Ribosomal Biogenesis and Heterogeneity in Development, Disease, and Aging
Abstract
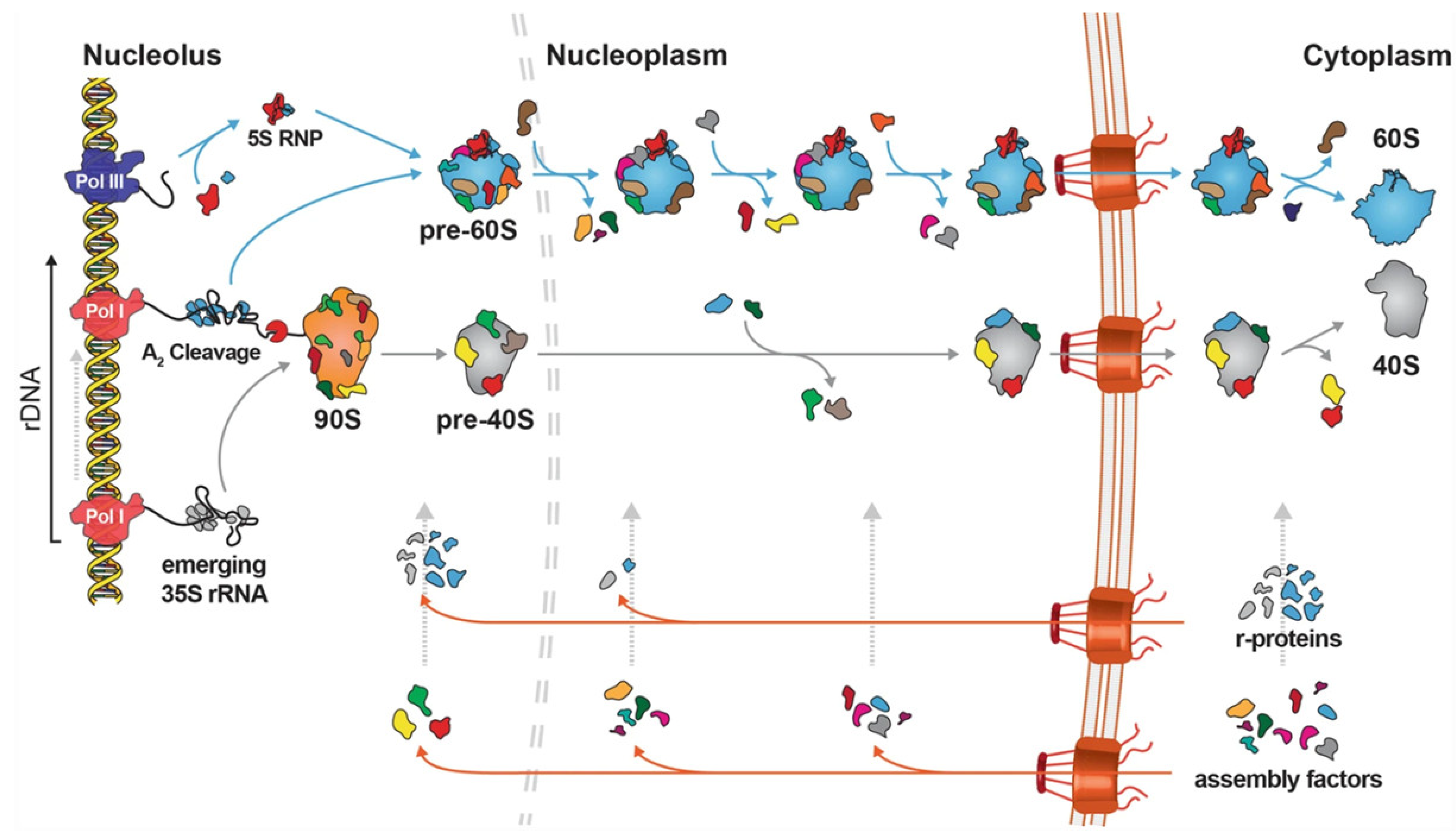
1. Introduction
2. Evolution of the Idea of Ribosome Heterogeneity and Its Key Factors
3. Ribosome Heterogeneity in Development
4. Diseases Associated with Ribosomes and Relevant Mutations
5. Ageing and the Ribosome
6. General Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- The Nobel Prize in Physiology or Medicine 1974. Available online: https://www.nobelprize.org/prizes/medicine/1974/ceremony-speech/ (accessed on 8 February 2023).
- Zhao, P. The 2009 Nobel Prize in Chemistry: Thomas A. Steitz and the Structure of the Ribosome. Yale J. Biol. Med. 2011, 84, 125–129. [Google Scholar] [PubMed]
- Genuth, N.R.; Barna, M. The Discovery of Ribosome Heterogeneity and Its Implications for Gene Regulation and Organismal Life. Mol. Cell 2018, 71, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Petrov, A.S.; Bernier, C.R.; Hsiao, C.; Norris, A.M.; Kovacs, N.A.; Waterbury, C.C.; Stepanov, V.G.; Harvey, S.C.; Fox, G.E.; Wartell, R.M.; et al. Evolution of the Ribosome at Atomic Resolution. Proc. Natl. Acad. Sci. USA 2014, 111, 10251–10256. [Google Scholar] [CrossRef] [PubMed]
- Guo, H. Specialized Ribosomes and the Control of Translation. Biochem. Soc. Trans. 2018, 46, 855–869. [Google Scholar] [CrossRef] [PubMed]
- Cutler, S.; Lee, L.J.; Tsukiyama, T. Chromatin Remodeling Factors Isw2 and Ino80 Regulate Chromatin, Replication, and Copy Number of the Saccharomyces Cerevisiae Ribosomal DNA Locus. Genetics 2018, 210, 1543–1556. [Google Scholar] [CrossRef]
- Kobayashi, T. Regulation of Ribosomal RNA Gene Copy Number and Its Role in Modulating Genome Integrity and Evolutionary Adaptability in Yeast. Cell. Mol. Life Sci. 2011, 68, 1395–1403. [Google Scholar] [CrossRef]
- Oborská-Oplová, M.; Fischer, U.; Altvater, M.; Panse, V.G. Eukaryotic Ribosome Assembly and Nucleocytoplasmic Transport. Methods Mol. Biol. 2022, 2533, 99–126. [Google Scholar] [CrossRef] [PubMed]
- Schächner, C.; Merkl, P.E.; Pilsl, M.; Schwank, K.; Hergert, K.; Kruse, S.; Milkereit, P.; Tschochner, H.; Griesenbeck, J. Establishment and Maintenance of Open Ribosomal RNA Gene Chromatin States in Eukaryotes. Methods Mol. Biol. 2022, 2533, 25–38. [Google Scholar] [CrossRef]
- Geiduschek, E.P.; Kassavetis, G.A. The RNA Polymerase III Transcription Apparatus. J. Mol. Biol. 2001, 310, 1–26. [Google Scholar] [CrossRef]
- Shigeoka, T.; Koppers, M.; Wong, H.H.-W.; Lin, J.Q.; Cagnetta, R.; Dwivedy, A.; de Freitas Nascimento, J.; van Tartwijk, F.W.; Ströhl, F.; Cioni, J.-M.; et al. On-Site Ribosome Remodeling by Locally Synthesized Ribosomal Proteins in Axons. Cell Rep. 2019, 29, 3605–3619.e10. [Google Scholar] [CrossRef]
- Temaj, G.; Chichiarelli, S.; Eufemi, M.; Altieri, F.; Hadziselimovic, R.; Farooqi, A.A.; Yaylim, I.; Saso, L. Ribosome-Directed Therapies in Cancer. Biomedicines 2022, 10, 2088. [Google Scholar] [CrossRef] [PubMed]
- Jüttner, M.; Ferreira-Cerca, S. A Comparative Perspective on Ribosome Biogenesis: Unity and Diversity Across the Tree of Life. Methods Mol. Biol. 2022, 2533, 3–22. [Google Scholar] [CrossRef]
- Li, D.; Wang, J. Ribosome Heterogeneity in Stem Cells and Development. J. Cell Biol. 2020, 219, e202001108. [Google Scholar] [CrossRef] [PubMed]
- Petrov, A.S.; Gulen, B.; Norris, A.M.; Kovacs, N.A.; Bernier, C.R.; Lanier, K.A.; Fox, G.E.; Harvey, S.C.; Wartell, R.M.; Hud, N.V.; et al. History of the Ribosome and the Origin of Translation. Proc. Natl. Acad. Sci. USA 2015, 112, 15396–15401. [Google Scholar] [CrossRef] [PubMed]
- Kozak, M. An Analysis of 5’-Noncoding Sequences from 699 Vertebrate Messenger RNAs. Nucleic Acids Res. 1987, 15, 8125–8148. [Google Scholar] [CrossRef]
- Weisser, M.; Ban, N. Extensions, Extra Factors, and Extreme Complexity: Ribosomal Structures Provide Insights into Eukaryotic Translation. Cold Spring Harb. Perspect. Biol. 2019, 11, a032367. [Google Scholar] [CrossRef]
- Ramakrishnan, V. Ribosome Structure and the Mechanism of Translation. Cell 2002, 108, 557–572. [Google Scholar] [CrossRef]
- Crick, F.H. On Protein Synthesis. Symp. Soc. Exp. Biol. 1958, 12, 138–163. [Google Scholar] [PubMed]
- Crick, F.H.; Barnett, L.; Brenner, S.; Watts-Tobin, R.J. General Nature of the Genetic Code for Proteins. Nature 1961, 192, 1227–1232. [Google Scholar] [CrossRef]
- Ramagopal, S.; Ennis, H.L. Regulation of Synthesis of Cell-Specific Ribosomal Proteins during Differentiation of Dictyostelium Discoideum. Proc. Natl. Acad. Sci. USA 1981, 78, 3083–3087. [Google Scholar] [CrossRef]
- Mauro, V.P.; Edelman, G.M. The Ribosome Filter Hypothesis. Proc. Natl. Acad. Sci. USA 2002, 99, 12031–12036. [Google Scholar] [CrossRef]
- Leppek, K.; Fujii, K.; Quade, N.; Susanto, T.T.; Boehringer, D.; Lenarčič, T.; Xue, S.; Genuth, N.R.; Ban, N.; Barna, M. Gene- and Species-Specific Hox MRNA Translation by Ribosome Expansion Segments. Mol. Cell 2020, 80, 980–995.e13. [Google Scholar] [CrossRef]
- Pelletier, J.; Sonenberg, N. Internal Initiation of Translation of Eukaryotic MRNA Directed by a Sequence Derived from Poliovirus RNA. Nature 1988, 334, 320–325. [Google Scholar] [CrossRef]
- Xue, S.; Tian, S.; Fujii, K.; Kladwang, W.; Das, R.; Barna, M. RNA Regulons in Hox 5’ UTRs Confer Ribosome Specificity to Gene Regulation. Nature 2015, 517, 33–38. [Google Scholar] [CrossRef]
- Carvajal, F.; Vallejos, M.; Walters, B.; Contreras, N.; Hertz, M.I.; Olivares, E.; Cáceres, C.J.; Pino, K.; Letelier, A.; Thompson, S.R.; et al. Structural Domains within the HIV-1 MRNA and the Ribosomal Protein S25 Influence Cap-Independent Translation Initiation. FEBS J. 2016, 283, 2508–2527. [Google Scholar] [CrossRef]
- LaFontaine, E.; Miller, C.M.; Permaul, N.; Martin, E.T.; Fuchs, G. Ribosomal Protein RACK1 Enhances Translation of Poliovirus and Other Viral IRESs. Virology 2020, 545, 53–62. [Google Scholar] [CrossRef]
- Akirtava, C.; May, G.E.; McManus, C.J. False-Positive IRESes from Hoxa9 and Other Genes Resulting from Errors in Mammalian 5’ UTR Annotations. Proc. Natl. Acad. Sci. USA 2022, 119, e2122170119. [Google Scholar] [CrossRef]
- Ivanov, I.P.; Saba, J.A.; Fan, C.-M.; Wang, J.; Firth, A.E.; Cao, C.; Green, R.; Dever, T.E. Evolutionarily Conserved Inhibitory UORFs Sensitize Hox MRNA Translation to Start Codon Selection Stringency. Proc. Natl. Acad. Sci. USA 2022, 119, e2117226119. [Google Scholar] [CrossRef]
- Chen, Y.-X.; Xu, Z.-Y.; Ge, X.; Sanyal, S.; Lu, Z.J.; Javid, B. Selective Translation by Alternative Bacterial Ribosomes. Proc. Natl. Acad. Sci. USA 2020, 117, 19487–19496. [Google Scholar] [CrossRef]
- Marcel, V.; Catez, F.; Diaz, J.-J. Ribosome Heterogeneity in Tumorigenesis: The RRNA Point of View. Mol. Cell Oncol. 2015, 2, e983755. [Google Scholar] [CrossRef] [PubMed]
- Kondrashov, N.; Pusic, A.; Stumpf, C.R.; Shimizu, K.; Hsieh, A.C.; Ishijima, J.; Shiroishi, T.; Barna, M. Ribosome-Mediated Specificity in Hox MRNA Translation and Vertebrate Tissue Patterning. Cell 2011, 145, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Fujii, K.; Kovary, K.M.; Genuth, N.R.; Röst, H.L.; Teruel, M.N.; Barna, M. Heterogeneous Ribosomes Preferentially Translate Distinct Subpools of MRNAs Genome-Wide. Mol. Cell 2017, 67, 71–83.e7. [Google Scholar] [CrossRef]
- Malik Ghulam, M.; Catala, M.; Reulet, G.; Scott, M.S.; Abou Elela, S. Duplicated Ribosomal Protein Paralogs Promote Alternative Translation and Drug Resistance. Nat. Commun. 2022, 13, 4938. [Google Scholar] [CrossRef]
- Higa-Nakamine, S.; Suzuki, T.; Uechi, T.; Chakraborty, A.; Nakajima, Y.; Nakamura, M.; Hirano, N.; Suzuki, T.; Kenmochi, N. Loss of Ribosomal RNA Modification Causes Developmental Defects in Zebrafish. Nucleic Acids Res. 2012, 40, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Sloan, K.E.; Warda, A.S.; Sharma, S.; Entian, K.-D.; Lafontaine, D.L.J.; Bohnsack, M.T. Tuning the Ribosome: The Influence of RRNA Modification on Eukaryotic Ribosome Biogenesis and Function. RNA Biol. 2017, 14, 1138–1152. [Google Scholar] [CrossRef] [PubMed]
- Schosserer, M.; Minois, N.; Angerer, T.B.; Amring, M.; Dellago, H.; Harreither, E.; Calle-Perez, A.; Pircher, A.; Gerstl, M.P.; Pfeifenberger, S.; et al. Methylation of Ribosomal RNA by NSUN5 Is a Conserved Mechanism Modulating Organismal Lifespan. Nat. Commun. 2015, 6, 6158. [Google Scholar] [CrossRef]
- Marygold, S.J.; Roote, J.; Reuter, G.; Lambertsson, A.; Ashburner, M.; Millburn, G.H.; Harrison, P.M.; Yu, Z.; Kenmochi, N.; Kaufman, T.C.; et al. The Ribosomal Protein Genes and Minute Loci of Drosophila Melanogaster. Genome Biol. 2007, 8, R216. [Google Scholar] [CrossRef] [PubMed]
- Polymenis, M. Ribosomal Proteins: Mutant Phenotypes by the Numbers and Associated Gene Expression Changes. Open Biol. 2020, 10, 200114. [Google Scholar] [CrossRef] [PubMed]
- Adjaye, J.; Huntriss, J.; Herwig, R.; BenKahla, A.; Brink, T.C.; Wierling, C.; Hultschig, C.; Groth, D.; Yaspo, M.-L.; Picton, H.M.; et al. Primary Differentiation in the Human Blastocyst: Comparative Molecular Portraits of Inner Cell Mass and Trophectoderm Cells. Stem Cells 2005, 23, 1514–1525. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Ji, S.-Y.; Dang, Y.-J.; Sha, Q.-Q.; Yuan, Y.-F.; Zhou, J.-J.; Yan, L.-Y.; Qiao, J.; Tang, F.; Fan, H.-Y. Oocyte-Expressed Yes-Associated Protein Is a Key Activator of the Early Zygotic Genome in Mouse. Cell Res. 2016, 26, 275–287. [Google Scholar] [CrossRef]
- Genuth, N.R.; Shi, Z.; Kunimoto, K.; Hung, V.; Xu, A.F.; Kerr, C.H.; Tiu, G.C.; Oses-Prieto, J.A.; Salomon-Shulman, R.E.A.; Axelrod, J.D.; et al. A Stem Cell Roadmap of Ribosome Heterogeneity Reveals a Function for RPL10A in Mesoderm Production. Nat. Commun. 2022, 13, 5491. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Duc, A.-C.E.; Rao, S.; Sun, X.-L.; Bilbee, A.N.; Rhodes, M.; Li, Q.; Kappes, D.J.; Rhodes, J.; Wiest, D.L. Control of Hematopoietic Stem Cell Emergence by Antagonistic Functions of Ribosomal Protein Paralogs. Dev. Cell 2013, 24, 411–425. [Google Scholar] [CrossRef]
- Zhang, Y.; O’Leary, M.N.; Peri, S.; Wang, M.; Zha, J.; Melov, S.; Kappes, D.J.; Feng, Q.; Rhodes, J.; Amieux, P.S.; et al. Ribosomal Proteins Rpl22 and Rpl22l1 Control Morphogenesis by Regulating Pre-MRNA Splicing. Cell Rep. 2017, 18, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Locati, M.D.; Pagano, J.F.B.; Girard, G.; Ensink, W.A.; van Olst, M.; van Leeuwen, S.; Nehrdich, U.; Spaink, H.P.; Rauwerda, H.; Jonker, M.J.; et al. Expression of Distinct Maternal and Somatic 5.8S, 18S, and 28S RRNA Types during Zebrafish Development. RNA 2017, 23, 1188–1199. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.; van Spaendonk, R.M.; Choudhuri, R.; Sinden, R.E.; Janse, C.J.; Waters, A.P. Heterogeneous Ribosome Populations Are Present in Plasmodium Berghei during Development in Its Vector. Mol. Microbiol. 1999, 31, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Diesend, J.; Birkedal, U.; Kjellin, J.; Zhang, J.; Jablonski, K.P.; Söderbom, F.; Nielsen, H.; Hammann, C. Fractional 2’-O-Methylation in the Ribosomal RNA of Dictyostelium Discoideum Supports Ribosome Heterogeneity in Amoebozoa. Sci. Rep. 2022, 12, 1952. [Google Scholar] [CrossRef]
- Ihara, M.; Tseng, H.; Schultz, R.M. Expression of Variant Ribosomal RNA Genes in Mouse Oocytes and Preimplantation Embryos. Biol. Reprod. 2011, 84, 944–946. [Google Scholar] [CrossRef] [PubMed]
- Delorme, J.; Wang, L.; Kodoth, V.; Wang, Y.; Ma, J.; Jiang, S.; Aton, S.J. Hippocampal Neurons’ Cytosolic and Membrane-Bound Ribosomal Transcript Profiles Are Differentially Regulated by Learning and Subsequent Sleep. Proc. Natl. Acad. Sci. USA 2021, 118, e2108534118. [Google Scholar] [CrossRef]
- Lyons, L.C.; Chatterjee, S.; Vanrobaeys, Y.; Gaine, M.E.; Abel, T. Translational Changes Induced by Acute Sleep Deprivation Uncovered by TRAP-Seq. Mol. Brain 2020, 13, 165. [Google Scholar] [CrossRef]
- Rozenbaum, M.; Rajman, M.; Rishal, I.; Koppel, I.; Koley, S.; Medzihradszky, K.F.; Oses-Prieto, J.A.; Kawaguchi, R.; Amieux, P.S.; Burlingame, A.L.; et al. Translatome Regulation in Neuronal Injury and Axon Regrowth. eNeuro 2018, 5. [Google Scholar] [CrossRef]
- Fusco, C.M.; Desch, K.; Dörrbaum, A.R.; Wang, M.; Staab, A.; Chan, I.C.W.; Vail, E.; Villeri, V.; Langer, J.D.; Schuman, E.M. Neuronal Ribosomes Exhibit Dynamic and Context-Dependent Exchange of Ribosomal Proteins. Nat. Commun. 2021, 12, 6127. [Google Scholar] [CrossRef]
- Jiao, J.; Kavdia, K.; Pagala, V.; Palmer, L.; Finkelstein, D.; Fan, Y.; Peng, J.; Demontis, F. An Age-Downregulated Ribosomal RpS28 Protein Variant Regulates the Muscle Proteome. G3 Bethesda 2021, 11, jkab165. [Google Scholar] [CrossRef]
- Maitra, N.; He, C.; Blank, H.M.; Tsuchiya, M.; Schilling, B.; Kaeberlein, M.; Aramayo, R.; Kennedy, B.K.; Polymenis, M. Translational Control of One-Carbon Metabolism Underpins Ribosomal Protein Phenotypes in Cell Division and Longevity. Elife 2020, 9, e53127. [Google Scholar] [CrossRef]
- McElreavey, K.; Pailhoux, E.; Bashamboo, A. DHX37 and 46,XY DSD: A New Ribosomopathy? Sex. Dev. 2022, 16, 194–206. [Google Scholar] [CrossRef] [PubMed]
- Orgebin, E.; Lamoureux, F.; Isidor, B.; Charrier, C.; Ory, B.; Lézot, F.; Baud’huin, M. Ribosomopathies: New Therapeutic Perspectives. Cells 2020, 9, 2080. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Su, Y.; Chen, L.; Lin, Y.; Ru, K. Identification of Novel Mutations in Patients with Diamond-Blackfan Anemia and Literature Review of RPS10 and RPS26 Mutations. Int. J. Lab. Hematol. 2023. ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Engidaye, G.; Melku, M.; Enawgaw, B. Diamond Blackfan Anemia: Genetics, Pathogenesis, Diagnosis and Treatment. EJIFCC 2019, 30, 67–81. [Google Scholar] [PubMed]
- Arbiv, O.A.; Cuvelier, G.; Klaassen, R.J.; Fernandez, C.V.; Robitaille, N.; Steele, M.; Breakey, V.; Abish, S.; Wu, J.; Sinha, R.; et al. Molecular Analysis and Genotype-Phenotype Correlation of Diamond-Blackfan Anemia. Clin. Genet. 2018, 93, 320–328. [Google Scholar] [CrossRef]
- Juli, G.; Gismondi, A.; Monteleone, V.; Caldarola, S.; Iadevaia, V.; Aspesi, A.; Dianzani, I.; Proud, C.G.; Loreni, F. Depletion of Ribosomal Protein S19 Causes a Reduction of RRNA Synthesis. Sci. Rep. 2016, 6, 35026. [Google Scholar] [CrossRef]
- Bohnsack, K.E.; Bohnsack, M.T. Uncovering the Assembly Pathway of Human Ribosomes and Its Emerging Links to Disease. EMBO J. 2019, 38, e100278. [Google Scholar] [CrossRef]
- Sulima, S.O.; Kampen, K.R.; De Keersmaecker, K. Cancer Biogenesis in Ribosomopathies. Cells 2019, 8, 229. [Google Scholar] [CrossRef]
- Groarke, E.M.; Calado, R.T.; Liu, J.M. Cell Senescence and Malignant Transformation in the Inherited Bone Marrow Failure Syndromes: Overlapping Pathophysiology with Therapeutic Implications. Semin. Hematol. 2022, 59, 30–37. [Google Scholar] [CrossRef]
- De Keersmaecker, K.; Sulima, S.O.; Dinman, J.D. Ribosomopathies and the Paradox of Cellular Hypo- to Hyperproliferation. Blood 2015, 125, 1377–1382. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Kermasson, L.; Hilcenko, C.; Kargas, V.; Traynor, D.; Boukerrou, A.Z.; Escudero-Urquijo, N.; Faille, A.; Bertrand, A.; Rossmann, M.; et al. Somatic Genetic Rescue of a Germline Ribosome Assembly Defect. Nat. Commun. 2021, 12, 5044. [Google Scholar] [CrossRef]
- Kennedy, A.L.; Myers, K.C.; Bowman, J.; Gibson, C.J.; Camarda, N.D.; Furutani, E.; Muscato, G.M.; Klein, R.H.; Ballotti, K.; Liu, S.; et al. Distinct Genetic Pathways Define Pre-Malignant versus Compensatory Clonal Hematopoiesis in Shwachman-Diamond Syndrome. Nat. Commun. 2021, 12, 1334. [Google Scholar] [CrossRef]
- Ebright, R.Y.; Lee, S.; Wittner, B.S.; Niederhoffer, K.L.; Nicholson, B.T.; Bardia, A.; Truesdell, S.; Wiley, D.F.; Wesley, B.; Li, S.; et al. Deregulation of Ribosomal Protein Expression and Translation Promotes Breast Cancer Metastasis. Science 2020, 367, 1468–1473. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.C.; MacDonald, C.C.; Kellogg, M.K.; Karamysheva, Z.N.; Karamyshev, A.L. Specialized Ribosomes in Health and Disease. Int. J. Mol. Sci. 2023, 24, 6334. [Google Scholar] [CrossRef] [PubMed]
- Larionova, T.D.; Bastola, S.; Aksinina, T.E.; Anufrieva, K.S.; Wang, J.; Shender, V.O.; Andreev, D.E.; Kovalenko, T.F.; Arapidi, G.P.; Shnaider, P.V.; et al. Alternative RNA Splicing Modulates Ribosomal Composition and Determines the Spatial Phenotype of Glioblastoma Cells. Nat. Cell Biol. 2022, 24, 1541–1557. [Google Scholar] [CrossRef]
- Wu, W.; Yu, N.; Li, F.; Gao, P.; Lin, S.; Zhu, Y. RPL35 Promotes Neuroblastoma Progression via the Enhanced Aerobic Glycolysis. Am. J. Cancer Res. 2021, 11, 5701–5714. [Google Scholar]
- Mills, E.W.; Green, R. Ribosomopathies: There’s Strength in Numbers. Science 2017, 358, eaan2755. [Google Scholar] [CrossRef]
- Aubert, M.; O’Donohue, M.-F.; Lebaron, S.; Gleizes, P.-E. Pre-Ribosomal RNA Processing in Human Cells: From Mechanisms to Congenital Diseases. Biomolecules 2018, 8, 123. [Google Scholar] [CrossRef]
- Gregory, B.; Rahman, N.; Bommakanti, A.; Shamsuzzaman, M.; Thapa, M.; Lescure, A.; Zengel, J.M.; Lindahl, L. The Small and Large Ribosomal Subunits Depend on Each Other for Stability and Accumulation. Life Sci. Alliance 2019, 2, e201800150. [Google Scholar] [CrossRef]
- Essers, P.; Tain, L.S.; Nespital, T.; Goncalves, J.; Froehlich, J.; Partridge, L. Reduced Insulin/Insulin-like Growth Factor Signaling Decreases Translation in Drosophila and Mice. Sci. Rep. 2016, 6, 30290. [Google Scholar] [CrossRef]
- Karunadharma, P.P.; Basisty, N.; Dai, D.-F.; Chiao, Y.A.; Quarles, E.K.; Hsieh, E.J.; Crispin, D.; Bielas, J.H.; Ericson, N.G.; Beyer, R.P.; et al. Subacute Calorie Restriction and Rapamycin Discordantly Alter Mouse Liver Proteome Homeostasis and Reverse Aging Effects. Aging Cell 2015, 14, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Bjedov, I.; Rallis, C. The Target of Rapamycin Signalling Pathway in Ageing and Lifespan Regulation. Genes 2020, 11, 1043. [Google Scholar] [CrossRef]
- Rallis, C.; Townsend, S.; Bähler, J. Genetic Interactions and Functional Analyses of the Fission Yeast Gsk3 and Amk2 Single and Double Mutants Defective in TORC1-Dependent Processes. Sci. Rep. 2017, 7, 44257. [Google Scholar] [CrossRef]
- Syntichaki, P.; Troulinaki, K.; Tavernarakis, N. Protein Synthesis Is a Novel Determinant of Aging in Caenorhabditis Elegans. Ann. N. Y. Acad. Sci. 2007, 1119, 289–295. [Google Scholar] [CrossRef]
- Pan, K.Z.; Palter, J.E.; Rogers, A.N.; Olsen, A.; Chen, D.; Lithgow, G.J.; Kapahi, P. Inhibition of MRNA Translation Extends Lifespan in Caenorhabditis Elegans. Aging Cell 2007, 6, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Chiocchetti, A.; Zhou, J.; Zhu, H.; Karl, T.; Haubenreisser, O.; Rinnerthaler, M.; Heeren, G.; Oender, K.; Bauer, J.; Hintner, H.; et al. Ribosomal Proteins Rpl10 and Rps6 Are Potent Regulators of Yeast Replicative Life Span. Exp. Gerontol. 2007, 42, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Gonskikh, Y.; Polacek, N. Alterations of the Translation Apparatus during Aging and Stress Response. Mech. Ageing Dev. 2017, 168, 30–36. [Google Scholar] [CrossRef]
- Anisimova, A.S.; Meerson, M.B.; Gerashchenko, M.V.; Kulakovskiy, I.V.; Dmitriev, S.E.; Gladyshev, V.N. Multifaceted Deregulation of Gene Expression and Protein Synthesis with Age. Proc. Natl. Acad. Sci. USA 2020, 117, 15581–15590. [Google Scholar] [CrossRef]
- Yang, H.W.; Kim, H.D.; Kim, T.-S.; Kim, J. Senescent Cells Differentially Translate Senescence-Related MRNAs Via Ribosome Heterogeneity. J. Gerontol. A Biol. Sci. Med. Sci. 2019, 74, 1015–1024. [Google Scholar] [CrossRef]
- Xiao, Y.; Cai, G.-P.; Feng, X.; Li, Y.-J.; Guo, W.-H.; Guo, Q.; Huang, Y.; Su, T.; Li, C.-J.; Luo, X.-H.; et al. Splicing Factor YBX1 Regulates Bone Marrow Stromal Cell Fate during Aging. EMBO J. 2023, 42, e111762. [Google Scholar] [CrossRef] [PubMed]
- Gay, D.M.; Lund, A.H.; Jansson, M.D. Translational Control through Ribosome Heterogeneity and Functional Specialization. Trends Biochem. Sci. 2022, 47, 66–81. [Google Scholar] [CrossRef] [PubMed]
- Haag, E.S.; Dinman, J.D. Still Searching for Specialized Ribosomes. Dev. Cell 2019, 48, 744–746. [Google Scholar] [CrossRef]
- Kelmer Sacramento, E.; Kirkpatrick, J.M.; Mazzetto, M.; Baumgart, M.; Bartolome, A.; Di Sanzo, S.; Caterino, C.; Sanguanini, M.; Papaevgeniou, N.; Lefaki, M.; et al. Reduced Proteasome Activity in the Aging Brain Results in Ribosome Stoichiometry Loss and Aggregation. Mol. Syst. Biol. 2020, 16, e9596. [Google Scholar] [CrossRef] [PubMed]
- Amirbeigiarab, S.; Kiani, P.; Velazquez Sanchez, A.; Krisp, C.; Kazantsev, A.; Fester, L.; Schlüter, H.; Ignatova, Z. Invariable Stoichiometry of Ribosomal Proteins in Mouse Brain Tissues with Aging. Proc. Natl. Acad. Sci. USA 2019, 116, 22567–22572. [Google Scholar] [CrossRef] [PubMed]
| Biological Process | Organism | Associated Ribosomal Protein | Reference | |
|---|---|---|---|---|
| Cross Domain Name | Human Name | |||
| Maintenance of germ-line cells | Drosophila | eS10 | RPS10 | [38] |
| eS28 | RPS28 (paralogues Rps28a and Rps28-like) | [53] | ||
| Maintenance of preimplantation embryo | mouse | eL13 | RPL13 | [41] |
| Mesoderm production | human embryonic stem cell | uL1 | RPL10A | [42] |
| mouse | ||||
| Eye development | mouse | eL24 | RPL24 | [39] |
| Neurocranium development | zebrafish | uL18 | RPL5 | [39] |
| Body plan/axial skeletal patterning | mouse | eL38 | RPL38 | [32] |
| Haematopoiesis and blood vessel formation | zebrafish | uS12 | RPS23 | [39] |
| uS14 | RPS29 | |||
| Haematopoiesis | zebrafish | eL22 | RPL22 (paralogue RPL22l1) | [44] |
| uL18 | RPL5 | [39] | ||
| Serine and methionine metabolism | yeast | eL22 | RPL22 (paralogue RPL22a) | [54] |
| Circadian regulation | mouse | es12 | RPS12 | [50] |
| eS19 | RPS19 | |||
| eS24 | RPS24 | |||
| uS4 | RPS9 | |||
| eL19 | RPL19 | |||
| eL33 | RPL35A | |||
| eL34 | RPL34 | |||
| Oxidative stress response | rat neuronal cells | eS30 | RPS30 | [52] |
| RACK1 | RACK1 | |||
| P2 | P2 | |||
| uL10 | P0 | |||
| uL16 | RPL10 | |||
| Learning and memory | mouse | es12 | RPS12 | [49] |
| eS21 | RPS21 | |||
| eS27 | RPS27 | |||
| eS28 | RPS28 | |||
| eS6 | RPS6 | |||
| uS10 | RPS20 | |||
| uS14 | RPS29 | |||
| eL21 | RPL21 | |||
| eL36 | RPL36 | |||
| eL37 | RPL37 | |||
| eL39 | RPL39 | |||
| eL41 | RPL41 | |||
| eL43 | RPL37A | |||
| Disease | Associated Ribosomal Proteins | References | |
|---|---|---|---|
| Diamond–Blackfan anemia | RP-SSU | RPS7, RPS8, RPS10, RPS15, RPS15A, RPS17, RPS19, RPS24, RPS26, RPS27, RPS27A, RPS28, RPS29 | [56,57,58] |
| RP-LSU | RPL3, RPL5, RPL7, RPL9, RPL11, RPL14, RPL15, RPL18, RPL19, RPL23A, RPL26, RPL27, RPL31, RPL35, RPL35A, RPL36 | ||
| Other proteins | TSR2, GATA1, EPO, ADA2 | [56] | |
| Refractory macrocytic anaemia | RPS14 | [39] | |
| Autism, susceptibility to X-linked 5 | RPL10 | [39] | |
| Isolated congenital asplenia | RPSA | [39] | |
| Brachycephaly, trichomegaly, and development delay | RPS23 | [39] | |
| Spondyloepimetaphyseal dysplasia | RPL13 | [39] | |
| Hypotrichosis | RPL21 | [39] | |
| Shwachman–Diamond syndrome | Shwachman–Bodian–Diamond Syndrome (SBDS) | [55] | |
| Treacher Collins syndrome | TCOF1 | [55] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, R.A.; Rallis, C. Ribosomal Biogenesis and Heterogeneity in Development, Disease, and Aging. Epigenomes 2023, 7, 17. https://doi.org/10.3390/epigenomes7030017
Islam RA, Rallis C. Ribosomal Biogenesis and Heterogeneity in Development, Disease, and Aging. Epigenomes. 2023; 7(3):17. https://doi.org/10.3390/epigenomes7030017
Chicago/Turabian StyleIslam, Rowshan Ara, and Charalampos Rallis. 2023. "Ribosomal Biogenesis and Heterogeneity in Development, Disease, and Aging" Epigenomes 7, no. 3: 17. https://doi.org/10.3390/epigenomes7030017
APA StyleIslam, R. A., & Rallis, C. (2023). Ribosomal Biogenesis and Heterogeneity in Development, Disease, and Aging. Epigenomes, 7(3), 17. https://doi.org/10.3390/epigenomes7030017





