DNA Hypomethylation May Contribute to Metabolic Recovery of Frozen Wood Frog Brains
Abstract
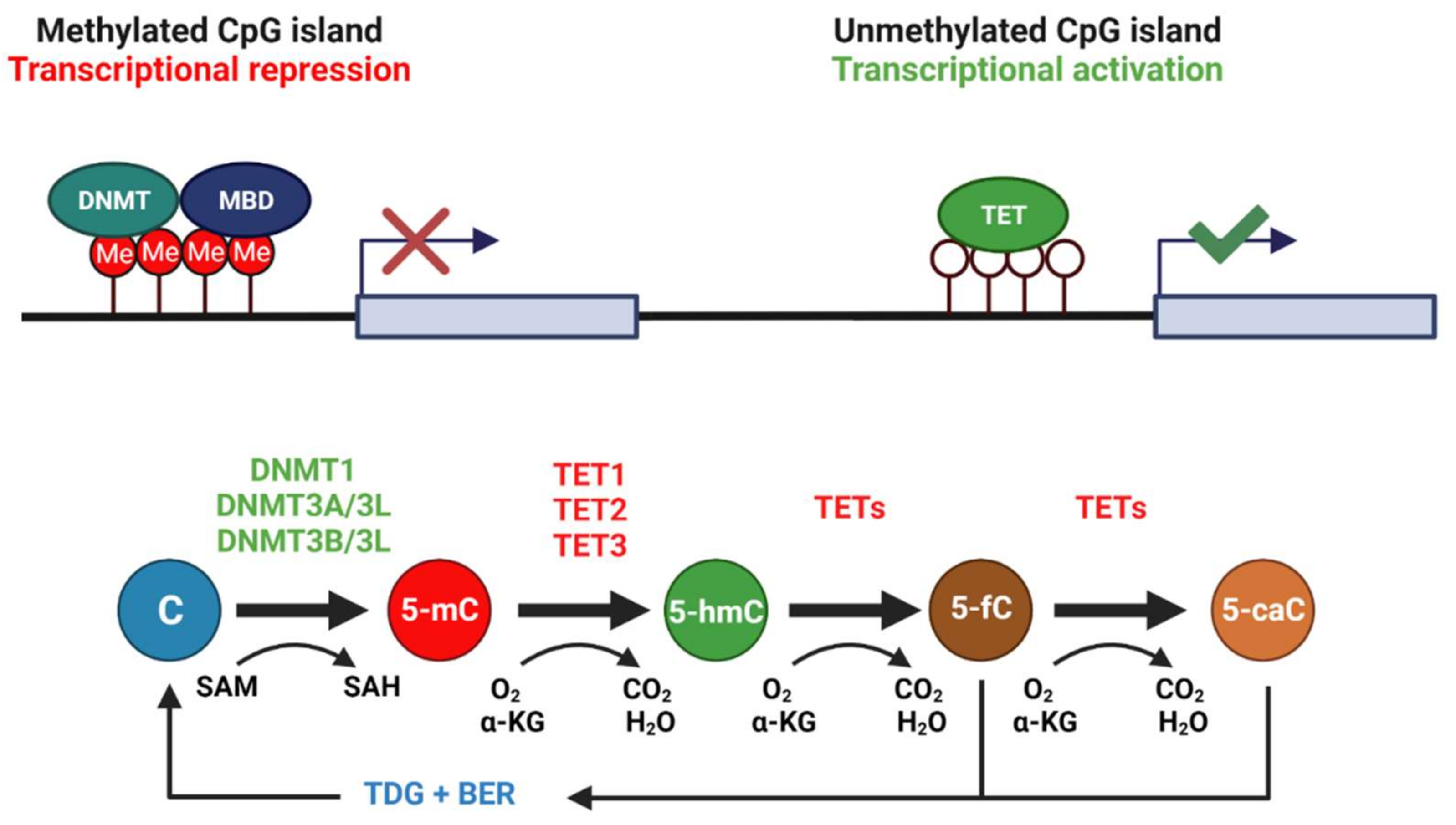
:1. Introduction
2. Results
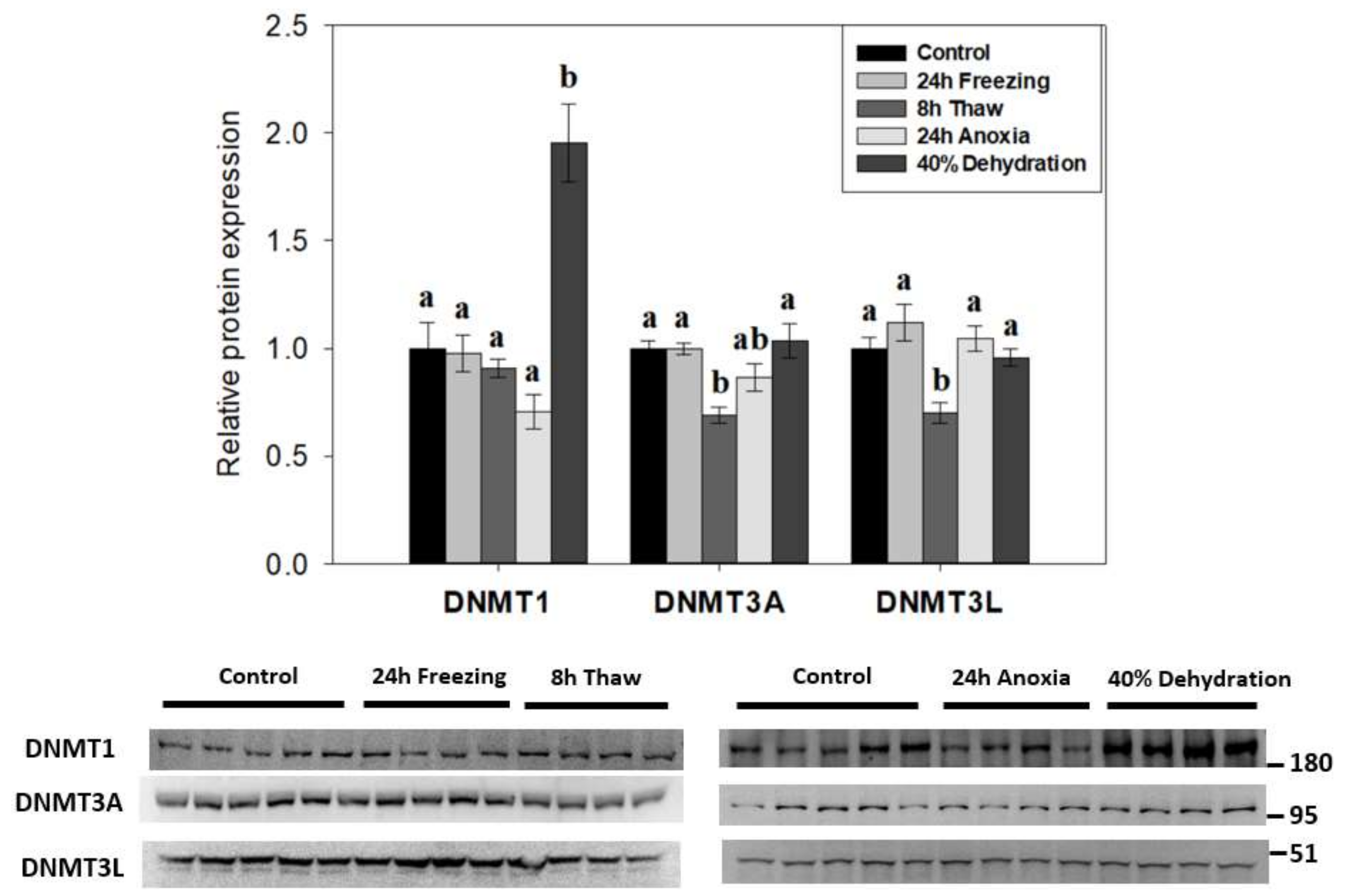
2.1. DNMT Expression Consistent with the Transcriptional State of Cells across the Wood Frog Freeze-Thaw Cycle
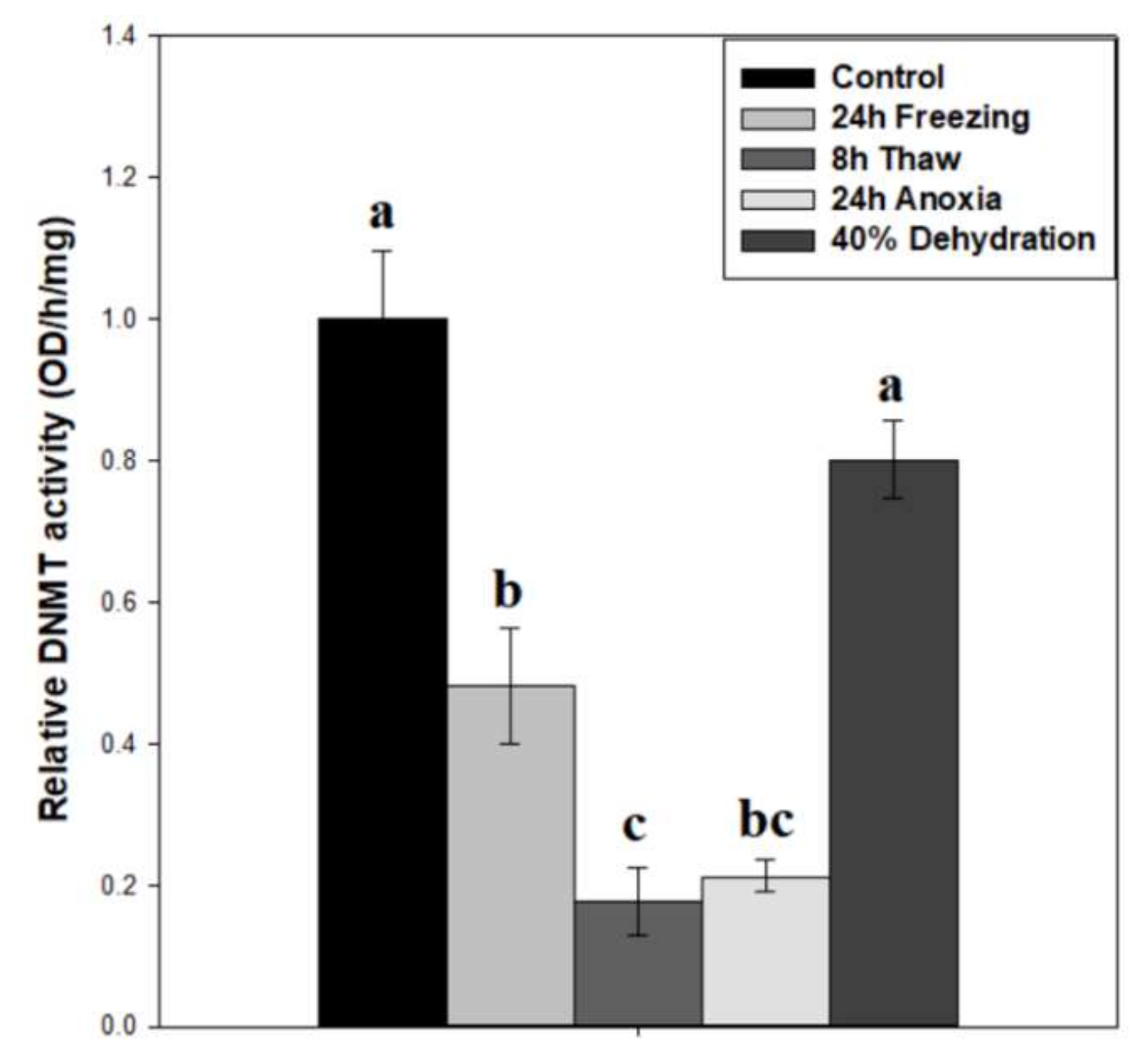
2.2. Noticeable Depression in DNA Methyltransferase Activity during Freezing, Freeze Recovery, and Anoxia
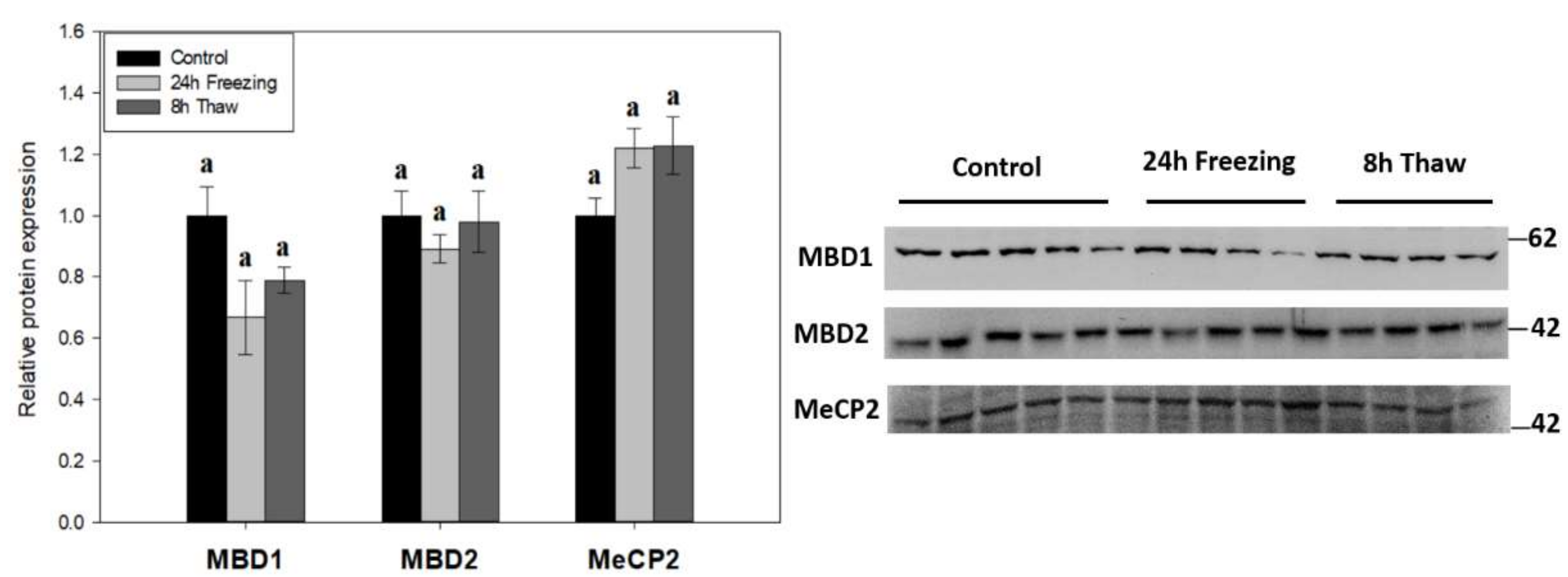
2.3. Methyl-CpG Binding Protein Expression Unchanged across the Freeze-Thaw Cycle
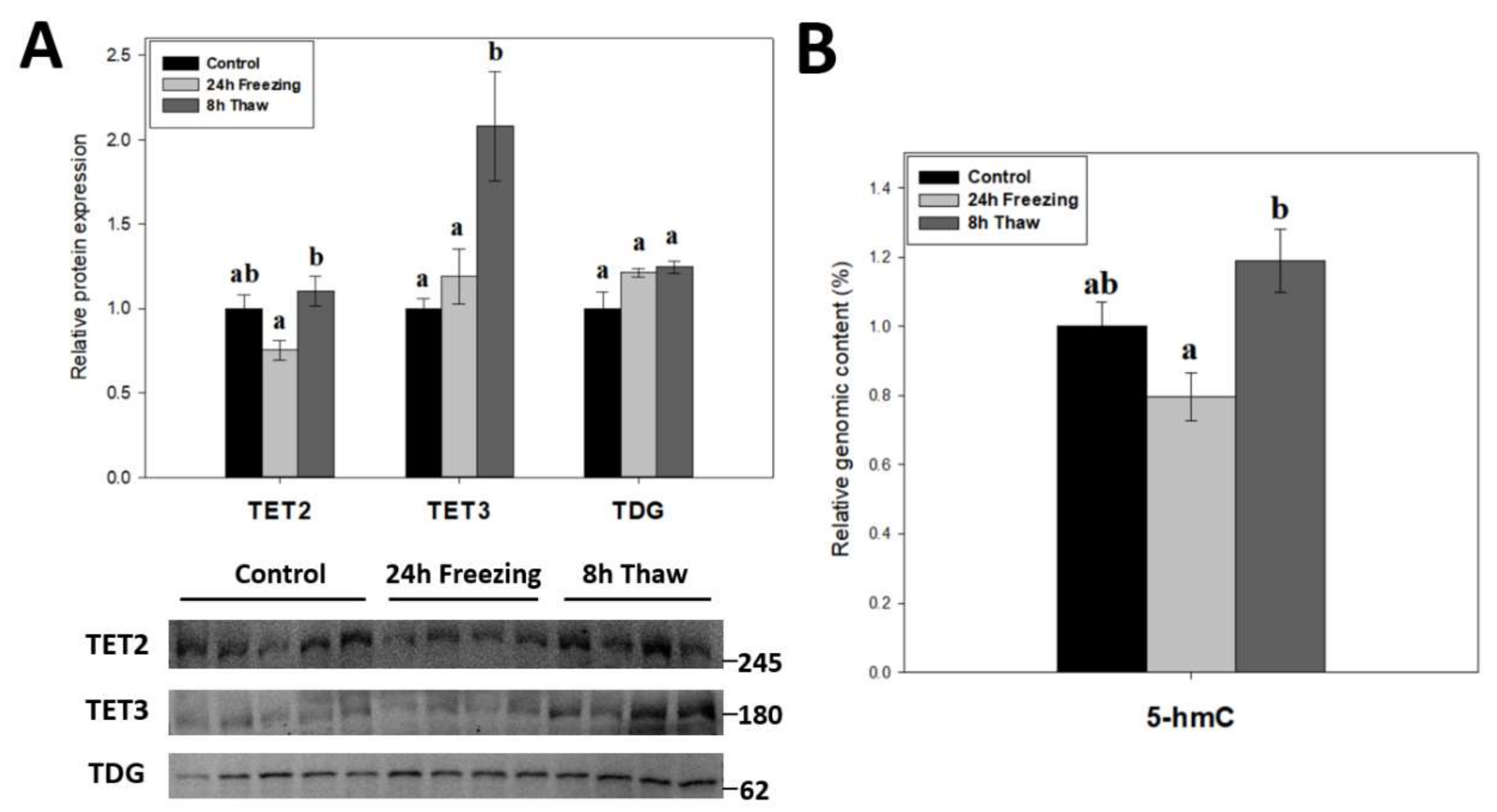
2.4. Multi-Faceted Support for Increased DNA Demethylation during Freeze Recovery
3. Discussion
3.1. Diverse Roles for DNA Methyltransferases in the Brain during Wood Frog Freeze Tolerance
3.2. Thaw-Mediated Hyperactive DNA Demethylation Relative to Freezing
4. Materials and Methods
4.1. Animal Experiments
4.2. Total Soluble Protein Isolation
4.3. Nuclear Protein Extraction
4.4. Immunoblotting
4.5. Quantification and Statistics
4.6. DNMT Activity ELISA
4.7. Genomic DNA Extraction
4.8. 5-Hydroxymethylcytosine ELISA
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, J.; Hawkins, L.J.; Storey, K.B. DNA Methylation and Regulation of DNA Methyltransferases in a Freeze-Tolerant Vertebrate. Biochem. Cell Biol. 2020, 98, 145–153. [Google Scholar] [CrossRef]
- Hawkins, L.J.; Storey, K.B. Histone Methylation in the Freeze-Tolerant Wood Frog (Rana Sylvatica). J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2018, 188, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Wijenayake, S.; Storey, K.B. The Role of DNA Methylation during Anoxia Tolerance in a Freshwater Turtle (Trachemys Scripta Elegans). J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2016, 186, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Wijenayake, S.; Hawkins, L.J.; Storey, K.B. Dynamic Regulation of Six Histone H3 Lysine (K) Methyltransferases in Response to Prolonged Anoxia Exposure in a Freshwater Turtle. Gene 2018, 649, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Morin, P.; Storey, K.B. Evidence for a Reduced Transcriptional State during Hibernation in Ground Squirrels. Cryobiology 2006, 53, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Watts, A.J.; Storey, K.B. Hibernation Impacts Lysine Methylation Dynamics in the 13-Lined Ground Squirrel, Ictidomys Tridecemlineatus. J. Exp. Zool. Part A Ecol. Integr. Physiol. 2019, 331, 234–244. [Google Scholar] [CrossRef]
- Biggar, Y.; Storey, K.B. Global DNA Modifications Suppress Transcription in Brown Adipose Tissue during Hibernation. Cryobiology 2014, 69, 333–338. [Google Scholar] [CrossRef]
- Tessier, S.N.; Ingelson-Filpula, W.A.; Storey, K.B. Epigenetic Regulation by DNA Methyltransferases during Torpor in the Thirteen-Lined Ground Squirrel Ictidomys Tridecemlineatus. Mol. Cell. Biochem. 2021, 476, 3975–3985. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, M.; Storey, K.B.; Sun, L.; Yang, H. DNA Methylation Levels Analysis in Four Tissues of Sea Cucumber Apostichopus Japonicus Based on Fluorescence-Labeled Methylation-Sensitive Amplified Polymorphism (F-MSAP) during Aestivation. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2015, 181, 26–32. [Google Scholar] [CrossRef]
- Rolfe, D.F.S.; Brown, G.C. Cellular Energy Utilization and Molecular Origin of Standard Metabolic Rate in Mammals. Physiol. Rev. 1997, 77, 731–758. [Google Scholar] [CrossRef] [Green Version]
- Van Breukelen, F.; Martin, S.L. Reversible Depression of Transcription during Hibernation. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2002, 172, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Bocharova, L.S.; Gordon, R.Y.; Arkhipov, V.I. Uridine Uptake and RNA Synthesis in the Brain of Torpid and Awakened Ground Squirrels. Comp. Biochem. Physiol. B 1992, 101, 189–192. [Google Scholar] [CrossRef]
- Bogdanović, O.; Veenstra, G.J.C. DNA Methylation and Methyl-CpG Binding Proteins: Developmental Requirements and Function. Chromosoma 2009, 118, 549–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nan, X.; Ng, H.H.; Johnson, C.A.; Laherty, C.D.; Turner, B.M.; Eisenman, R.N.; Bird, A. Transcriptional Repression by the Methyl-CpG-Binding Protein MeCP2 Involves a Histone Deacetylase Complex. Nature 1998, 393, 386–389. [Google Scholar] [CrossRef]
- Lyko, F. The DNA Methyltransferase Family: A Versatile Toolkit for Epigenetic Regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [CrossRef]
- Feng, J.; Zhou, Y.; Campbell, S.L.; Le, T.; Li, E.; Sweatt, J.D.; Silva, A.J.; Fan, G. Dnmt1 and Dnmt3a Maintain DNA Methylation and Regulate Synaptic Function in Adult Forebrain Neurons. Nat. Neurosci. 2010, 13, 423–430. [Google Scholar] [CrossRef]
- Dodge, J.E.; Okano, M.; Dick, F.; Tsujimoto, N.; Chen, T.; Wang, S.; Ueda, Y.; Dyson, N.; Li, E. Inactivation of Dnmt3b in Mouse Embryonic Fibroblasts Results in DNA Hypomethylation, Chromosomal Instability, and Spontaneous Immortalization. J. Biol. Chem. 2005, 280, 17986–17991. [Google Scholar] [CrossRef] [Green Version]
- Jair, K.W.; Bachman, K.E.; Suzuki, H.; Ting, A.H.; Rhee, I.; Yen, R.W.C.; Baylin, S.B.; Schuebel, K.E. De Novo CpG Island Methylation in Human Cancer Cells. Cancer Res. 2006, 66, 682–692. [Google Scholar] [CrossRef] [Green Version]
- Chédin, F.; Lieber, M.R.; Hsieh, C.L. The DNA Methyltransferase-like Protein DNMT3L Stimulates de Novo Methylation by Dnmt3a. Proc. Natl. Acad. Sci. USA 2002, 99, 16916–16921. [Google Scholar] [CrossRef] [Green Version]
- Suetake, I.; Shinozaki, F.; Miyagawa, J.; Takeshima, H.; Tajima, S. DNMT3L Stimulates the DNA Methylation Activity of Dnmt3a and Dnmt3b through a Direct Interaction. J. Biol. Chem. 2004, 279, 27816–27823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, L.; Wu, H.; Diep, D.; Yamaguchi, S.; D’Alessio, A.C.; Fung, H.L.; Zhang, K.; Zhang, Y. Genome-Wide Analysis Reveals TET- and TDG-Dependent 5-Methylcytosine Oxidation Dynamics. Cell 2013, 153, 692–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Storey, K.B.; Storey, J.M. Molecular Physiology of Freeze Tolerance in Vertebrates. Physiol. Rev. 2017, 97, 623–665. [Google Scholar] [CrossRef] [Green Version]
- Dieni, C.A.; Storey, K.B. Regulation of Hexokinase by Reversible Phosphorylation in Skeletal Muscle of a Freeze-Tolerant Frog. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2011, 159, 236–243. [Google Scholar] [CrossRef]
- Dieni, C.A.; Storey, K.B. Creatine Kinase Regulation by Reversible Phosphorylation in Frog Muscle. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2009, 152, 405–412. [Google Scholar] [CrossRef]
- Dieni, C.A.; Storey, K.B. Regulation of Glucose-6-Phosphate Dehydrogenase by Reversible Phosphorylation in Liver of a Freeze Tolerant Frog. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2010, 180, 1133–1142. [Google Scholar] [CrossRef]
- Dawson, N.J.; Storey, K.B. A Hydrogen Peroxide Safety Valve: The Reversible Phosphorylation of Catalase from the Freeze-Tolerant North American Wood Frog, Rana Sylvatica. Biochim. Biophys. Acta Gen. Subj. 2016, 1860, 476–485. [Google Scholar] [CrossRef]
- Biggar, K.K.; Storey, K.B. Functional Impact of MicroRNA Regulation in Models of Extreme Stress Adaptation. J. Mol. Cell Biol. 2018, 10, 93–101. [Google Scholar] [CrossRef] [Green Version]
- Biggar, K.K.; Storey, K.B. Insight into Post-Transcriptional Gene Regulation: Stress-Responsive MicroRNAs and Their Role in the Environmental Stress Survival of Tolerant Animals. J. Exp. Biol. 2015, 218, 1281–1289. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Jurkowska, R.; Soeroes, S.; Rajavelu, A.; Dhayalan, A.; Bock, I.; Rathert, P.; Brandt, O.; Reinhardt, R.; Fischle, W.; et al. Chromatin Methylation Activity of Dnmt3a and Dnmt3a/3L Is Guided by Interaction of the ADD Domain with the Histone H3 Tail. Nucleic Acids Res. 2010, 38, 4246–4253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hermann, A.; Goyal, R.; Jeltsch, A. The Dnmt1 DNA-(Cytosine-C5)-Methyltransferase Methylates DNA Processively with High Preference for Hemimethylated Target Sites. J. Biol. Chem. 2004, 279, 48350–48359. [Google Scholar] [CrossRef] [Green Version]
- Jeltsch, A.; Jurkowska, R.Z. New Concepts in DNA Methylation. Trends Biochem. Sci. 2014, 39. [Google Scholar] [CrossRef] [PubMed]
- Laget, S.; Miotto, B.; Chin, H.G.; Estève, P.O.; Roberts, R.J.; Pradhan, S.; Defossez, P.A. MBD4 Cooperates with DNMT1 to Mediate Methyl-DNA Repression and Protects Mammalian Cells from Oxidative Stress. Epigenetics 2014, 9, 546–556. [Google Scholar] [CrossRef] [PubMed]
- Lavoie, G.; Estève, P.O.; Laulan, N.B.; Pradhan, S.; St-Pierre, Y. PKC Isoforms Interact with and Phosphorylate DNMT1. BMC Biol. 2011, 9, 31. [Google Scholar] [CrossRef] [Green Version]
- Holden, C.P.; Storey, K.B. Signal Transduction, Second Messenger, and Protein Kinase Responses during Freezing Exposures in Wood Frogs. Am. J. Physiol. Regul. Integr. Comp. Physiol. 1996, 271. [Google Scholar] [CrossRef]
- Deplus, R.; Blanchon, L.; Rajavelu, A.; Boukaba, A.; Defrance, M.; Luciani, J.; Rothé, F.; Dedeurwaerder, S.; Denis, H.; Brinkman, A.B.; et al. Regulation of DNA Methylation Patterns by CK2-Mediated Phosphorylation of Dnmt3a. Cell Rep. 2014, 8, 743–753. [Google Scholar] [CrossRef]
- Litchfield, D.W. Protein Kinase CK2: Structure, Regulation and Role in Cellular Decisions of Life and Death. Biochem. J. 2003, 369, 1–15. [Google Scholar] [CrossRef]
- Storey, K.B. Regulation of Hypometabolism: Insights into Epigenetic Controls. J. Exp. Biol. 2015, 218, 150–159. [Google Scholar] [CrossRef] [Green Version]
- Burr, S.; Caldwell, A.; Chong, M.; Beretta, M.; Metcalf, S.; Hancock, M.; Arno, M.; Balu, S.; Kropf, V.L.; Mistry, R.K.; et al. Oxygen Gradients Can Determine Epigenetic Asymmetry and Cellular Differentiation via Differential Regulation of Tet Activity in Embryonic Stem Cells. Nucleic Acids Res. 2018, 46, 1210. [Google Scholar] [CrossRef] [Green Version]
- Morin, P.J.; McMullen, D.C.; Storey, K.B. HIF-1 α Involvement in Low Temperature and Anoxia Survival by a Freeze Tolerant Insect. Mol. Cell. Biochem. 2005, 280, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.P.; Miles, S.L. Silencing HIF-1α Induces TET2 Expression and Augments Ascorbic Acid Induced 5-Hydroxymethylation of DNA in Human Metastatic Melanoma Cells. Biochem. Biophys. Res. Commun. 2017, 490, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Zhang, Y.; Hart, R.P.; Chen, J.; Herrup, K.; Li, J. Alteration in 5-Hydroxymethylcytosine-Mediated Epigenetic Regulation Leads to Purkinje Cell Vulnerability in ATM Deficiency. Brain 2015, 138, 3520–3536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, D.; Wei, S.; Chen, F.; Zhang, Y.; Li, J. TET3-mediated DNA Oxidation Promotes ATR-dependent DNA Damage Response. EMBO Rep. 2017, 18, 781–796. [Google Scholar] [CrossRef] [Green Version]
- Lung, Z.D.; Storey, K.B. DNA Damage and Repair Responses to Freezing and Anoxia Stresses in Wood Frogs, Rana Sylvatica. J. Therm. Biol. 2022, 107, 103274. [Google Scholar] [CrossRef]
- Lv, X.; Jiang, H.; Liu, Y.; Lei, X.; Jiao, J. Micro RNA -15b Promotes Neurogenesis and Inhibits Neural Progenitor Proliferation by Directly Repressing TET 3 during Early Neocortical Development. EMBO Rep. 2014, 15, 1305–1314. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Storey, K.B. Cell Cycle Regulation in the Freeze-Tolerant Wood Frog, Rana Sylvatica. Cell Cycle 2012, 11, 1727–1742. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.; Guo, S.; Chen, S.; Mastriano, S.J.; Liu, C.; D’Alessio, A.C.; Hysolli, E.; Guo, Y.; Yao, H.; Megyola, C.M.; et al. An Extensive Network of TET2-Targeting MicroRNAs Regulates Malignant Hematopoiesis. Cell Rep. 2013, 5, 471–481. [Google Scholar] [CrossRef] [Green Version]
- Song, S.J.; Ito, K.; Ala, U.; Kats, L.; Webster, K.; Sun, S.M.; Jongen-Lavrencic, M.; Manova-Todorova, K.; Teruya-Feldstein, J.; Avigan, D.E.; et al. The Oncogenic MicroRNA MiR-22 Targets the TET2 Tumor Suppressor to Promote Hematopoietic Stem Cell Self-Renewal and Transformation. Cell Stem Cell 2013, 13, 87–101. [Google Scholar] [CrossRef] [Green Version]
- Hadj-Moussa, H.; Storey, K.B. Micromanaging Freeze Tolerance: The Biogenesis and Regulation of Neuroprotective MicroRNAs in Frozen Brains. Cell. Mol. Life Sci. 2018, 75, 3635–3647. [Google Scholar] [CrossRef]
- Gerber, V.E.M.; Wijenayake, S.; Storey, K.B. Anti-Apoptotic Response during Anoxia and Recovery in a Freeze-Tolerant Wood Frog (Rana Sylvatica). PeerJ 2016, 2016, e1834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Churchill, T.A.; Storey, K.B. Metabolic Responses to Dehydration by Liver of the Wood Frog, Rana Sylvatica. Can. J. Zool. 1994, 72, 1420–1425. [Google Scholar] [CrossRef] [Green Version]
| Target | Company | Catalogue # |
|---|---|---|
| DNMT1 | Abclonal | #A16729 |
| DNMT3A | Genetex | #GTX128157 |
| DNMT3B * | Genetex | #GTX129127 |
| DNMT3L | Abgent | #AP1040a |
| MBD1 | Active Motif | #39858 |
| MBD2 | Active Motif | #39548 |
| MeCP2 | Active Motif | #39189 |
| TET1 * | Genetex | #GTX124207 |
| TET2 | Abclonal | #A5682 |
| TET3 | Genetex | #GTX121453 |
| TDG | Genetex | #GTX110473 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bloskie, T.; Storey, K.B. DNA Hypomethylation May Contribute to Metabolic Recovery of Frozen Wood Frog Brains. Epigenomes 2022, 6, 17. https://doi.org/10.3390/epigenomes6030017
Bloskie T, Storey KB. DNA Hypomethylation May Contribute to Metabolic Recovery of Frozen Wood Frog Brains. Epigenomes. 2022; 6(3):17. https://doi.org/10.3390/epigenomes6030017
Chicago/Turabian StyleBloskie, Tighe, and Kenneth B. Storey. 2022. "DNA Hypomethylation May Contribute to Metabolic Recovery of Frozen Wood Frog Brains" Epigenomes 6, no. 3: 17. https://doi.org/10.3390/epigenomes6030017
APA StyleBloskie, T., & Storey, K. B. (2022). DNA Hypomethylation May Contribute to Metabolic Recovery of Frozen Wood Frog Brains. Epigenomes, 6(3), 17. https://doi.org/10.3390/epigenomes6030017







