Pulmonary Pathogen-Induced Epigenetic Modifications
Abstract
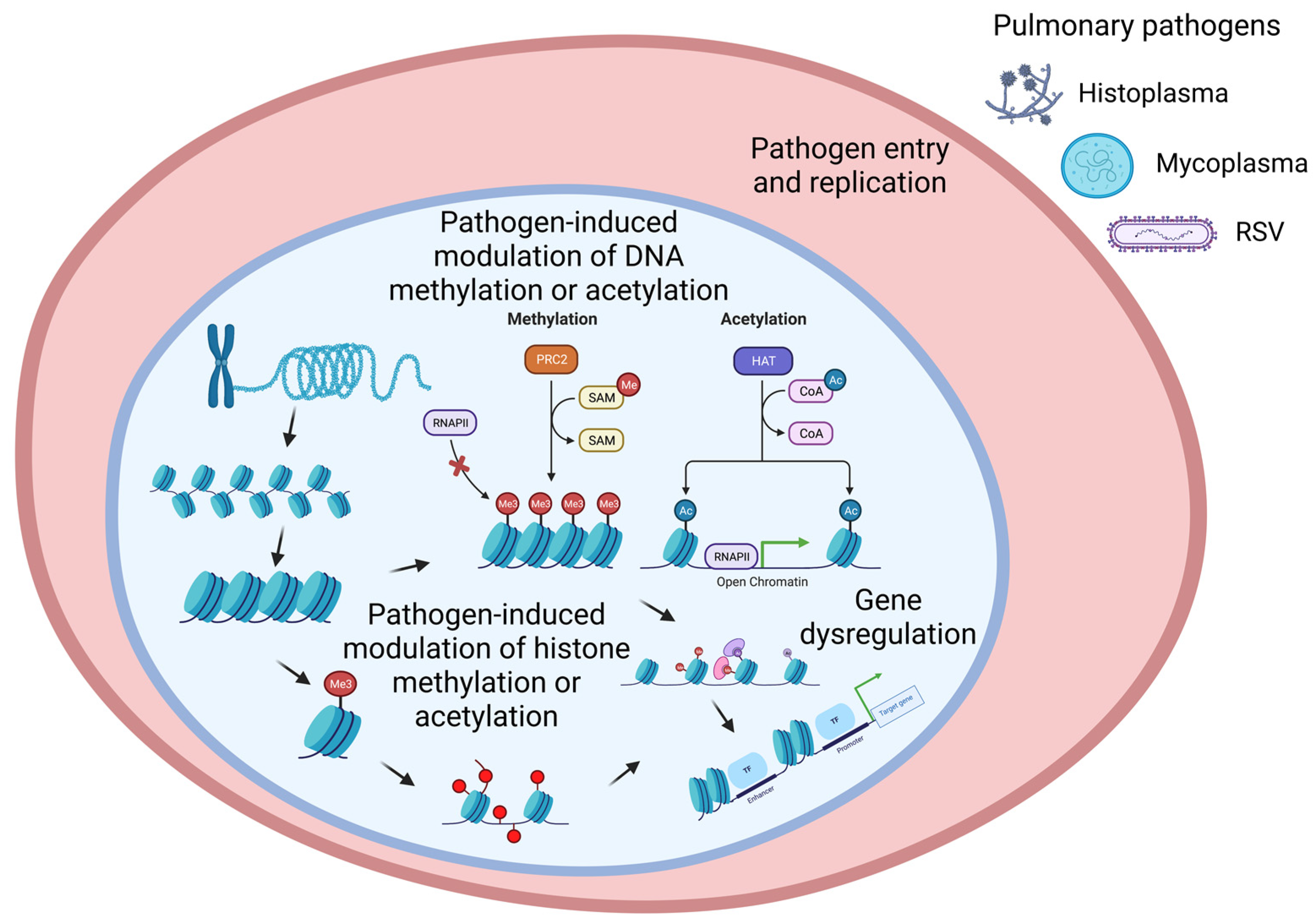
1. Introduction
2. Epigenetic Mechanisms Regulate Gene Expression
2.1. Histone Modification
2.2. DNA Methylation
3. Bacterial Infection-Mediated Pulmonary Epigenetics
4. Viral Infection-Mediated Pulmonary Epigenetics
4.1. Adenovirus
4.2. RSV
4.3. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2)
| Pulmonary Pathogen | Epigenetic Changes | Cellular Impact | References |
|---|---|---|---|
| Adenovirus | Histone deacetylase inhibitor suppresses host HDAC proteins | Increased global acetylation, transcription modulation | [68,72] |
| Respiratory syncytial virus (RSV) | Chromatin modifications inducing nucleosome-free regions Demethylation of Nodal promoter Increased TGFβ expression | Impacts tyrosine kinase growth factor signaling, affects extracellular matrix secretory pathways Reduced pro-inflammatory cytokines release | [74,75,76] |
| Severe acute respiratory syndrome coronavirus 2 (SARS-CoV2) | Hypermethylation of IFN related Hypomethylation of inflammatory genes Perturbation of epigenetic clock IRF1 and IRF7 were upregulated Epi-factors HDAC9 and SIRT1 were deregulated. | Increased level of ACE2 receptor expression Increased cytokine release | [77,78,79,80] |
| Influenza virus | Post translational histone modification Decrease in histone acetylation Demethylation on CREB1 binding region | Hypercytokinemia | [81,82,83] |
| Rhino virus | Modifications in DNA methylation | [64] | |
| Middle East respiratory syndrome-related corona virus (MERS) | H3K27 methylation | Downregulate antigen-presenting molecules | [13,77,83] |
| Types of Epigenetic Change | Adenovirus | SARS-CoV-2 | RSV | Influenza Virus | Rhino Virus | MERS |
|---|---|---|---|---|---|---|
| Chromatin remodeling | Not Identified | Not Identified | Yes [75] | Yes [83] | Not Identified | Not Identified |
| Changes in DNA methylation | Yes [68] | Yes [77,78,79] | Yes [74] | Yes [81,82] | Yes [64] | Yes [83] |
| Non-coding RNA | Yes [80] | Yes [74] | ||||
| Changes in DNA acetylation | Yes [68] | Not Identified | Not Identified | Yes [81] | Not Identified | Not Identified |
5. Fungal Epigenetics
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rajeev, R.; Dwivedi, A.P.; Sinha, A.; Agarwaal, V.; Dev, R.R.; Kar, A.; Khosla, S. Epigenetic interaction of microbes with their mammalian hosts. J. Biosci. 2021, 46, 94. [Google Scholar] [CrossRef] [PubMed]
- Indrio, F.; Martini, S.; Francavilla, R.; Corvaglia, L.; Cristofori, F.; Mastrolia, S.A.; Neu, J.; Rautava, S.; Russo Spena, G.; Raimondi, F.; et al. Epigenetic Matters: The Link between Early Nutrition, Microbiome, and Long-term Health Development. Front. Pediatr. 2017, 5, 178. [Google Scholar] [CrossRef] [PubMed]
- Maselli, D.J.; Bhatt, S.P.; Anzueto, A.; Bowler, R.P.; DeMeo, D.L.; Diaz, A.A.; Dransfield, M.T.; Fawzy, A.; Foreman, M.G.; Hanania, N.A.; et al. Clinical Epidemiology of COPD: Insights From 10 Years of the COPDGene Study. Chest 2019, 156, 228–238. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, C.J.; Batai, K.; Bleda, M.; Haimel, M.; Southgate, L.; Germain, M.; Pauciulo, M.W.; Hadinnapola, C.; Aman, J.; Girerd, B.; et al. Genetic determinants of risk in pulmonary arterial hypertension: International genome-wide association studies and meta-analysis. Lancet Respir. Med. 2019, 7, 227–238. [Google Scholar] [CrossRef]
- Sakornsakolpat, P.; Prokopenko, D.; Lamontagne, M.; Reeve, N.F.; Guyatt, A.L.; Jackson, V.E.; Shrine, N.; Qiao, D.; Bartz, T.M.; Kim, D.K.; et al. Genetic landscape of chronic obstructive pulmonary disease identifies heterogeneous cell-type and phenotype associations. Nat. Genet. 2019, 51, 494–505. [Google Scholar] [CrossRef]
- Benincasa, G.; DeMeo, D.L.; Glass, K.; Silverman, E.K.; Napoli, C. Epigenetics and pulmonary diseases in the horizon of precision medicine: A review. Eur. Respir. J. 2021, 57, 2003406. [Google Scholar] [CrossRef]
- Hoang, T.T.; Sikdar, S.; Xu, C.J.; Lee, M.K.; Cardwell, J.; Forno, E.; Imboden, M.; Jeong, A.; Madore, A.M.; Qi, C.; et al. Epigenome-wide association study of DNA methylation and adult asthma in the Agricultural Lung Health Study. Eur. Respir. J. 2020, 56, 2000217. [Google Scholar] [CrossRef]
- Napoli, C.; Benincasa, G.; Loscalzo, J. Epigenetic Inheritance Underlying Pulmonary Arterial Hypertension. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 653–664. [Google Scholar] [CrossRef]
- Regan, E.A.; Hersh, C.P.; Castaldi, P.J.; DeMeo, D.L.; Silverman, E.K.; Crapo, J.D.; Bowler, R.P. Omics and the Search for Blood Biomarkers in Chronic Obstructive Pulmonary Disease. Insights from COPDGene. Am. J. Respir. Cell Mol. Biol. 2019, 61, 143–149. [Google Scholar] [CrossRef]
- DeVries, A.; Vercelli, D. Epigenetic Mechanisms in Asthma. Ann. Am. Thorac. Soc. 2016, 13 (Suppl. S1), S48–S50. [Google Scholar] [CrossRef]
- Bierne, H.; Hamon, M.; Cossart, P. Epigenetics and bacterial infections. Cold Spring Harb. Perspect. Med. 2012, 2, a010272. [Google Scholar] [CrossRef] [PubMed]
- Youssef, N.; Budd, A.; Bielawski, J.P. Introduction to Genome Biology and Diversity. Methods Mol. Biol. 2019, 1910, 3–31. [Google Scholar] [CrossRef] [PubMed]
- Salgado-Albarran, M.; Navarro-Delgado, E.I.; Del Moral-Morales, A.; Alcaraz, N.; Baumbach, J.; Gonzalez-Barrios, R.; Soto-Reyes, E. Comparative transcriptome analysis reveals key epigenetic targets in SARS-CoV-2 infection. NPJ Syst. Biol. Appl. 2021, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Denzer, L.; Schroten, H.; Schwerk, C. From gene to protein—How bacterial virulence factors manipulate host gene expression during infection. Int. J. Mol. Sci. 2020, 21, 3730. [Google Scholar] [CrossRef] [PubMed]
- De Monerri, N.C.S.; Kim, K. Pathogens hijack the epigenome: A new twist on host-pathogen interactions. Am. J. Pathol. 2014, 184, 897–911. [Google Scholar] [CrossRef]
- Jaenisch, R.; Bird, A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003, 33 (Suppl. S3), 245–254. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef]
- Niller, H.H.; Masa, R.; Venkei, A.; Mészáros, S.; Minarovits, J. Pathogenic mechanisms of intracellular bacteria. Curr. Opin. Infect. Dis. 2017, 30, 309–315. [Google Scholar] [CrossRef]
- Zhao, Y.; Garcia, B.A. Comprehensive Catalog of Currently Documented Histone Modifications. Cold Spring Harb. Perspect. Biol. 2015, 7, a025064. [Google Scholar] [CrossRef]
- Cavalieri, V. The Expanding Constellation of Histone Post-Translational Modifications in the Epigenetic Landscape. Genes 2021, 12, 1596. [Google Scholar] [CrossRef]
- Hamon, M.A.; Batsché, E.; Régnault, B.; Tham, T.N.; Seveau, S.; Muchardt, C.; Cossart, P. Histone modifications induced by a family of bacterial toxins. Proc. Natl. Acad. Sci. USA 2007, 104, 13467–13472. [Google Scholar] [CrossRef]
- Pennini, M.E.; Perrinet, S.; Dautry-Varsat, A.; Subtil, A. Histone methylation by NUE, a novel nuclear effector of the intracellular pathogen Chlamydia trachomatis. PLoS Pathog. 2010, 6, e1000995. [Google Scholar] [CrossRef]
- Wellen, K.E.; Hatzivassiliou, G.; Sachdeva, U.M.; Bui, T.V.; Cross, J.R.; Thompson, C.B. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 2009, 324, 1076–1080. [Google Scholar] [CrossRef]
- Rennoll-Bankert, K.E.; Garcia-Garcia, J.C.; Sinclair, S.H.; Dumler, J.S. Chromatin-bound bacterial effector ankyrin A recruits histone deacetylase 1 and modifies host gene expression. Cell. Microbiol. 2015, 17, 1640–1652. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Upadhyay, S.; Srilalitha, M.; Nandicoori, V.K.; Khosla, S. The interaction of mycobacterial protein Rv2966c with host chromatin is mediated through non-CpG methylation and histone H3/H4 binding. Nucleic Acids Res. 2015, 43, 3922–3937. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Sowpati, D.T.; Singh, P.; Khan, M.Z.; Ganji, R.; Upadhyay, S.; Banerjee, S.; Nandicoori, V.K.; Khosla, S. Genome-wide non-CpG methylation of the host genome during M. tuberculosis infection. Sci. Rep. 2016, 6, 25006. [Google Scholar] [CrossRef]
- Ding, S.-Z.; Fischer, W.; Kaparakis-Liaskos, M.; Liechti, G.; Merrell, D.S.; Grant, P.A.; Ferrero, R.L.; Crowe, S.E.; Haas, R.; Hatakeyama, M. Helicobacter pylori-induced histone modification, associated gene expression in gastric epithelial cells, and its implication in pathogenesis. PLoS ONE 2010, 5, e9875. [Google Scholar] [CrossRef] [PubMed]
- Nardone, G.; Compare, D.; De Colibus, P.; De Nucci, G.; Rocco, A. Helicobacter pylori and epigenetic mechanisms underlying gastric carcinogenesis. Dig. Dis. 2007, 25, 225–229. [Google Scholar] [CrossRef]
- Santos, J.C.; Ribeiro, M.L. Epigenetic regulation of DNA repair machinery in Helicobacter pylori-induced gastric carcinogenesis. World J. Gastroenterol. WJG 2015, 21, 9021. [Google Scholar] [CrossRef] [PubMed]
- Soto, D.; Song, C.; McLaughlin-Drubin, M.E. Epigenetic alterations in human papillomavirus-associated cancers. Viruses 2017, 9, 248. [Google Scholar] [CrossRef] [PubMed]
- Al Akeel, R. Role of epigenetic reprogramming of host genes in bacterial pathogenesis. Saudi J. Biol. Sci. 2013, 20, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Ebenezer, D.L.; Berdyshev, E.V.; Bronova, I.A.; Liu, Y.; Tiruppathi, C.; Komarova, Y.; Benevolenskaya, E.V.; Suryadevara, V.; Ha, A.W.; Harijith, A.; et al. Pseudomonas aeruginosa stimulates nuclear sphingosine-1-phosphate generation and epigenetic regulation of lung inflammatory injury. Thorax 2019, 74, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Haller, D.; Holt, L.; Kim, S.C.; Schwabe, R.F.; Sartor, R.B.; Jobin, C. Transforming growth factor-beta 1 inhibits non-pathogenic Gram negative bacteria-induced NF-kappa B recruitment to the interleukin-6 gene promoter in intestinal epithelial cells through modulation of histone acetylation. J. Biol. Chem. 2003, 278, 23851–23860. [Google Scholar] [CrossRef] [PubMed]
- Slevogt, H.; Schmeck, B.; Jonatat, C.; Zahlten, J.; Beermann, W.; van Laak, V.; Opitz, B.; Dietel, S.; N’Guessan, P.D.; Hippenstiel, S.; et al. Moraxella catarrhalis induces inflammatory response of bronchial epithelial cells via MAPK and NF-κB activation and histone deacetylase activity reduction. Am. J. Physiol. Lung Cell. Mol. Physiol. 2006, 290, L818–L826. [Google Scholar] [CrossRef]
- Dupont, C.; Armant, D.R.; Brenner, C.A. Epigenetics: Definition, mechanisms and clinical perspective. In Seminars in Reproductive Medicine; Thieme Medical Publishers: New York, NY, USA, 2009. [Google Scholar]
- Blair, J.M.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Motta, S.S.; Cluzel, P.; Aldana, M. Adaptive resistance in bacteria requires epigenetic inheritance, genetic noise, and cost of efflux pumps. PLoS ONE 2015, 10, e0118464. [Google Scholar] [CrossRef]
- Cohen, N.R.; Lobritz, M.A.; Collins, J.J. Microbial persistence and the road to drug resistance. Cell Host Microbe 2013, 13, 632–642. [Google Scholar] [CrossRef]
- Handy, D.E.; Castro, R.; Loscalzo, J. Epigenetic modifications: Basic mechanisms and role in cardiovascular disease. Circulation 2011, 123, 2145–2156. [Google Scholar] [CrossRef]
- Varghese, T.; Jayasri, M.; Suthindhiran, K. Marine A ctinomycetes as potential source for histone deacetylase inhibitors and epigenetic modulation. Lett. Appl. Microbiol. 2015, 61, 69–76. [Google Scholar] [CrossRef]
- Li, T.; Lu, Q.; Wang, G.; Xu, H.; Huang, H.; Cai, T.; Kan, B.; Ge, J.; Shao, F. SET-domain bacterial effectors target heterochromatin protein 1 to activate host rDNA transcription. EMBO Rep. 2013, 14, 733–740. [Google Scholar] [CrossRef]
- Krishnananthasivam, S.; Jayathilaka, N.; Sathkumara, H.D.; Corea, E.; Natesan, M.; De Silva, A.D. Host gene expression analysis in Sri Lankan melioidosis patients. PLoS Neglected Trop. Dis. 2017, 11, e0005643. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Curry, H.M.; Zwilling, B.S.; Lafuse, W.P. Mycobacteria inhibition of IFN-γ induced HLA-DR gene expression by up-regulating histone deacetylation at the promoter region in human THP-1 monocytic cells. J. Immunol. 2005, 174, 5687–5694. [Google Scholar] [CrossRef] [PubMed]
- Yaseen, I.; Kaur, P.; Nandicoori, V.K.; Khosla, S. Mycobacteria modulate host epigenetic machinery by Rv1988 methylation of a non-tail arginine of histone H3. Nat. Commun. 2015, 6, 8922. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Yi, M.; Chen, J.; Li, S.; Chen, W. Mycobacterium tuberculosis EIS gene inhibits macrophage autophagy through up-regulation of IL-10 by increasing the acetylation of histone H3. Biochem. Biophys. Res. Commun. 2016, 473, 1229–1234. [Google Scholar] [CrossRef] [PubMed]
- Jose, L.; Ramachandran, R.; Bhagavat, R.; Gomez, R.L.; Chandran, A.; Raghunandanan, S.; Omkumar, R.V.; Chandra, N.; Mundayoor, S.; Kumar, R.A. Hypothetical protein Rv3423. 1 of Mycobacterium tuberculosis is a histone acetyltransferase. FEBS J. 2016, 283, 265–281. [Google Scholar] [CrossRef] [PubMed]
- Komar, D.; Juszczynski, P. Rebelled epigenome: Histone H3S10 phosphorylation and H3S10 kinases in cancer biology and therapy. Clin. Epigenet. 2020, 12, 147. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Feng, C.; Zhai, Y.Z.; Zhou, X.; Li, B.; Wang, L.L.; Chen, W.; Lv, F.Q.; Li, T.S. Identification of miRNA biomarkers of pneumonia using RNA-sequencing and bioinformatics analysis. Exp. Ther. Med. 2017, 13, 1235–1244. [Google Scholar] [CrossRef]
- Li, H.; Xu, H.; Zhou, Y.; Zhang, J.; Long, C.; Li, S.; Chen, S.; Zhou, J.-M.; Shao, F. The phosphothreonine lyase activity of a bacterial type III effector family. Science 2007, 315, 1000–1003. [Google Scholar] [CrossRef]
- Rolando, M.; Sanulli, S.; Rusniok, C.; Gomez-Valero, L.; Bertholet, C.; Sahr, T.; Margueron, R.; Buchrieser, C. Legionella pneumophila effector RomA uniquely modifies host chromatin to repress gene expression and promote intracellular bacterial replication. Cell Host Microbe 2013, 13, 395–405. [Google Scholar] [CrossRef]
- Schuelein, R.; Spencer, H.; Dagley, L.F.; Li, P.F.; Luo, L.; Stow, J.L.; Abraham, G.; Naderer, T.; Gomez-Valero, L.; Buchrieser, C. Targeting of RNA Polymerase II by a nuclear Legionella pneumophila Dot/Icm effector SnpL. Cell. Microbiol. 2018, 20, e12852. [Google Scholar] [CrossRef]
- Von Dwingelo, J.; Chung, I.Y.W.; Price, C.T.; Li, L.; Jones, S.; Cygler, M.; Abu Kwaik, Y. Interaction of the Ankyrin H core effector of legionella with the host LARP7 component of the 7SK snRNP complex. mBio 2019, 10, e01942-19. [Google Scholar] [CrossRef] [PubMed]
- Mujtaba, S.; Winer, B.Y.; Jaganathan, A.; Patel, J.; Sgobba, M.; Schuch, R.; Gupta, Y.K.; Haider, S.; Wang, R.; Fischetti, V.A. Anthrax SET protein: A potential virulence determinant that epigenetically represses NF-κB activation in infected macrophages. J. Biol. Chem. 2013, 288, 23458–23472. [Google Scholar] [CrossRef] [PubMed]
- Raymond, B.; Ravaux, L.; Mémet, S.; Wu, Y.; Sturny-Leclère, A.; Leduc, D.; Denoyelle, C.; Goossens, P.L.; Payá, M.; Raymondjean, M. Anthrax lethal toxin down-regulates type-IIA secreted phospholipase A2 expression through MAPK/NF-κB inactivation. Biochem. Pharmacol. 2010, 79, 1149–1155. [Google Scholar] [CrossRef]
- Ha, S.-D.; Reid, C.; Meshkibaf, S.; Kim, S.O. Inhibition of interleukin 1β (IL-1β) expression by anthrax lethal toxin (LeTx) is reversed by histone deacetylase 8 (HDAC8) inhibition in murine macrophages. J. Biol. Chem. 2016, 291, 8745–8755. [Google Scholar] [CrossRef]
- Murata, M.; Azuma, Y.; Miura, K.; Rahman, M.A.; Matsutani, M.; Aoyama, M.; Suzuki, H.; Sugi, K.; Shirai, M. Chlamydial SET domain protein functions as a histone methyltransferase. Microbiology 2007, 153, 585–592. [Google Scholar] [CrossRef]
- Mojica, S.A.; Hovis, K.M.; Frieman, M.B.; Tran, B.; Hsia, R.-c.; Ravel, J.; Jenkins-Houk, C.; Wilson, K.L.; Bavoil, P.M. SINC, a type III secreted protein of Chlamydia psittaci, targets the inner nuclear membrane of infected cells and uninfected neighbors. Mol. Biol. Cell 2015, 26, 1918–1934. [Google Scholar] [CrossRef]
- Choung, H.-K.; Kim, Y.A.; Lee, M.J.; Kim, N.; Khwarg, S.I. Multigene methylation analysis of ocular adnexal MALT lymphoma and their relationship to Chlamydophila psittaci infection and clinical characteristics in South Korea. Investig. Ophthalmol. Vis. Sci. 2012, 53, 1928–1935. [Google Scholar] [CrossRef] [PubMed]
- Vdovikova, S.; Gilfillan, S.; Wang, S.; Dongre, M.; Wai, S.N.; Hurtado, A. Modulation of gene transcription and epigenetics of colon carcinoma cells by bacterial membrane vesicles. Sci. Rep. 2018, 8, 7434. [Google Scholar] [CrossRef]
- Bandyopadhaya, A.; Tsurumi, A.; Maura, D.; Jeffrey, K.L.; Rahme, L.G. A quorum-sensing signal promotes host tolerance training through HDAC1-mediated epigenetic reprogramming. Nat. Microbiol. 2016, 1, 16174. [Google Scholar] [CrossRef]
- Dortet, L.; Lombardi, C.; Cretin, F.; Dessen, A.; Filloux, A. Pore-forming activity of the Pseudomonas aeruginosa type III secretion system translocon alters the host epigenome. Nat. Microbiol. 2018, 3, 378–386. [Google Scholar] [CrossRef]
- Zughaier, S.M.; Rouquette-Loughlin, C.E.; Shafer, W.M. Identification of a Neisseria gonorrhoeae histone deacetylase: Epigenetic impact on host gene expression. Pathogens 2020, 9, 132. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Hartert, T.V. Evidence for a causal relationship between respiratory syncytial virus infection and asthma. Expert Rev. Anti-Infect. Ther. 2011, 9, 731–745. [Google Scholar] [CrossRef] [PubMed]
- Pech, M.; Weckmann, M.; Konig, I.R.; Franke, A.; Heinsen, F.A.; Oliver, B.; Ricklefs, I.; Fuchs, O.; Rabe, K.; Hansen, G.; et al. Rhinovirus infections change DNA methylation and mRNA expression in children with asthma. PLoS ONE 2018, 13, e0205275. [Google Scholar] [CrossRef]
- Britto, C.J.; Brady, V.; Lee, S.; Dela Cruz, C.S. Respiratory Viral Infections in Chronic Lung Diseases. Clin. Chest Med. 2017, 38, 87–96. [Google Scholar] [CrossRef]
- Jackson, D.J.; Gangnon, R.E.; Evans, M.D.; Roberg, K.A.; Anderson, E.L.; Pappas, T.E.; Printz, M.C.; Lee, W.M.; Shult, P.A.; Reisdorf, E.; et al. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am. J. Respir. Crit. Care Med. 2008, 178, 667–672. [Google Scholar] [CrossRef]
- Kusel, M.M.; de Klerk, N.H.; Kebadze, T.; Vohma, V.; Holt, P.G.; Johnston, S.L.; Sly, P.D. Early-life respiratory viral infections, atopic sensitization, and risk of subsequent development of persistent asthma. J. Allergy Clin. Immunol. 2007, 119, 1105–1110. [Google Scholar] [CrossRef] [PubMed]
- Milavetz, B.I.; Balakrishnan, L. Viral epigenetics. In Cancer Epigenetics; Humana Press: New York, NY, USA, 2015; pp. 569–596. [Google Scholar]
- Tooze, J.; Acheson, N. DNA Tumor Viruses; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1980. [Google Scholar]
- Ferrari, R.; Pellegrini, M.; Horwitz, G.A.; Xie, W.; Berk, A.J.; Kurdistani, S.K. Epigenetic reprogramming by adenovirus e1a. Science 2008, 321, 1086–1088. [Google Scholar] [CrossRef]
- Ferrari, R.; Su, T.; Li, B.; Bonora, G.; Oberai, A.; Chan, Y.; Sasidharan, R.; Berk, A.J.; Pellegrini, M.; Kurdistani, S.K. Reorganization of the host epigenome by a viral oncogene. Genome Res. 2012, 22, 1212–1221. [Google Scholar] [CrossRef]
- Lynch, K.L.; Gooding, L.R.; Garnett-Benson, C.; Ornelles, D.A.; Avgousti, D.C. Epigenetics and the dynamics of chromatin during adenovirus infections. FEBS Lett. 2019, 593, 3551–3570. [Google Scholar] [CrossRef]
- Avgousti, D.C.; Herrmann, C.; Kulej, K.; Pancholi, N.J.; Sekulic, N.; Petrescu, J.; Molden, R.C.; Blumenthal, D.; Paris, A.J.; Reyes, E.D. A core viral protein binds host nucleosomes to sequester immune danger signals. Nature 2016, 535, 173–177. [Google Scholar] [CrossRef]
- Caixia, L.; Yang, X.; Yurong, T.; Xiaoqun, Q. Involvement of epigenetic modification in epithelial immune responses during respiratory syncytial virus infection. Microb. Pathog. 2019, 130, 186–189. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Qiao, D.; Mann, M.; Garofalo, R.P.; Brasier, A.R. Respiratory syncytial virus infection induces chromatin remodeling to activate growth factor and extracellular matrix secretion pathways. Viruses 2020, 12, 804. [Google Scholar] [CrossRef]
- Fonseca, W.; Lukacs, N.W.; Ptaschinski, C. Factors affecting the immunity to respiratory syncytial virus: From epigenetics to microbiome. Front. Immunol. 2018, 9, 226. [Google Scholar] [CrossRef] [PubMed]
- Chlamydas, S.; Papavassiliou, A.G.; Piperi, C. Epigenetic mechanisms regulating COVID-19 infection. Epigenetics 2021, 16, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Ozturkler, Z.; Kalkan, R. A New Perspective of COVID-19 Infection: An Epigenetics Point of View. Glob. Med. Genet. 2022, 9, 004–006. [Google Scholar] [CrossRef] [PubMed]
- Corley, M.J.; Pang, A.P.; Dody, K.; Mudd, P.A.; Patterson, B.K.; Seethamraju, H.; Bram, Y.; Peluso, M.J.; Torres, L.; Iyer, N.S. Genome-wide DNA methylation profiling of peripheral blood reveals an epigenetic signature associated with severe COVID-19. J. Leukoc. Biol. 2021, 110, 21–26. [Google Scholar] [CrossRef]
- Khan, M.A.-A.-K.; Sany, M.R.U.; Islam, M.S.; Islam, A.B.M.M.K. Epigenetic regulator miRNA pattern differences among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 world-wide isolates delineated the mystery behind the epic pathogenicity and distinct clinical characteristics of pandemic COVID-19. Front. Genet. 2020, 11, 765. [Google Scholar] [CrossRef]
- Marcos-Villar, L.; Diaz-Colunga, J.; Sandoval, J.; Zamarreno, N.; Landeras-Bueno, S.; Esteller, M.; Falcon, A.; Nieto, A. Epigenetic control of influenza virus: Role of H3K79 methylation in interferon-induced antiviral response. Sci. Rep. 2018, 8, 1230. [Google Scholar] [CrossRef]
- Mukherjee, S.; Vipat, V.C.; Chakrabarti, A.K. Infection with influenza A viruses causes changes in promoter DNA methylation of inflammatory genes. Influenza Other Respir. Viruses 2013, 7, 979–986. [Google Scholar] [CrossRef]
- Menachery, V.D.; Schafer, A.; Burnum-Johnson, K.E.; Mitchell, H.D.; Eisfeld, A.J.; Walters, K.B.; Nicora, C.D.; Purvine, S.O.; Casey, C.P.; Monroe, M.E.; et al. MERS-CoV and H5N1 influenza virus antagonize antigen presentation by altering the epigenetic landscape. Proc. Natl. Acad. Sci. USA 2018, 115, E1012–E1021. [Google Scholar] [CrossRef]
- Doehlemann, G.; Okmen, B.; Zhu, W.; Sharon, A. Plant Pathogenic Fungi. Microbiol. Spectr. 2017, 5, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Lu, G.; Meng, G. Pathogenic Fungal Infection in the Lung. Front. Immunol. 2019, 10, 1524. [Google Scholar] [CrossRef] [PubMed]
- Bellocchio, S.; Moretti, S.; Perruccio, K.; Fallarino, F.; Bozza, S.; Montagnoli, C.; Mosci, P.; Lipford, G.B.; Pitzurra, L.; Romani, L. TLRs govern neutrophil activity in aspergillosis. J. Immunol. 2004, 173, 7406–7415. [Google Scholar] [CrossRef] [PubMed]
- Jose, R.J.; Brown, J.S. Opportunistic and fungal infections of the lung. Medicine 2012, 40, 335–339. [Google Scholar] [CrossRef]
- José, R.J.; Periselneris, J.N.; Brown, J.S. Opportunistic bacterial, viral and fungal infections of the lung. Medicine 2020, 48, 366–372. [Google Scholar] [CrossRef]
- Henriques-Normark, B.; Tuomanen, E.I. The pneumococcus: Epidemiology, microbiology, and pathogenesis. Cold Spring Harb. Perspect. Med. 2013, 3, a010215. [Google Scholar] [CrossRef]
- Rayens, E.; Norris, K.A. Prevalence and healthcare burden of fungal infections in the United States, 2018. In Open Forum Infectious Diseases; Oxford University Press: New York, NY, USA, 2022; p. ofab593. [Google Scholar]
- Saco, T.V.; Breitzig, M.T.; Lockey, R.F.; Kolliputi, N. Epigenetics of mucus hypersecretion in chronic respiratory diseases. Am. J. Respir. Cell Mol. Biol. 2018, 58, 299–309. [Google Scholar] [CrossRef]
- Pérez, F.J.; Ponce, C.A.; Rojas, D.A.; Iturra, P.A.; Bustamante, R.I.; Gallo, M.; Hananias, K.; Vargas, S.L. Fungal colonization with Pneumocystis correlates to increasing chloride channel accessory 1 (hCLCA1) suggesting a pathway for up-regulation of airway mucus responses, in infant lungs. Results Immunol. 2014, 4, 58–61. [Google Scholar] [CrossRef]
- Hoenigl, M. Invasive Fungal Disease Complicating Coronavirus Disease 2019: When It Rains, It Spores; Oxford University Press: New York, NY, USA, 2021; pp. e1645–e1648. [Google Scholar]
| Pulmonary Pathogen | Epigenetic Changes | Cellular Impact | References |
|---|---|---|---|
| Actinomyces spp. | Histone deacetylase inhibitor suppresses host HDAC proteins | Increased global acetylation, transcription modulation | [1,40] |
| Bordetella bronchiseptica | bbSET17 methylates histones associated with rRNA genes | rRNA transcription modulation | [1,41] |
| Burkholderia spp. | btSET methylates H3 | Methylation of H3K4 at rDNA promoter regions | [1,41] |
| Suppression of HDAC1/2 activity | Histone hyperacetylation, suppression of DNMT3B | [1,42] | |
| Mycobacterium tuberculosis | Activation of HDAC1, global deacetylation of H3 | Suppresses IL-12B, IFN-Y | [14,43] |
| Rv1988 methylates H3R42 | Suppresses innate immunity genes | [1,44] | |
| Rv2416c acetylates H3 at IL-10 promoter. | Inhibits Th-1, host immune attenuation, protects m. tuberculosis from autophagy | [1,45] | |
| Rv2699c activates DNA methyltransferase activity at cytosine residues and histones H3, H4 | Non-CpG methylation, global transcription suppression | [1,25] | |
| Rv3423.1 activates histone acetyltransferase | Activation of host anti-inflammatory cascades, protects infection | [1,46] | |
| Streptococcus pneumoniae | Pneumolysin dephosphorylates H3S10 | May impair cell proliferation, tumor suppression | [1,14,47] |
| Pneumolysin activates miRNA-200b | Blocks KALRN, enhances pneumonia pathology | [1,48] | |
| Legionella pneumophila | LegAS4 methylates H3K4 | Increased transcription of rRNA/ribosomal protein | [1,14,49] |
| RomA globally methylates H3K14 | Global transcription suppression | [1,14,50] | |
| Snpl targets DSIF complex | Inhibits RNA polymerase II | [1,51] | |
| AnkH targets LARP7 (snRNP) | Inhibits transcription elongation and splicing | [1,52] | |
| Bacillus anthracis | BaSET directly trimethylates the NFkB gene at H1 lysine | Shuts down NFkB proinflammatory cascade | [1,14,53] |
| LT targets IL-8 promoter | Condensation of chromatin at H3S10ph and H3K14ac | [1,54] | |
| LT targets IL-1B enhancer region (HDAC8) | Deacetylation of H3K27ac | [1,55] | |
| Burkholderia thaliadensis | BtSET methylates H3K4 at NFkB gene | Suppresses NFkB cascade; activates rRNA transcription | [14,41,53] |
| Chlamydia trachomatis | NUE methylation of H2B, H3, H4 | Global transcription suppression | [1,14,22] |
| Chlamydia Pneumoniae | cpnSET methylates H3 | Global transcription modulation/suppression | [1,56] |
| Chlamydia psittaci | SINC targets MAN1, LAMP1 | Modulates chromatin anchoring on inner nuclear membrane. | [1,57] |
| Excess methylation on CpG islands, CDH1 gene | Inactivation of E-cadherin expression | [1,58] | |
| Escherichia coli | Membrane vesicles target H3 (methylation) | Increase in transcription of H3K4me3 genes | [1,59] |
| Moraxella catarrhalis | Phosphorylation of H3S10, acetylation of H3K14 at IL-8 gene | Induces inflammatory response, MAPK, NFkB activation, release of IL-8 | [1,34] |
| Pseudomonas aeruginosa | 2-amnoacetophone induces HDAC1 activity | H3K18 deacetylation, heterochromatin state, reduced expression of cytokines/chemokines | [1,14,60] |
| 2-amnoacetophone indirectly dephosphorylates H3 | Global dephosphorylation of H3S10, transcription modulation | [1,61] | |
| miRNA-93 targets IL-8 transcripts | Suppression of IL-8 response | [1,61] | |
| Neisseria gonorrhea | Gc-HDAC targets HDAC1, deacetylates at H3/Mir-146a | Condensation of chromatin at H2K9ac/suppression of immune response | [1,62] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wrede, D.; Bordak, M.; Abraham, Y.; Mehedi, M. Pulmonary Pathogen-Induced Epigenetic Modifications. Epigenomes 2023, 7, 13. https://doi.org/10.3390/epigenomes7030013
Wrede D, Bordak M, Abraham Y, Mehedi M. Pulmonary Pathogen-Induced Epigenetic Modifications. Epigenomes. 2023; 7(3):13. https://doi.org/10.3390/epigenomes7030013
Chicago/Turabian StyleWrede, Dylan, Mika Bordak, Yeabtsega Abraham, and Masfique Mehedi. 2023. "Pulmonary Pathogen-Induced Epigenetic Modifications" Epigenomes 7, no. 3: 13. https://doi.org/10.3390/epigenomes7030013
APA StyleWrede, D., Bordak, M., Abraham, Y., & Mehedi, M. (2023). Pulmonary Pathogen-Induced Epigenetic Modifications. Epigenomes, 7(3), 13. https://doi.org/10.3390/epigenomes7030013






