Induction of DNA Demethylation: Strategies and Consequences
Abstract
1. Introduction
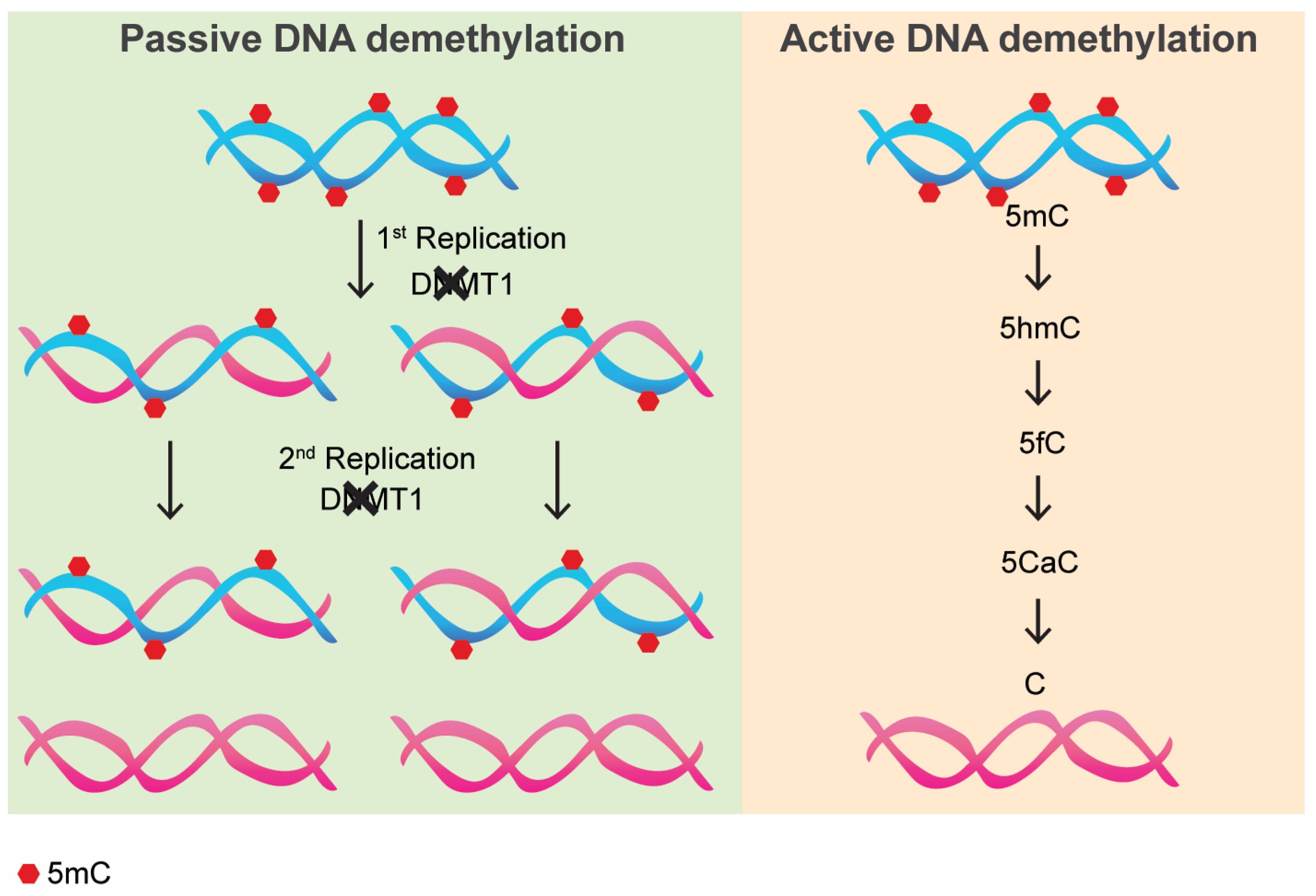
2. Passive DNA Methylation: How It Works and Its Consequences
3. Active DNA Demethylation: How It Works and Its Consequences
4. The Role of Non-Coding RNAs in DNMTs’ Activity: Impact on DNA Methylation
5. Pharmacological Inhibition of DNA Methylation in Biomedical Research: Pros and Cons
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Illingworth, R.S.; Bird, A.P. CpG islands—‘A rough guide’. FEBS Lett. 2009, 583, 1713–1720. [Google Scholar] [CrossRef]
- Choy, J.S.; Wei, S.; Lee, J.Y.; Tan, S.; Chu, S.; Lee, T.-H. DNA methylation increases nucleosome compaction and rigidity. J. Am. Chem. Soc. 2010, 132, 1782–1783. [Google Scholar] [CrossRef] [PubMed]
- Sharp, A.J.; Stathaki, E.; Migliavacca, E.; Brahmachary, M.; Montgomery, S.B.; Dupre, Y.; Antonarakis, S.E. DNA methylation profiles of human active and inactive X chromosomes. Genome Res. 2011, 21, 1592–1600. [Google Scholar] [CrossRef]
- Sheaffer, K.L.; Elliott, E.N.; Kaestner, K.H. DNA hypomethylation contributes to genomic instability and intestinal cancer initiation. Cancer Prev. Res. 2016, 9, 534–546. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Kagiampakis, I.; Pan, L.; Zhang, Y.W.; Murphy, L.; Tao, Y.; Kong, X.; Xia, L.; Carvalho, F.L.; Sen, S.; et al. DNA methylation patterns separate senescence from transformation potential and indicate cancer risk. Cancer Cell 2018, 33, 309–321.e5. [Google Scholar] [CrossRef]
- Tucci, V.; Isles, A.R.; Kelsey, G.; Ferguson-Smith, A.C.; Tucci, V.; Bartolomei, M.S.; Benvenisty, N.; Bourc’his, D.; Charalambous, M.; Dulac, C.; et al. Genomic imprinting and physiological processes in mammals. Cell 2019, 176, 952–965. [Google Scholar] [CrossRef]
- Barra, V.; Chiavetta, R.F.; Titoli, S.; Provenzano, I.M.; Carollo, P.S.; Di Leonardo, A. Specific irreversible cell-cycle arrest and depletion of cancer cells obtained by combining curcumin and the flavonoids quercetin and fisetin. Genes 2022, 13, 1125. [Google Scholar] [CrossRef] [PubMed]
- Crouch, J.; Shvedova, M.; Thanapaul, R.J.R.S.; Botchkarev, V.; Roh, D. Epigenetic regulation of cellular senescence. Cells 2022, 11, 672. [Google Scholar] [CrossRef]
- Carollo, P.S.; Barra, V. Chromatin epigenetics and nuclear lamina keep the nucleus in shape: Examples from natural and accelerated aging. Biol. Cell 2023, 115, e2200023. [Google Scholar] [CrossRef]
- Martino, S.; Carollo, P.S.; Barra, V. A glimpse into chromatin organization and nuclear lamina contribution in neuronal differentiation. Genes 2023, 14, 1046. [Google Scholar] [CrossRef]
- Chang, W.; Zhao, Y.; Rayêe, D.; Xie, Q.; Suzuki, M.; Zheng, D.; Cvekl, A. Dynamic changes in whole genome DNA methylation, chromatin and gene expression during mouse lens differentiation. Epigenetics Chromatin 2023, 16, 4. [Google Scholar] [CrossRef]
- Lyko, F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [CrossRef]
- Yano, N.; Fedulov, A.V. Targeted DNA demethylation: Vectors, effectors and perspectives. Biomedicines 2023, 11, 1334. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, A.; Nakanishi, M. Navigating the DNA methylation landscape of cancer. Trends Genet. 2021, 37, 1012–1027. [Google Scholar] [CrossRef]
- Hollenbach, P.W.; Nguyen, A.N.; Brady, H.; Williams, M.; Ning, Y.; Richard, N.; Krushel, L.; Aukerman, S.L.; Heise, C.; MacBeth, K.J. A Comparison of azacitidine and decitabine activities in acute myeloid leukemia cell lines. PLoS ONE 2010, 5, e9001. [Google Scholar] [CrossRef] [PubMed]
- Costa, G.; Barra, V.; Lentini, L.; Cilluffo, D.; Leonardo, A.D. DNA demethylation caused by 5-Aza-2’-deoxycytidine induces mitotic alterations and aneuploidy. Oncotarget 2016, 7, 3726–3739. [Google Scholar] [CrossRef] [PubMed]
- Pappalardi, M.B.; Keenan, K.; Cockerill, M.; Kellner, W.A.; Stowell, A.; Sherk, C.; Wong, K.; Pathuri, S.; Briand, J.; Steidel, M.; et al. Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia. Nat. Cancer 2021, 2, 1002–1017. [Google Scholar] [CrossRef]
- Chen, Q.; Liu, B.; Zeng, Y.; Hwang, J.W.; Dai, N.; Corrêa, I.R.; Estecio, M.R.; Zhang, X.; Santos, M.A.; Chen, T.; et al. GSK-3484862 targets DNMT1 for degradation in cells. NAR Cancer 2023, 5, zcad022. [Google Scholar] [CrossRef]
- Laranjeira, A.B.A.; Hollingshead, M.G.; Nguyen, D.; Kinders, R.J.; Doroshow, J.H.; Yang, S.X. DNA damage, demethylation and anticancer activity of DNA methyltransferase (DNMT) inhibitors. Sci. Rep. 2023, 13, 5964. [Google Scholar] [CrossRef]
- Patel, K.; Dickson, J.; Din, S.; Macleod, K.; Jodrell, D.; Ramsahoye, B. Targeting of 5-aza-2′-deoxycytidine residues by chromatin-associated DNMT1 induces proteasomal degradation of the free enzyme. Nucleic Acids Res. 2010, 38, 4313–4324. [Google Scholar] [CrossRef]
- Stresemann, C.; Brueckner, B.; Musch, T.; Stopper, H.; Lyko, F. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res. 2006, 66, 2794–2800. [Google Scholar] [CrossRef]
- Čihák, A. biological effects of 5-azacytidine in eukaryotes: A review. Oncology 2009, 30, 405–422. [Google Scholar] [CrossRef]
- Rhee, I.; Jair, K.-W.; Yen, R.-W.C.; Lengauer, C.; Herman, J.G.; Kinzler, K.W.; Vogelstein, B.; Baylin, S.B.; Schuebel, K.E. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature 2000, 404, 1003–1007. [Google Scholar] [CrossRef]
- Chen, T.; Hevi, S.; Gay, F.; Tsujimoto, N.; He, T.; Zhang, B.; Ueda, Y.; Li, E. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat. Genet. 2007, 39, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, F.; Hodgson, J.G.; Eden, A.; Jackson-Grusby, L.; Dausman, J.; Gray, J.W.; Leonhardt, H.; Jaenisch, R. Induction of tumors in mice by genomic hypomethylation. Science 2003, 300, 489–492. [Google Scholar] [CrossRef]
- Barra, V.; Schillaci, T.; Lentini, L.; Costa, G.; Di Leonardo, A. Bypass of cell cycle arrest induced by transient DNMT1 post-transcriptional silencing triggers aneuploidy in human cells. Cell Div. 2012, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Besselink, N.; Keijer, J.; Vermeulen, C.; Boymans, S.; de Ridder, J.; van Hoeck, A.; Cuppen, E.; Kuijk, E. The genome-wide mutational consequences of DNA hypomethylation. Sci. Rep. 2023, 13, 6874. [Google Scholar] [CrossRef]
- Scelfo, A.; Barra, V.; Abdennur, N.; Spracklin, G.; Busato, F.; Salinas-Luypaert, C.; Bonaiti, E.; Velasco, G.; Bonhomme, F.; Chipont, A.; et al. Tunable DNMT1 degradation reveals DNMT1/DNMT3B synergy in DNA methylation and genome organization. J. Cell Biol. 2024, 223, e202307026. [Google Scholar] [CrossRef]
- Martino, S.; Gargano, S.; Carollo, P.S.; Di Leonardo, A.; Barra, V. DNMT1 prolonged absence is a tunable cellular stress that triggers cell proliferation arrest to protect from major DNA methylation loss. Cell. Mol. Life Sci. 2024, 82, 7. [Google Scholar] [CrossRef]
- Kohli, R.M.; Zhang, Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 2013, 502, 472–479. [Google Scholar] [CrossRef]
- Hendrich, B.; Hardeland, U.; Ng, H.-H.; Jiricny, J.; Bird, A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature 1999, 401, 301–304. [Google Scholar] [CrossRef]
- Sanders, M.A.; Chew, E.; Flensburg, C.; Zeilemaker, A.; Miller, S.E.; al Hinai, A.S.; Bajel, A.; Luiken, B.; Rijken, M.; Mclennan, T.; et al. MBD4 guards against methylation damage and germ line deficiency predisposes to clonal hematopoiesis and early-onset AML. Blood 2018, 132, 1526–1534. [Google Scholar] [CrossRef]
- Cortellino, S.; Xu, J.; Sannai, M.; Moore, R.; Caretti, E.; Cigliano, A.; Le Coz, M.; Devarajan, K.; Wessels, A.; Soprano, D.; et al. Thymine DNA glycosylase is essential for active DNA Demethylation by Linked Deamination-Base Excision Repair. Cell 2011, 146, 67–79. [Google Scholar] [CrossRef]
- Maeder, M.L.; Angstman, J.F.; Richardson, M.E.; Linder, S.J.; Cascio, V.M.; Tsai, S.Q.; Ho, Q.H.; Sander, J.D.; Reyon, D.; Bernstein, B.E.; et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat. Biotechnol. 2013, 31, 1137–1142. [Google Scholar] [CrossRef]
- Chen, H.; Kazemier, H.G.; de Groote, M.L.; Ruiters, M.H.J.; Xu, G.-L.; Rots, M.G. Induced DNA demethylation by targeting Ten-Eleven Translocation 2 to the human ICAM-1 promoter. Nucleic Acids Res. 2014, 42, 1563–1574. [Google Scholar] [CrossRef]
- Xu, X.; Tao, Y.; Gao, X.; Zhang, L.; Li, X.; Zou, W.; Ruan, K.; Wang, F.; Xu, G.; Hu, R. A CRISPR-based approach for targeted DNA demethylation. Cell Discov. 2016, 2, 16009. [Google Scholar] [CrossRef]
- Oka, Y.; Nakajima, K.; Nagao, K.; Miura, K.; Ishii, N.; Kobayashi, H. 293FT cells transduced with four transcription factors (OCT4, SOX2, NANOG, and LIN28) generate aberrant ES-like cells. J. Stem Cells Regen. Med. 2010, 6, 149–156. [Google Scholar]
- Wu, W.; Hill, S.E.; Nathan, W.J.; Paiano, J.; Callen, E.; Wang, D.; Shinoda, K.; van Wietmarschen, N.; Colón-Mercado, J.M.; Zong, D.; et al. Neuronal enhancers are hotspots for DNA single-strand break repair. Nature 2021, 593, 440–444. [Google Scholar] [CrossRef]
- Wang, D.; Wu, W.; Callen, E.; Pavani, R.; Zolnerowich, N.; Kodali, S.; Zong, D.; Wong, N.; Noriega, S.; Nathan, W.J.; et al. Active DNA demethylation promotes cell fate specification and the DNA damage response. Science 2022, 378, 983–989. [Google Scholar] [CrossRef]
- Nemeth, K.; Bayraktar, R.; Ferracin, M.; Calin, G.A. Non-coding RNAs in disease: From mechanisms to therapeutics. Nat. Rev. Genet. 2024, 25, 211–232. [Google Scholar] [CrossRef]
- Fabbri, M.; Garzon, R.; Cimmino, A.; Liu, Z.; Zanesi, N.; Callegari, E.; Liu, S.; Alder, H.; Costinean, S.; Fernandez-Cymering, C.; et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. USA 2007, 104, 15805–15810. [Google Scholar] [CrossRef]
- Garzon, R.; Liu, S.; Fabbri, M.; Liu, Z.; Heaphy, C.E.A.; Callegari, E.; Schwind, S.; Pang, J.; Yu, J.; Muthusamy, N.; et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood 2009, 113, 6411–6418. [Google Scholar] [CrossRef]
- Déjardin, T.; Carollo, P.S.; Sipieter, F.; Davidson, P.M.; Seiler, C.; Cuvelier, D.; Cadot, B.; Sykes, C.; Gomes, E.R.; Borghi, N. Nesprins are mechanotransducers that discriminate epithelial-mesenchymal transition programs. J. Cell Biol. 2020, 219, e201908036. [Google Scholar] [CrossRef]
- Canever, H.; Carollo, P.S.; Fleurisson, R.; Girard, P.P.; Borghi, N. Molecular Tension Microscopy of E-Cadherin During Epithelial-Mesenchymal Transition. In The Epithelial-to Mesenchymal Transition: Methods and Protocols; Campbell, K., Theveneau, E., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2021; pp. 289–299. ISBN 978-1-07-160779-4. [Google Scholar]
- Chen, K.-C.; Wang, Y.-S.; Hu, C.-Y.; Chang, W.-C.; Liao, Y.-C.; Dai, C.-Y.; Juo, S.-H.H. OxLDL up-regulates microRNA-29b, leading to epigenetic modifications of MMP-2/MMP-9 genes: A novel mechanism for cardiovascular diseases. FASEB J. 2011, 25, 1718–1728. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, S.; Tian, L.; Wu, M.; Ai, F.; Tang, W.; Zhao, L.; Ding, J.; Zhang, L.; Tang, A. miR-124 and miR-506 inhibit colorectal cancer progression by targeting DNMT3B and DNMT1. Oncotarget 2015, 6, 38139–38150. [Google Scholar] [CrossRef]
- Di Ruscio, A.; Ebralidze, A.K.; Benoukraf, T.; Amabile, G.; Goff, L.A.; Terragni, J.; Figueroa, M.E.; De Figueiredo Pontes, L.L.; Alberich-Jorda, M.; Zhang, P.; et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature 2013, 503, 371–376. [Google Scholar] [CrossRef]
- Esposito, C.L.; Autiero, I.; Sandomenico, A.; Li, H.; Bassal, M.A.; Ibba, M.L.; Wang, D.; Rinaldi, L.; Ummarino, S.; Gaggi, G.; et al. Targeted systematic evolution of an RNA platform neutralizing DNMT1 function and controlling DNA methylation. Nat. Commun. 2023, 14, 99. [Google Scholar] [CrossRef]
- Jones, R.; Wijesinghe, S.; Wilson, C.; Halsall, J.; Liloglou, T.; Kanhere, A. A long intergenic non-coding RNA regulates nuclear localization of DNA methyl transferase-1. iScience 2021, 24, 102273. [Google Scholar] [CrossRef]
- DeZern, A.E. Nine years without a new FDA-approved therapy for MDS: How can we break through the impasse? Hematology 2015, 2015, 308–316. [Google Scholar] [CrossRef][Green Version]
- Mehdipour, P.; Murphy, T.; De Carvalho, D.D. The role of DNA-demethylating agents in cancer therapy. Pharmacol. Ther. 2020, 205, 107416. [Google Scholar] [CrossRef]
- Döhner, H.; Wei, A.H.; Appelbaum, F.R.; Craddock, C.; DiNardo, C.D.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Godley, L.A.; Hasserjian, R.P.; et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood 2022, 140, 1345–1377. [Google Scholar] [CrossRef] [PubMed]
- Silverman, L.R.; Demakos, E.P.; Peterson, B.L.; Kornblith, A.B.; Holland, J.C.; Odchimar-Reissig, R.; Stone, R.M.; Nelson, D.; Powell, B.L.; DeCastro, C.M.; et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: A study of the cancer and leukemia group B. J. Clin. Oncol. 2002, 20, 2429–2440. [Google Scholar] [CrossRef] [PubMed]
- Kantarjian, H.; Issa, J.-P.J.; Rosenfeld, C.S.; Bennett, J.M.; Albitar, M.; DiPersio, J.; Klimek, V.; Slack, J.; de Castro, C.; Ravandi, F.; et al. Decitabine improves patient outcomes in myelodysplastic syndromes. Cancer 2006, 106, 1794–1803. [Google Scholar] [CrossRef] [PubMed]
- Fenaux, P.; Mufti, G.J.; Hellstrom-Lindberg, E.; Santini, V.; Finelli, C.; Giagounidis, A.; Schoch, R.; Gattermann, N.; Sanz, G.; List, A.; et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: A randomised, open-label, phase III study. Lancet Oncol. 2009, 10, 223–232. [Google Scholar] [CrossRef]
- Lübbert, M.; Suciu, S.; Baila, L.; Rüter, B.H.; Platzbecker, U.; Giagounidis, A.; Selleslag, D.; Labar, B.; Germing, U.; Salih, H.R.; et al. Low-dose decitabine versus best supportive care in elderly patients with intermediate- or high-risk myelodysplastic syndrome (MDS) ineligible for intensive chemotherapy: Final results of the randomized phase III study of the european organisation for research and treatment of cancer leukemia group and the German MDS study group. J. Clin. Oncol. 2011, 29, 1987–1996. [Google Scholar] [CrossRef]
- Jackson-Grusby, L.; Laird, P.W.; Magge, S.N.; Moeller, B.J.; Jaenisch, R. Mutagenicity of 5-aza-2′-deoxycytidine is mediated by the mammalian DNA methyltransferase. Proc. Natl. Acad. Sci. USA 1997, 94, 4681–4685. [Google Scholar] [CrossRef]
- Jüttermann, R.; Li, E.; Jaenisch, R. Toxicity of 5-aza-2’-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc. Natl. Acad. Sci. USA 1994, 91, 11797–11801. [Google Scholar] [CrossRef]
- Tsai, H.-C.; Li, H.; Van Neste, L.; Cai, Y.; Robert, C.; Rassool, F.V.; Shin, J.J.; Harbom, K.M.; Beaty, R.; Pappou, E.; et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell 2012, 21, 430–446. [Google Scholar] [CrossRef]
- Herman, J.G. Hypermethylation of tumor suppressor genes in cancer. Semin. Cancer Biol. 1999, 9, 359–367. [Google Scholar] [CrossRef]
- Alshammari, E.; Zhang, Y.; Sobota, J.; Yang, Z. Aberrant DNA methylation of tumor suppressor genes and oncogenes as cancer biomarkers. In Genomic and Epigenomic Biomarkers of Toxicology and Disease; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2022; pp. 251–271. ISBN 978-1-119-80770-4. [Google Scholar]
- Zhang, Z.; Jia, Y.; Xv, F.; Song, L.; Shi, L.; Guo, J.; Chang, C. Decitabine induces change of biological traits in myelodysplastic syndromes via FOXO1 activation. Front. Genet. 2021, 11, 603956. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, J.; Geng, Y.; Liu, L.; Li, D. Azacitidine regulates DNA methylation of GADD45γ in myelodysplastic syndromes. J. Clin. Lab. Anal. 2021, 35, e23597. [Google Scholar] [CrossRef] [PubMed]
- Cokus, S.J.; Feng, S.; Zhang, X.; Chen, Z.; Merriman, B.; Haudenschild, C.D.; Pradhan, S.; Nelson, S.F.; Pellegrini, M.; Jacobsen, S.E. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 2008, 452, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Ball, M.P.; Li, J.B.; Gao, Y.; Lee, J.-H.; LeProust, E.M.; Park, I.-H.; Xie, B.; Daley, G.Q.; Church, G.M. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009, 27, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Maunakea, A.K.; Nagarajan, R.P.; Bilenky, M.; Ballinger, T.J.; D’Souza, C.; Fouse, S.D.; Johnson, B.E.; Hong, C.; Nielsen, C.; Zhao, Y.; et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 2010, 466, 253–257. [Google Scholar] [CrossRef]
- Yang, X.; Han, H.; De Carvalho, D.D.; Lay, F.D.; Jones, P.A.; Liang, G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell 2014, 26, 577–590. [Google Scholar] [CrossRef]
- Le, L.; Qipeng, W.; Chunmeng, M.; Hasnat, M.; Luyong, Z.; Zhenzhou, J.; Qinwei, Y. 5-Azacytidine promotes HCC cell metastasis by up-regulating RDH16 expression. Eur. J. Pharmacol. 2023, 950, 175736. [Google Scholar] [CrossRef]
- Poplineau, M.; Schnekenburger, M.; Dufer, J.; Kosciarz, A.; Brassart-Pasco, S.; Antonicelli, F.; Diederich, M.; Trussardi-Régnier, A. The DNA hypomethylating agent, 5-aza-2′-deoxycytidine, enhances tumor cell invasion through a transcription-dependent modulation of MMP-1 expression in human fibrosarcoma cells. Mol. Carcinog. 2015, 54, 24–34. [Google Scholar] [CrossRef]
- Bernal, T.; Moncada-Pazos, A.; Soria-Valles, C.; Gutiérrez-Fernández, A. Effects of azacitidine on matrix metalloproteinase-9 in acute myeloid leukemia and myelodysplasia. Exp. Hematol. 2013, 41, 172–179. [Google Scholar] [CrossRef]
| DNA Demethylating Drugs | Malignancies | Type |
|---|---|---|
| 5-Azacitidine (AZA) | AML, MDS, Solid tumors | Nucleoside analog |
| 5-aza-2′-deoxycytidine (DAC or decitabine) | AML, MDS | Nucleoside analog |
| GSK3685032 | AML | Non-nucleoside |
| Guadecitabine (SGI-110) | AML, MDS | Nucleoside analog |
| 5-azacytidine-5′-elaidate (CP-4200) | AML | Nucleoside analog |
| MG98 | Solid tumors | Oligonucleotide Antisense |
| NSC309132 (or Zebularine) | Solid tumors | Nucleoside analog |
| Procainamide | Solid tumors | Non-nucleoside |
| Epigallocatechin-3-gallate (EGCG) | Solid tumors | Non-nucleoside |
| SGI-1027 | Leukemia | Non-nucleoside |
| Hydralazine | Breast, ovarian, cervical cancers | Non-nucleoside |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carollo, P.S.; Barra, V. Induction of DNA Demethylation: Strategies and Consequences. Epigenomes 2025, 9, 11. https://doi.org/10.3390/epigenomes9020011
Carollo PS, Barra V. Induction of DNA Demethylation: Strategies and Consequences. Epigenomes. 2025; 9(2):11. https://doi.org/10.3390/epigenomes9020011
Chicago/Turabian StyleCarollo, Pietro Salvatore, and Viviana Barra. 2025. "Induction of DNA Demethylation: Strategies and Consequences" Epigenomes 9, no. 2: 11. https://doi.org/10.3390/epigenomes9020011
APA StyleCarollo, P. S., & Barra, V. (2025). Induction of DNA Demethylation: Strategies and Consequences. Epigenomes, 9(2), 11. https://doi.org/10.3390/epigenomes9020011








