Epigenetic Landscape of DNA Methylation in Pancreatic Ductal Adenocarcinoma
Abstract
1. Introduction
2. Regulation of DNA Methylation
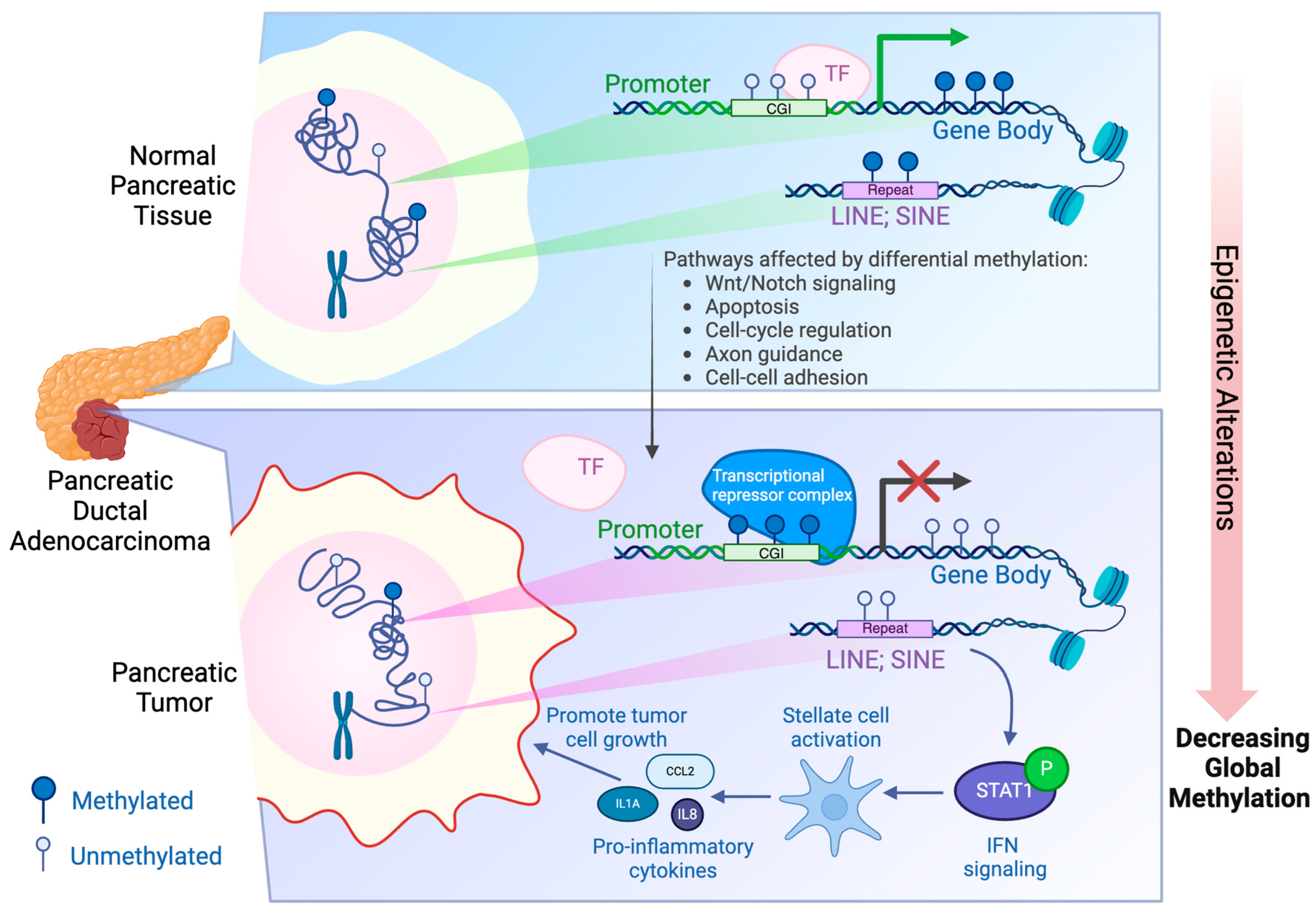
3. Aberrant DNA Methylation in PDAC
4. Recent WGBS Studies on PDAC Patients
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef]
- Wang, S.; Zheng, Y.; Yang, F.; Zhu, L.; Zhu, X.Q.; Wang, Z.F.; Wu, X.L.; Zhou, C.H.; Yan, J.Y.; Hu, B.Y.; et al. The molecular biology of pancreatic adenocarcinoma: Translational challenges and clinical perspectives. Signal Transduct. Target. Ther. 2021, 6, 249. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.; Pottler, M.; Lan, B.; Grutzmann, R.; Pilarsky, C.; Yang, H. Chemoresistance in Pancreatic Cancer. Int. J. Mol. Sci. 2019, 20, 4504. [Google Scholar] [CrossRef] [PubMed]
- Waddell, N.; Pajic, M.; Patch, A.M.; Chang, D.K.; Kassahn, K.S.; Bailey, P.; Johns, A.L.; Miller, D.; Nones, K.; Quek, K.; et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015, 518, 495–501. [Google Scholar] [CrossRef]
- Kanda, M.; Matthaei, H.; Wu, J.; Hong, S.M.; Yu, J.; Borges, M.; Hruban, R.H.; Maitra, A.; Kinzler, K.; Vogelstein, B.; et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology 2012, 142, 730–733 e9. [Google Scholar] [CrossRef]
- Waters, A.M.; Der, C.J. KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb. Perspect. Med. 2018, 8, a031435. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2017, 32, 185–203 e13. [Google Scholar] [CrossRef]
- Halbrook, C.J.; Lyssiotis, C.A.; Pasca di Magliano, M.; Maitra, A. Pancreatic cancer: Advances and challenges. Cell 2023, 186, 1729–1754. [Google Scholar] [CrossRef]
- Jones, S.; Zhang, X.; Parsons, D.W.; Lin, J.C.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Kamiyama, H.; Jimeno, A.; et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008, 321, 1801–1806. [Google Scholar] [CrossRef]
- Xu, J.; Roe, J.S.; Lee, E.; Tonelli, C.; Ji, K.Y.; Younis, O.W.; Somervile, T.D.D.; Yao, M.; Milazzo, J.P.; Tiriac, H.; et al. Engrailed-1 Promotes Pancreatic Cancer Metastasis. Adv. Sci. 2024, 11, e2308537. [Google Scholar] [CrossRef] [PubMed]
- Roe, J.S.; Hwang, C.I.; Somerville, T.D.D.; Milazzo, J.P.; Lee, E.J.; Da Silva, B.; Maiorino, L.; Tiriac, H.; Young, C.M.; Miyabayashi, K.; et al. Enhancer Reprogramming Promotes Pancreatic Cancer Metastasis. Cell 2017, 170, 875–888 e20. [Google Scholar] [CrossRef]
- Kim, H.R.; Yim, J.; Yoo, H.B.; Lee, S.E.; Oh, S.; Jung, S.; Hwang, C.I.; Shin, D.M.; Kim, T.; Yoo, K.H.; et al. EVI1 activates tumor-promoting transcriptional enhancers in pancreatic cancer. NAR Cancer 2021, 3, zcab023. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.S.; Xu, J.; Ji, K.Y.; Hwang, C.I. Epigenetic Alterations in Pancreatic Cancer Metastasis. Biomolecules 2021, 11, 1082. [Google Scholar] [CrossRef]
- Saghafinia, S.; Mina, M.; Riggi, N.; Hanahan, D.; Ciriello, G. Pan-Cancer Landscape of Aberrant DNA Methylation across Human Tumors. Cell Rep. 2018, 25, 1066–1080 e8. [Google Scholar] [CrossRef] [PubMed]
- Paz, M.F.; Fraga, M.F.; Avila, S.; Guo, M.; Pollan, M.; Herman, J.G.; Esteller, M. A systematic profile of DNA methylation in human cancer cell lines. Cancer Res. 2003, 63, 1114–1121. [Google Scholar]
- Bergman, Y.; Cedar, H. DNA methylation dynamics in health and disease. Nat. Struct. Mol. Biol. 2013, 20, 274–281. [Google Scholar] [CrossRef]
- Lomberk, G.; Blum, Y.; Nicolle, R.; Nair, A.; Gaonkar, K.S.; Marisa, L.; Mathison, A.; Sun, Z.; Yan, H.; Elarouci, N.; et al. Distinct epigenetic landscapes underlie the pathobiology of pancreatic cancer subtypes. Nat. Commun. 2018, 9, 1978. [Google Scholar] [CrossRef]
- Spainhour, J.C.; Lim, H.S.; Yi, S.V.; Qiu, P. Correlation Patterns Between DNA Methylation and Gene Expression in The Cancer Genome Atlas. Cancer Inform. 2019, 18, 1176935119828776. [Google Scholar] [CrossRef]
- Smith, J.; Sen, S.; Weeks, R.J.; Eccles, M.R.; Chatterjee, A. Promoter DNA Hypermethylation and Paradoxical Gene Activation. Trends Cancer 2020, 6, 392–406. [Google Scholar] [CrossRef]
- Wang, S.S.; Hall, M.L.; Lee, E.; Kim, S.-C.; Ramesh, N.; Lee, S.H.; Jang, J.-Y.; Ku, J.-L.; Hwang, C.-I. Whole genome bisulfite sequencing identifies stage- and subtype-specific DNA methylation signatures in pancreatic cancer. iScience 2024, 27, 109414. [Google Scholar] [CrossRef] [PubMed]
- Espinet, E.; Gu, Z.; Imbusch, C.D.; Giese, N.A.; Buscher, M.; Safavi, M.; Weisenburger, S.; Klein, C.; Vogel, V.; Falcone, M.; et al. Aggressive PDACs Show Hypomethylation of Repetitive Elements and the Execution of an Intrinsic IFN Program Linked to a Ductal Cell of Origin. Cancer Discov. 2021, 11, 638–659. [Google Scholar] [CrossRef]
- Robertson, K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005, 6, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Jabbari, K.; Bernardi, G. Cytosine methylation and CpG, TpG (CpA) and TpA frequencies. Gene 2004, 333, 143–149. [Google Scholar] [CrossRef]
- Li, E.; Zhang, Y. DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a019133. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Morales Valencia, M.; Yu, Y.; Ouchi, Y.; Takahashi, K.; Shokhirev, M.N.; Lande, K.; Williams, A.E.; Fresia, C.; Kurita, M.; et al. Transgenerational inheritance of acquired epigenetic signatures at CpG islands in mice. Cell 2023, 186, 715–731 e719. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Rao, C.M. Epigenetic tools (The Writers, The Readers and The Erasers) and their implications in cancer therapy. Eur. J. Pharmacol. 2018, 837, 8–24. [Google Scholar] [CrossRef]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef]
- Bestor, T.H.; Ingram, V.M. Two DNA methyltransferases from murine erythroleukemia cells: Purification, sequence specificity, and mode of interaction with DNA. Proc. Natl. Acad. Sci. USA 1983, 80, 5559–5563. [Google Scholar] [CrossRef]
- Probst, A.V.; Dunleavy, E.; Almouzni, G. Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 2009, 10, 192–206. [Google Scholar] [CrossRef]
- Jin, B.; Robertson, K.D. DNA methyltransferases, DNA damage repair, and cancer. Adv. Exp. Med. Biol. 2013, 754, 3–29. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.C.; Zhang, Y. Active DNA demethylation: Many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 2010, 11, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhuang, Z.; Wang, W.; He, L.; Wu, H.; Cao, Y.; Pan, F.; Zhao, J.; Hu, Z.; Sekhar, C.; et al. OGG1 is essential in oxidative stress induced DNA demethylation. Cell Signal 2016, 28, 1163–1171. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yang, M.; Wang, S.; Abbas, S.J.; Zhang, J.; Li, Y.; Shao, R.; Liu, Y. An Overview of Epigenetic Methylation in Pancreatic Cancer Progression. Front. Oncol. 2022, 12, 854773. [Google Scholar] [CrossRef] [PubMed]
- Angeloni, A.; Bogdanovic, O. Enhancer DNA methylation: Implications for gene regulation. Essays Biochem. 2019, 63, 707–715. [Google Scholar] [CrossRef]
- Brenet, F.; Moh, M.; Funk, P.; Feierstein, E.; Viale, A.J.; Socci, N.D.; Scandura, J.M. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS ONE 2011, 6, e14524. [Google Scholar] [CrossRef]
- Du, Q.; Luu, P.L.; Stirzaker, C.; Clark, S.J. Methyl-CpG-binding domain proteins: Readers of the epigenome. Epigenomics 2015, 7, 1051–1073. [Google Scholar] [CrossRef]
- Skoulidis, F.; Cassidy, L.D.; Pisupati, V.; Jonasson, J.G.; Bjarnason, H.; Eyfjord, J.E.; Karreth, F.A.; Lim, M.; Barber, L.M.; Clatworthy, S.A.; et al. Germline Brca2 heterozygosity promotes Kras(G12D) -driven carcinogenesis in a murine model of familial pancreatic cancer. Cancer Cell 2010, 18, 499–509. [Google Scholar] [CrossRef]
- Watt, F.; Molloy, P.L. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988, 2, 1136–1143. [Google Scholar] [CrossRef]
- Monteagudo-Sanchez, A.; Noordermeer, D.; Greenberg, M.V.C. The impact of DNA methylation on CTCF-mediated 3D genome organization. Nat. Struct. Mol. Biol. 2024, 31, 404–412. [Google Scholar] [CrossRef]
- Wang, H.; Maurano, M.T.; Qu, H.; Varley, K.E.; Gertz, J.; Pauli, F.; Lee, K.; Canfield, T.; Weaver, M.; Sandstrom, R.; et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012, 22, 1680–1688. [Google Scholar] [CrossRef] [PubMed]
- Ball, M.P.; Li, J.B.; Gao, Y.; Lee, J.H.; LeProust, E.M.; Park, I.H.; Xie, B.; Daley, G.Q.; Church, G.M. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009, 27, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. The DNA methylation paradox. Trends Genet. 1999, 15, 34–37. [Google Scholar] [CrossRef] [PubMed]
- Neri, F.; Rapelli, S.; Krepelova, A.; Incarnato, D.; Parlato, C.; Basile, G.; Maldotti, M.; Anselmi, F.; Oliviero, S. Intragenic DNA methylation prevents spurious transcription initiation. Nature 2017, 543, 72–77. [Google Scholar] [CrossRef]
- Yoder, J.A.; Walsh, C.P.; Bestor, T.H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997, 13, 335–340. [Google Scholar] [CrossRef]
- Ibrahim, J.; Op de Beeck, K.; Fransen, E.; Peeters, M.; Van Camp, G. Genome-wide DNA methylation profiling and identification of potential pan-cancer and tumor-specific biomarkers. Mol. Oncol. 2022, 16, 2432–2447. [Google Scholar] [CrossRef]
- Frommer, M.; McDonald, L.E.; Millar, D.S.; Collis, C.M.; Watt, F.; Grigg, G.W.; Molloy, P.L.; Paul, C.L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA 1992, 89, 1827–1831. [Google Scholar] [CrossRef]
- Wang, R.Y.; Gehrke, C.W.; Ehrlich, M. Comparison of bisulfite modification of 5-methyldeoxycytidine and deoxycytidine residues. Nucleic Acids Res. 1980, 8, 4777–4790. [Google Scholar] [CrossRef] [PubMed]
- Umer, M.; Herceg, Z. Deciphering the epigenetic code: An overview of DNA methylation analysis methods. Antioxid. Redox Signal. 2013, 18, 1972–1986. [Google Scholar] [CrossRef]
- Warnecke, P.M.; Stirzaker, C.; Melki, J.R.; Millar, D.S.; Paul, C.L.; Clark, S.J. Detection and measurement of PCR bias in quantitative methylation analysis of bisulphite-treated DNA. Nucleic Acids Res. 1997, 25, 4422–4426. [Google Scholar] [CrossRef]
- Ehrich, M.; Zoll, S.; Sur, S.; van den Boom, D. A new method for accurate assessment of DNA quality after bisulfite treatment. Nucleic Acids Res. 2007, 35, e29. [Google Scholar] [CrossRef] [PubMed]
- Meissner, A.; Gnirke, A.; Bell, G.W.; Ramsahoye, B.; Lander, E.S.; Jaenisch, R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005, 33, 5868–5877. [Google Scholar] [CrossRef] [PubMed]
- Harrison, A.; Parle-McDermott, A. DNA methylation: A timeline of methods and applications. Front. Genet. 2011, 2, 74. [Google Scholar] [CrossRef] [PubMed]
- Weichenhan, D.; Wang, Q.; Adey, A.; Wolf, S.; Shendure, J.; Eils, R.; Plass, C. Tagmentation-Based Library Preparation for Low DNA Input Whole Genome Bisulfite Sequencing. Methods Mol. Biol. 2018, 1708, 105–122. [Google Scholar] [CrossRef]
- Tan, A.C.; Jimeno, A.; Lin, S.H.; Wheelhouse, J.; Chan, F.; Solomon, A.; Rajeshkumar, N.V.; Rubio-Viqueira, B.; Hidalgo, M. Characterizing DNA methylation patterns in pancreatic cancer genome. Mol. Oncol. 2009, 3, 425–438. [Google Scholar] [CrossRef]
- Ueki, T.; Toyota, M.; Sohn, T.; Yeo, C.J.; Issa, J.P.; Hruban, R.H.; Goggins, M. Hypermethylation of multiple genes in pancreatic adenocarcinoma. Cancer Res. 2000, 60, 1835–1839. [Google Scholar]
- Das, P.M.; Singal, R. DNA methylation and cancer. J. Clin. Oncol. 2004, 22, 4632–4642. [Google Scholar] [CrossRef]
- Chen, X.; Yu, S.; Chen, J.; Chen, X. Analysis of PD-L1 promoter methylation combined with immunogenic context in pancreatic ductal adenocarcinoma. Cancer Immunol. Immunother. 2024, 73, 149. [Google Scholar] [CrossRef]
- Hatziapostolou, M.; Koutsioumpa, M.; Zaitoun, A.M.; Polytarchou, C.; Edderkaoui, M.; Mahurkar-Joshi, S.; Vadakekolathu, J.; D’Andrea, D.; Lay, A.R.; Christodoulou, N.; et al. Promoter Methylation Leads to Hepatocyte Nuclear Factor 4A Loss and Pancreatic Cancer Aggressiveness. Gastro Hep Adv. 2024, 3, 687–702. [Google Scholar] [CrossRef]
- Yao, Y.; Lv, H.; Zhang, M.; Li, Y.; Herman, J.G.; Brock, M.V.; Gao, A.; Wang, Q.; Fuks, F.; Zhang, L.; et al. Epigenetic silencing of BEND4, a novel DNA damage repair gene, is a synthetic lethal marker for ATM inhibitor in pancreatic cancer. Front. Med. 2024, 18, 721–734. [Google Scholar] [CrossRef]
- Chatterjee, A.; Bararia, A.; Ganguly, D.; Mondal, P.K.; Roy, P.; Banerjee, S.; Ghosh, S.; Gulati, S.; Ghatak, S.; Chattopadhay, B.K.; et al. DNA methylome in pancreatic cancer identified novel promoter hyper-methylation in NPY and FAIM2 genes associated with poor prognosis in Indian patient cohort. Cancer Cell Int. 2022, 22, 334. [Google Scholar] [CrossRef] [PubMed]
- Sakai, M.; Hibi, K.; Koshikawa, K.; Inoue, S.; Takeda, S.; Kaneko, T.; Nakao, A. Frequent promoter methylation and gene silencing of CDH13 in pancreatic cancer. Cancer Sci. 2004, 95, 588–591. [Google Scholar] [CrossRef] [PubMed]
- Matsubayashi, H.; Sato, N.; Fukushima, N.; Yeo, C.J.; Walter, K.M.; Brune, K.; Sahin, F.; Hruban, R.H.; Goggins, M. Methylation of cyclin D2 is observed frequently in pancreatic cancer but is also an age-related phenomenon in gastrointestinal tissues. Clin. Cancer Res. 2003, 9, 1446–1452. [Google Scholar] [PubMed]
- Sato, N.; Fukushima, N.; Maehara, N.; Matsubayashi, H.; Koopmann, J.; Su, G.H.; Hruban, R.H.; Goggins, M. SPARC/osteonectin is a frequent target for aberrant methylation in pancreatic adenocarcinoma and a mediator of tumor-stromal interactions. Oncogene 2003, 22, 5021–5030. [Google Scholar] [CrossRef]
- Weng, Y.C.; Ma, J.; Zhang, J.; Wang, J.C. Long non-coding RNA LINC01133 silencing exerts antioncogenic effect in pancreatic cancer through the methylation of DKK1 promoter and the activation of Wnt signaling pathway. Cancer Biol. Ther. 2019, 20, 368–380. [Google Scholar] [CrossRef]
- Yokoyama, S.; Iwaya, H.; Akahane, T.; Hamada, T.; Higashi, M.; Hashimoto, S.; Tanoue, S.; Ohtsuka, T.; Ido, A.; Tanimoto, A. Sequential evaluation of MUC promoter methylation using next-generation sequencing-based custom-made panels in liquid-based cytology specimens of pancreatic cancer. Diagn. Cytopathol. 2022, 50, 499–507. [Google Scholar] [CrossRef]
- Funel, N.; Morelli, M.; Giovannetti, E.; Del Chiaro, M.; Pollina, L.E.; Mosca, F.; Boggi, U.; Cavazzana, A.; Campani, D. Loss of heterozygosity status of D9S105 marker is associated with downregulation of Kruppel-like factor 4 expression in pancreatic ductal adenocarcinoma and pancreatic intraepithelial lesions. Pancreatology 2011, 11, 30–42. [Google Scholar] [CrossRef]
- Xie, V.K.; Li, Z.; Yan, Y.; Jia, Z.; Zuo, X.; Ju, Z.; Wang, J.; Du, J.; Xie, D.; Xie, K.; et al. DNA-Methyltransferase 1 Induces Dedifferentiation of Pancreatic Cancer Cells through Silencing of Kruppel-Like Factor 4 Expression. Clin. Cancer Res. 2017, 23, 5585–5597. [Google Scholar] [CrossRef]
- Mishra, N.K.; Guda, C. Genome-wide DNA methylation analysis reveals molecular subtypes of pancreatic cancer. Oncotarget 2017, 8, 28990–29012. [Google Scholar] [CrossRef]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.M.; Gingras, M.C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef]
- Boj, S.F.; Hwang, C.I.; Baker, L.A.; Chio, I.I.C.; Engle, D.D.; Corbo, V.; Jager, M.; Ponz-Sarvise, M.; Tiriac, H.; Spector, M.S.; et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 2015, 160, 324–338. [Google Scholar] [CrossRef] [PubMed]
- Boj, S.F.; Hwang, C.I.; Baker, L.A.; Engle, D.D.; Tuveson, D.A.; Clevers, H. Model organoids provide new research opportunities for ductal pancreatic cancer. Mol. Cell Oncol. 2016, 3, e1014757. [Google Scholar] [CrossRef]
- Hwang, C.I.; Boj, S.F.; Clevers, H.; Tuveson, D.A. Preclinical models of pancreatic ductal adenocarcinoma. J. Pathol. 2016, 238, 197–204. [Google Scholar] [CrossRef]
- Romero-Calvo, I.; Weber, C.R.; Ray, M.; Brown, M.; Kirby, K.; Nandi, R.K.; Long, T.M.; Sparrow, S.M.; Ugolkov, A.; Qiang, W.; et al. Human Organoids Share Structural and Genetic Features with Primary Pancreatic Adenocarcinoma Tumors. Mol. Cancer Res. 2019, 17, 70–83. [Google Scholar] [CrossRef]
- Sharick, J.T.; Jeffery, J.J.; Karim, M.R.; Walsh, C.M.; Esbona, K.; Cook, R.S.; Skala, M.C. Cellular Metabolic Heterogeneity In Vivo Is Recapitulated in Tumor Organoids. Neoplasia 2019, 21, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Tiriac, H.; Belleau, P.; Engle, D.D.; Plenker, D.; Deschenes, A.; Somerville, T.D.D.; Froeling, F.E.M.; Burkhart, R.A.; Denroche, R.E.; Jang, G.H.; et al. Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer. Cancer Discov. 2018, 8, 1112–1129. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, M.; Liu, L.; Wang, G.; Wang, L.; Zhong, C.; Gao, C.; Wu, W.; Li, L. Ultrasound-guided fine-needle aspiration/biopsy-based pancreatic organoids establishment: An alternative model for basic and preclinical research. Gastroenterol. Rep. 2023, 11, goad019. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, H.; Lee, S.H.; Ku, J.L.; Chun, J.W.; Seo, H.Y.; Kim, S.C.; Paik, W.H.; Ryu, J.K.; Lee, S.K.; et al. Establishment of Patient-Derived Pancreatic Cancer Organoids from Endoscopic Ultrasound-Guided Fine-Needle Aspiration Biopsies. Gut Liver 2022, 16, 625–636. [Google Scholar] [CrossRef]
- Vilgelm, A.E.; Bergdorf, K.; Wolf, M.; Bharti, V.; Shattuck-Brandt, R.; Blevins, A.; Jones, C.; Phifer, C.; Lee, M.; Lowe, C.; et al. Fine-Needle Aspiration-Based Patient-Derived Cancer Organoids. iScience 2020, 23, 101408. [Google Scholar] [CrossRef]
- Collisson, E.A.; Sadanandam, A.; Olson, P.; Gibb, W.J.; Truitt, M.; Gu, S.; Cooc, J.; Weinkle, J.; Kim, G.E.; Jakkula, L.; et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 2011, 17, 500–503. [Google Scholar] [CrossRef]
- Collisson, E.A.; Bailey, P.; Chang, D.K.; Biankin, A.V. Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Brunton, H.; Caligiuri, G.; Cunningham, R.; Upstill-Goddard, R.; Bailey, U.M.; Garner, I.M.; Nourse, C.; Dreyer, S.; Jones, M.; Moran-Jones, K.; et al. HNF4A and GATA6 Loss Reveals Therapeutically Actionable Subtypes in Pancreatic Cancer. Cell Rep. 2020, 31, 107625. [Google Scholar] [CrossRef] [PubMed]
- O’Kane, G.M.; Grunwald, B.T.; Jang, G.H.; Masoomian, M.; Picardo, S.; Grant, R.C.; Denroche, R.E.; Zhang, A.; Wang, Y.; Lam, B.; et al. GATA6 Expression Distinguishes Classical and Basal-like Subtypes in Advanced Pancreatic Cancer. Clin. Cancer Res. 2020, 26, 4901–4910. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ortiz, M.V.; Cano-Ramirez, P.; Toledano-Fonseca, M.; Aranda, E.; Rodriguez-Ariza, A. Diagnosing and monitoring pancreatic cancer through cell-free DNA methylation: Progress and prospects. Biomark. Res. 2023, 11, 88. [Google Scholar] [CrossRef]
| Gene Promoter | Hypermethylated or Hypomethylated in PDAC | Affected Gene Expression Level | Molecular Function of the Gene | Functional Phenotype in PDAC | Method | Reference |
|---|---|---|---|---|---|---|
| PD-L1 promoter | Hypomethylated | PD-L1 ↑ | Programmed cell death ligand | Aggressive clinical phenotypes ↑, immune cell infiltration ↓, expression of immune checkpoint genes ↓, overall survival ↓ | Microarray | [58] |
| HNF4A promoter | Hypermethylated | HNF4A ↓ | Controls genes that are especially important for development and function of beta cells in the pancreas; regulate cell proliferation through the Wnt/beta-Catenin Pathway | Patient survival ↓, cell growth, colony formation, and invasiveness ↑, expression decreases during early PDAC stages | Targeted bisulfite sequencing | [59] |
| BEND4 promoter | Hypermethylated | BEND4 ↓ | BEN domain-containing protein 4 involves in non-homologous end-joining signaling by interacting with Ku80 and promotes DNA damage repair; inhibits cell growth, migration and invasion | Prognosis ↓, synthetic lethality towards ATM inhibitor | MSP | [60] |
| NPY and FAIM2 promoters | Hypermethylated | NPY ↓, FAIM2 ↓ | NPY is involved in cell motion, invasion, and proliferation, and its hypermethylation is observed in certain cancers. FAIM2 is associated with apoptosis inhibition. | Prognosis ↓, progress to later stages ↑ | Microarray | [61] |
| CDH13 promoter | Hypermethylated | CDH13 ↓ | Cadherin, involved in cell-cell adhesion and contact inhibition of cell growth | Invasion and metastasis ↑ | MSP | [62] |
| Cyclin D2 promoter | Hypermethylated | Cyclin D2 ↓ | Involved in cell cycle regulation, cell differentiation, and neoplastic transformation | Unknown in PDAC context | MSP | [63] |
| SPARC promoter | Hypermethylated | SPARC ↓ | A multifunctional glycoprotein, involved in cell proliferation, cell spreading, adhesion, motility, and invasion | Proliferation ↑ | MSP, microarray | [64] |
| DKK1 promoter | Hypermethylated | DKK1 ↓ | Regulates Wnt signaling by binding to the Wnt coreceptor lipoprotein-related protein-5 (LRP5)/Arrow | Proliferation ↑, migration & invasion ↑ | MSP | [65] |
| MUC Promoter | Hypomethylated | Not tested | Mucins, bind to pathogens as part of the immune system; renewal and differentiation of the epithelium; involved in cell adhesion and signaling | Unknown in PDAC context | Microarray | [66] |
| KLF4 Promoter | Hypermethylated | KLF4 ↓ | Zinc transcription factor, involved in cell proliferation and terminal differentiation of epithelial cells | Tumor cell differentiation ↓, proliferation ↑, patient survival ↓ | MSP, targeted bisulfite sequencing | [67,68] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, P.; Jacques, J.; Hwang, C.-I. Epigenetic Landscape of DNA Methylation in Pancreatic Ductal Adenocarcinoma. Epigenomes 2024, 8, 41. https://doi.org/10.3390/epigenomes8040041
Liu P, Jacques J, Hwang C-I. Epigenetic Landscape of DNA Methylation in Pancreatic Ductal Adenocarcinoma. Epigenomes. 2024; 8(4):41. https://doi.org/10.3390/epigenomes8040041
Chicago/Turabian StyleLiu, Peiyi, Juliette Jacques, and Chang-Il Hwang. 2024. "Epigenetic Landscape of DNA Methylation in Pancreatic Ductal Adenocarcinoma" Epigenomes 8, no. 4: 41. https://doi.org/10.3390/epigenomes8040041
APA StyleLiu, P., Jacques, J., & Hwang, C.-I. (2024). Epigenetic Landscape of DNA Methylation in Pancreatic Ductal Adenocarcinoma. Epigenomes, 8(4), 41. https://doi.org/10.3390/epigenomes8040041






