunol. 2020, 205, 767–775. [Google Scholar] [CrossRef]
- Ronning, K.E.; Dechelle-Marquet, P.A.; Che, Y.; Guillonneau, X.; Sennlaub, F.; Delarasse, C. The P2X7 Receptor, a Multifaceted Receptor in Alzheimer’s Disease. Int. J. Mol. Sci. 2023, 24, 11747. [Google Scholar] [CrossRef] [PubMed]
- Vande Walle, L.; Lamkanfi, M. Drugging the NLRP3 inflammasome: From signalling mechanisms to therapeutic targets. Nat. Rev. Drug Discov. 2024, 23, 43–66. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, K.; Richez, C.; Uccellini, M.B.; Richards, R.J.; Bonegio, R.G.; Akira, S.; Monestier, M.; Corley, R.B.; Viglianti, G.A.; Marshak-Rothstein, A.; et al. Requirement for DNA CpG content in TLR9-dependent dendritic cell activation induced by DNA-containing immune complexes. J. Immunol. 2009, 183, 3109–3117. [Google Scholar] [CrossRef] [PubMed]
- Amadio, R.; Piperno, G.M.; Benvenuti, F. Self-DNA Sensing by cGAS-STING and TLR9 in Autoimmunity: Is the Cytoskeleton in Control? Front. Immunol. 2021, 12, 657344. [Google Scholar] [CrossRef]
- Liu, R.; Zhao, E.; Yu, H.; Yuan, C.; Abbas, M.N.; Cui, H. Methylation across the central dogma in health and diseases: New therapeutic strategies. Signal Transduct. Target. Ther. 2023, 8, 310. [Google Scholar] [CrossRef]
- Perez, R.F.; Tejedor, J.R.; Bayon, G.F.; Fernandez, A.F.; Fraga, M.F. Distinct chromatin signatures of DNA hypomethylation in aging and cancer. Aging Cell 2018, 17, e12744. [Google Scholar] [CrossRef]
- Chen, L.; Ganz, P.A.; Sehl, M.E. DNA Methylation, Aging, and Cancer Risk: A Mini-Review. Front. Bioinform. 2022, 2, 847629. [Google Scholar] [CrossRef]
- Wu, J.; Sun, L.; Chen, X.; Du, F.; Shi, H.; Chen, C.; Chen, Z.J. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 2013, 339, 826–830. [Google Scholar] [CrossRef]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013, 339, 786–791. [Google Scholar] [CrossRef]
- Beck, M.A.; Fischer, H.; Grabner, L.M.; Groffics, T.; Winter, M.; Tangermann, S.; Meischel, T.; Zaussinger-Haas, B.; Wagner, P.; Fischer, C.; et al. DNA hypomethylation leads to cGAS-induced autoinflammation in the epidermis. EMBO J. 2021, 40, e108234. [Google Scholar] [CrossRef]
- Lu, Y.; Zhao, M.; Chen, L.; Wang, Y.; Liu, T.; Liu, H. cGAS: Action in the nucleus. Front. Immunol. 2024, 15, 1380517. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Liu, F. Nuclear cGAS: Sequestration and beyond. Protein Cell 2022, 13, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Kim, S.J.; Lei, Y.; Wang, S.; Wang, H.; Huang, H.; Zhang, H.; Tsung, A. Neutrophil extracellular traps in homeostasis and disease. Signal Transduct. Target. Ther. 2024, 9, 235. [Google Scholar] [CrossRef]
- Thiam, H.R.; Wong, S.L.; Wagner, D.D.; Waterman, C.M. Cellular Mechanisms of NETosis. Annu. Rev. Cell Dev. Biol. 2020, 36, 191–218. [Google Scholar] [CrossRef]
- Allam, R.; Darisipudi, M.N.; Tschopp, J.; Anders, H.J. Histones trigger sterile inflammation by activating the NLRP3 inflammasome. Eur. J. Immunol. 2013, 43, 3336–3342. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Chen, H.W.; Evankovich, J.; Yan, W.; Rosborough, B.R.; Nace, G.W.; Ding, Q.; Loughran, P.; Beer-Stolz, D.; Billiar, T.R.; et al. Histones activate the NLRP3 inflammasome in Kupffer cells during sterile inflammatory liver injury. J. Immunol. 2013, 191, 2665–2679. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Lv, S.; Li, H.; Cui, W.; Wang, L. Enhancing the Anticancer Efficacy of Immunotherapy through Combination with Histone Modification Inhibitors. Genes 2018, 9, 633. [Google Scholar] [CrossRef]
- Park, J.; Lee, K.; Kim, K.; Yi, S.J. The role of histone modifications: From neurodevelopment to neurodiseases. Signal Transduct. Target. Ther. 2022, 7, 217. [Google Scholar] [CrossRef]
- Leshner, M.; Wang, S.; Lewis, C.; Zheng, H.; Chen, X.A.; Santy, L.; Wang, Y. PAD4 mediated histone hypercitrullination induces heterochromatin decondensation and chromatin unfolding to form neutrophil extracellular trap-like structures. Front. Immunol. 2012, 3, 307. [Google Scholar] [CrossRef]
- Wong, S.L.; Wagner, D.D. Peptidylarginine deiminase 4: A nuclear button triggering neutrophil extracellular traps in inflammatory diseases and aging. FASEB J. 2018, 32, 6358. [Google Scholar] [CrossRef]
- Peng, W.; Wu, S.; Wang, W. Correlation of serum citrullinated histone H3 levels with disease activity in patients with rheumatoid arthritis. Clin. Exp. Rheumatol. 2023, 41, 1792–1800. [Google Scholar] [CrossRef]
- Schwartz, S. Cracking the epitranscriptome. RNA 2016, 22, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Yoon, K.J. Voyages to map unexplored parts of the epitranscriptomic world. Exp. Mol. Med. 2022, 54, 1599–1600. [Google Scholar] [CrossRef] [PubMed]
- Kariko, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef]
- Kim, G.W.; Imam, H.; Khan, M.; Siddiqui, A. N(6)-Methyladenosine modification of hepatitis B and C viral RNAs attenuates host innate immunity via RIG-I signaling. J. Biol. Chem. 2020, 295, 13123–13133. [Google Scholar] [CrossRef]
- Lu, M.; Zhang, Z.; Xue, M.; Zhao, B.S.; Harder, O.; Li, A.; Liang, X.; Gao, T.Z.; Xu, Y.; Zhou, J.; et al. N(6)-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat. Microbiol. 2020, 5, 584–598. [Google Scholar] [CrossRef]
- Zhao, B.; Wang, W.; Zhao, Y.; Qiao, H.; Gao, Z.; Chuai, X. Regulation of Antiviral Immune Response by N (6)-Methyladenosine of mRNA. Front. Microbiol. 2021, 12, 789605. [Google Scholar] [CrossRef]
- Li, N.; Hui, H.; Bray, B.; Gonzalez, G.M.; Zeller, M.; Anderson, K.G.; Knight, R.; Smith, D.; Wang, Y.; Carlin, A.F.; et al. METTL3 regulates viral m6A RNA modification and host cell innate immune responses during SARS-CoV-2 infection. Cell Rep. 2021, 35, 109091. [Google Scholar] [CrossRef]
- Jansz, N.; Faulkner, G.J. Endogenous retroviruses in the origins and treatment of cancer. Genome Biol. 2021, 22, 147. [Google Scholar] [CrossRef]
- Yu, J.; Qiu, P.; Ai, J.; Liu, B.; Han, G.Z.; Zhu, F.; Zhang, W.; Cui, J. Endogenous retrovirus activation: Potential for immunology and clinical applications. Natl. Sci. Rev. 2024, 11, nwae034. [Google Scholar] [CrossRef] [PubMed]
- Dias Junior, A.G.; Sampaio, N.G.; Rehwinkel, J. A Balancing Act: MDA5 in Antiviral Immunity and Autoinflammation. Trends Microbiol. 2019, 27, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Rehwinkel, J.; Mehdipour, P. ADAR1: From basic mechanisms to inhibitors. Trends Cell Biol. 2025, 35, 59–73. [Google Scholar] [CrossRef]
- Wang, R.; Dong, X.; Zhang, X.; Liao, J.; Cui, W.; Li, W. Exploring viral mimicry combined with epigenetics and tumor immunity: New perspectives in cancer therapy. Int. J. Biol. Sci. 2025, 21, 958–973. [Google Scholar] [CrossRef]
- Liu, K.; Li, Y.; Shen, M.; Xu, W.; Wu, S.; Yang, X.; Zhang, B.; Lin, N. Epigenetic Regulation of Stromal and Immune Cells and Therapeutic Targets in the Tumor Microenvironment. Biomolecules 2025, 15, 71. [Google Scholar] [CrossRef]
- Xie, Z.; Zhou, Z.; Yang, S.; Zhang, S.; Shao, B. Epigenetic regulation and therapeutic targets in the tumor microenvironment. Mol. Biomed. 2023, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Chiappinelli, K.B.; Strissel, P.L.; Desrichard, A.; Li, H.; Henke, C.; Akman, B.; Hein, A.; Rote, N.S.; Cope, L.M.; Snyder, A.; et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 2015, 162, 974–986. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Zhang, B.; Li, B.; Wu, H.; Jiang, M. Cold and hot tumors: From molecular mechanisms to targeted therapy. Signal Transduct. Target. Ther. 2024, 9, 274. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Cui, G.; Liu, H.; Han, Y.; Cai, C.; Feng, Z.; Shen, H.; Zeng, S. Converting “cold” to “hot”: Epigenetics strategies to improve immune therapy effect by regulating tumor-associated immune suppressive cells. Cancer Commun. 2024, 44, 601–636. [Google Scholar] [CrossRef]
- Chen, M.; Linstra, R.; van Vugt, M. Genomic instability, inflammatory signaling and response to cancer immunotherapy. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188661. [Google Scholar] [CrossRef]
- Hosea, R.; Hillary, S.; Naqvi, S.; Wu, S.; Kasim, V. The two sides of chromosomal instability: Drivers and brakes in cancer. Signal Transduct. Target. Ther. 2024, 9, 75. [Google Scholar] [CrossRef]
- Kwon, J.; Bakhoum, S.F. The Cytosolic DNA-Sensing cGAS-STING Pathway in Cancer. Cancer Discov. 2020, 10, 26–39. [Google Scholar] [CrossRef]
- Tripathi, R.; Modur, V.; Senovilla, L.; Kroemer, G.; Komurov, K. Suppression of tumor antigen presentation during aneuploid tumor evolution contributes to immune evasion. Oncoimmunology 2019, 8, 1657374. [Google Scholar] [CrossRef]
- MacDonald, K.M.; Benguerfi, S.; Harding, S.M. Alerting the immune system to DNA damage: Micronuclei as mediators. Essays Biochem. 2020, 64, 753–764. [Google Scholar] [CrossRef]
- Bharti, V.; Kumar, A.; Wang, Y.; Roychowdhury, N.; de Lima Bellan, D.; Kassaye, B.B.; Watkins, R.; Capece, M.; Chung, C.G.; Hilinski, G.; et al. TTK inhibitor OSU13 promotes immunotherapy responses by activating tumor STING. JCI Insight 2024, 9, e177523. [Google Scholar] [CrossRef]
- Chan, C.Y.; Chiu, D.K.; Yuen, V.W.; Law, C.T.; Wong, B.P.; Thu, K.L.; Cescon, D.W.; Soria-Bretones, I.; Cheu, J.W.; Lee, D.; et al. CFI-402257, a TTK inhibitor, effectively suppresses hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 2022, 119, e2119514119. [Google Scholar] [CrossRef]
- Hu, Y.; Manasrah, B.K.; McGregor, S.M.; Lera, R.F.; Norman, R.X.; Tucker, J.B.; Scribano, C.M.; Yan, R.E.; Humayun, M.; Wisinski, K.B.; et al. Paclitaxel Induces Micronucleation and Activates Pro-Inflammatory cGAS-STING Signaling in Triple-Negative Breast Cancer. Mol. Cancer Ther. 2021, 20, 2553–2567. [Google Scholar] [CrossRef]
- Yue, B.; Gao, W.; Lovell, J.F.; Jin, H.; Huang, J. The cGAS-STING pathway in cancer immunity: Dual roles, therapeutic strategies, and clinical challenges. Essays Biochem. 2025, 69, EBC20253006. [Google Scholar] [CrossRef]
- Naik, A.; Lattab, B.; Qasem, H.; Decock, J. Cancer testis antigens: Emerging therapeutic targets leveraging genomic instability in cancer. Mol. Ther. Oncol. 2024, 32, 200768. [Google Scholar] [CrossRef]
- Ahmed, A.; Tait, S.W.G. Targeting immunogenic cell death in cancer. Mol. Oncol. 2020, 14, 2994–3006. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Huang, M.; Yao, Y.M. The Effect and Regulatory Mechanism of High Mobility Group Box-1 Protein on Immune Cells in Inflammatory Diseases. Cells 2021, 10, 1044. [Google Scholar] [CrossRef] [PubMed]
- Rivas-Yanez, E.; Barrera-Avalos, C.; Parra-Tello, B.; Briceno, P.; Rosemblatt, M.V.; Saavedra-Almarza, J.; Rosemblatt, M.; Acuna-Castillo, C.; Bono, M.R.; Sauma, D. P2X7 Receptor at the Crossroads of T Cell Fate. Int. J. Mol. Sci. 2020, 21, 4937. [Google Scholar] [CrossRef] [PubMed]
- Dhatchinamoorthy, K.; Colbert, J.D.; Rock, K.L. Cancer Immune Evasion Through Loss of MHC Class I Antigen Presentation. Front. Immunol. 2021, 12, 636568. [Google Scholar] [CrossRef]
- Wang, Y.; Bedford, M.T. Effectors and effects of arginine methylation. Biochem. Soc. Trans. 2023, 51, 725–734. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, W.; Shen, F.; Yang, X.; Liu, H.; Dai, S.; Sun, X.; Huang, J.; Guo, Q. YTH Domain Proteins: A Family of m(6)A Readers in Cancer Progression. Front. Oncol. 2021, 11, 629560. [Google Scholar] [CrossRef] [PubMed]
- Kumari, P.; Russo, A.J.; Shivcharan, S.; Rathinam, V.A. AIM2 in health and disease: Inflammasome and beyond. Immunol. Rev. 2020, 297, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Boulias, K.; Greer, E.L. Means, mechanisms and consequences of adenine methylation in DNA. Nat. Rev. Genet. 2022, 23, 411–428. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Lyu, Z.; Chen, J.; Chen, G. 5-Hydroxymethylcytosine: Far Beyond the Intermediate of DNA Demethylation. Int. J. Mol. Sci. 2024, 25, 11780. [Google Scholar] [CrossRef]
| Category of Signal | Source | Recognized by PRRs (Examples) |
|---|---|---|
| Pathogen-Associated Molecular Patterns (PAMPs) | Microbial molecules | TLR4 recognizes LPS on bacteria [25]. TLR5 recognizes flagellin [26]. RIG-I/MDA5 sense viral RNA [27]. TLR9 recognizes microbial DNA [14]. |
| Damage/Danger- Associated Molecular Patterns (DAMPs) | Host molecules released due to damage/danger | TLR4 and RAGE bind HMGB1 [28]. P2 × 7 receptor senses ATP [29]. NLRP3 inflammasome is activated by diverse DAMP-induced stress [30]. |
| Epigenetic alteration– Associated Molecular Patterns (EAMPs) | Epigenetic modifications that produce abnormal or pathogen-mimicking patterns without altering the DNA sequence. | TLR9 can respond to host DNA containing unmethylated CpG motifs (normally suppressed by methylation) [31]. extracellular DNA–Histone complexes engage TLR2/4 [18]. cGAS detects any cytosolic DNA (self or viral) leading to STING activation [32]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y. Innate Immune Surveillance and Recognition of Epigenetic Marks. Epigenomes 2025, 9, 33. https://doi.org/10.3390/epigenomes9030033
Wang Y. Innate Immune Surveillance and Recognition of Epigenetic Marks. Epigenomes. 2025; 9(3):33. https://doi.org/10.3390/epigenomes9030033
Chicago/Turabian StyleWang, Yalong. 2025. "Innate Immune Surveillance and Recognition of Epigenetic Marks" Epigenomes 9, no. 3: 33. https://doi.org/10.3390/epigenomes9030033
APA StyleWang, Y. (2025). Innate Immune Surveillance and Recognition of Epigenetic Marks. Epigenomes, 9(3), 33. https://doi.org/10.3390/epigenomes9030033






