Epigenomics of Plant Responses to Environmental Stress
Abstract
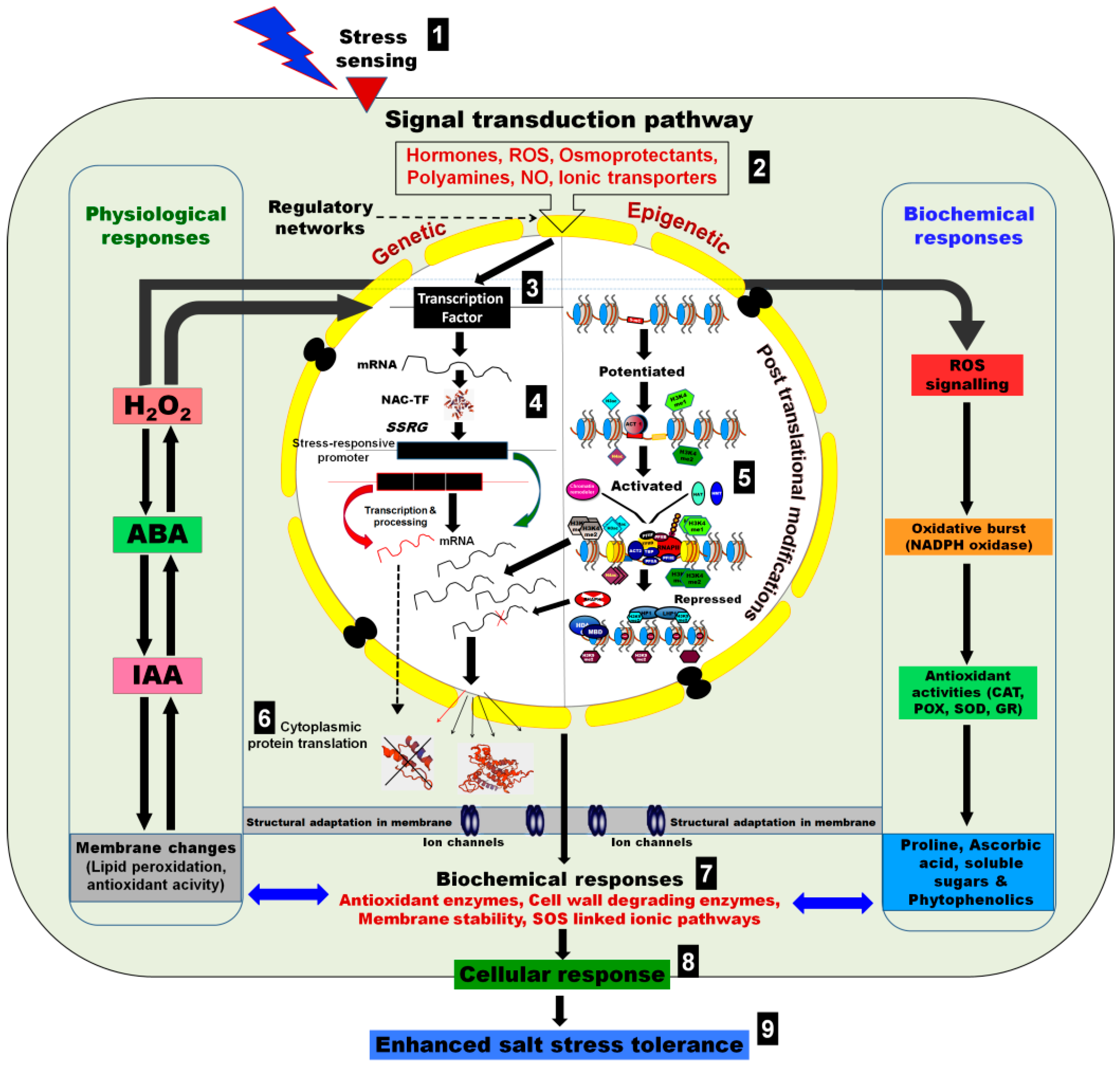
1. Introduction
2. Epigenetics of DNA Base Modification
3. Epigenetic Regulation of Chromatin Structure
4. Regulation of Gene Expression and Genome Stability
5. Salt-Induced Epigenetic Changes in Crop Plants
6. Transgenerational Inheritance of Epimarks
7. Future Perspectives of Epigenomic Studies
Author Contributions
Conflicts of Interest
References
- Cantara, W.A.; Crain, P.F.; Rozenski, J.; McCloskey, J.A.; Harris, K.A.; Zhang, X.; Vendeix, F.A.P.; Fabris, D.; Agris, P.F. The RNA modification database, RNAMDB. Nucleic Acids Res. 2011, 39, D195–D201. [Google Scholar] [CrossRef] [PubMed]
- Squires, J.E.; Patel, H.R.; Nousch, M.; Sibbritt, T.; Humphreys, D.T.; Parker, B.J.; Suter, C.M.; Preiss, T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012, 40, 5023–5033. [Google Scholar] [CrossRef] [PubMed]
- Agris, P.F. Bringing order to translation: The contributions of transfer RNA anticodon-domain modifications. EMBO Rep. 2008, 9, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Motorin, Y.; Helm, M. tRNA stabilization by modified nucleotides. Biochemistry 2010, 49, 4934–4944. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.; Pollex, T.; Hanna, K.; Tuorto, F.; Meusburger, M.; Helm, M.; Lyko, F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010, 24, 1590–1595. [Google Scholar] [CrossRef] [PubMed]
- Laland, K.; Uller, T.; Feldman, M.; Sterelny, K.; Müller, G.B.; Mozcek, A.; Jablonka, E.; Odling-Smee, J.; Wray, G.A.; Hoekstra, H.E.; et al. Does evolutionary theory need a rethink? Nature 2014, 514, 161–164. [Google Scholar] [CrossRef] [PubMed]
- Ho, W.C.; Zhang, J. The genotype-phenotype map of yeast complex traits: basic parameters and the role of natural selection. Mol. Biol. Evol. 2014, 31, 1568–1580. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S. Epigenetic control of apomixis: A new perspective of an old enigma. Adv. Plants Agric. Res. 2017, 7, 00243. [Google Scholar] [CrossRef]
- Skinner, M.K. Environmental epigenetics and a unified theory of the molecular aspects of evolution: A neo-Lamarckian concept that facilitates neo-Darwinian evolution. Genome Biol. Evol. 2015, 7, 1296–1302. [Google Scholar] [CrossRef] [PubMed]
- Farias, N.; Ho, N.; Butler, S.; Delaney, L.; Morrison, J.; Shahrzad, S.; Coomber, B.L. The effects of folic acid on global DNA methylation and colonosphere formation in colon cancer cell lines. J. Nutr. Biochem. 2015, 26, 818–826. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Oliver, V.F.; Wang, G.; Zhu, H.; Zack, D.J.; Merbs, S.L.; Qian, J. Characterization of tissue-specific differential DNA methylation suggests distinct modes of positive and negative gene expression regulation. BMC Genom. 2015, 16, 49. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Zhang, J.; Guo, Z.; Ma, Q.; Xu, Z.; Zhou, Y.; Xu, Z.; Li, Z.; Liu, Y.; Yuan, B.; et al. Loss of 5-hydroxymethylcytosine is linked to gene body hypermethylation in kidney cancer. Cell Res. 2016, 26, 103–118. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Morales, R.; Agís-Balboa, R.C.; Esteller, M.; Berdasco, M. Epigenetic mechanisms during ageing and neurogenesis as novel therapeutic avenues in human brain disorders. Clin. Epigenet. 2017, 9, 67. [Google Scholar] [CrossRef] [PubMed]
- Behera, P. Epigenetics changes in breast cancer: Current aspects in India. J. Bioeng. Biomed. Sci. 2017, 7, 223. [Google Scholar] [CrossRef]
- Kumar, S.; Beena, A.S.; Awana, M.; Singh, A. Salt-induced tissue-specific cytosine methylation downregulates expression of HKT genes in contrasting wheat (Triticum aestivum L.) genotypes. DNA Cell Biol. 2017, 36, 283–394. [Google Scholar] [CrossRef] [PubMed]
- Eichten, S.R.; Schmitz, R.J.; Springer, N.M. Epigenetics: Beyond chromatin modifications and complex genetic regulation. Plant Physiol. 2014, 165, 933–947. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Singh, A. Epigenetic regulation of abiotic stress tolerance in plants. Adv. Plants Agric. Res. 2016, 5, 00179. [Google Scholar] [CrossRef]
- Avramova, Z. Transcriptional “memory” of a stress: Transient chromatin and memory (epigenetic) marks at stress-response genes. Plant J. 2015, 83, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Lauria, M.; Rossi, V. Epigenetic control of gene regulation in plants. Biochim. Biophys. Acta 2011, 1809, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Pikaard, C.S.; MittelstenScheid, O. Epigenetic regulation in plants. Cold Spring Harb. Perspect. Biol. 2014, 6, a019315. [Google Scholar] [CrossRef] [PubMed]
- Gallusci, P.; Hodgman, C.; Teyssier, E.; Seymour, G.B. DNA methylation and chromatin regulation during fleshy fruit development and ripening. Front. Plant Sci. 2016, 7, 807. [Google Scholar] [CrossRef] [PubMed]
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Lanciano, S.; Mirouze, M. DNA methylation in rice and relevance for breeding. Epigenomes 2017, 1, 10. [Google Scholar] [CrossRef]
- Li, Y.; Kumar, S.; Qian, W. Active DNA demethylation: Mechanism and role in plant development. Plant Cell Rep. 2017. [Google Scholar] [CrossRef] [PubMed]
- Springer, N.M.; Schmitz, R.J. Exploiting induced and natural epigenetic variation for crop improvement. Nat. Rev. Genet. 2017. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Singh, A.K.; Mohapatra, T. Epigenetics: History, present status and future perspective. Indian J. Genet. Plant Breed. 2017, 77, 445–463. [Google Scholar]
- Breiling, A.; Lyko, F. Epigenetic regulatory functions of DNA modifications: 5-methylcytosine and beyond. Epigenet. Chromatin 2015, 8, 24. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Lister, R.; O’Malley, R.C.; Tonti-Filippini, J.; Gregory, B.D.; Berry, C.C.; Millar, A.H.; Ecker, J.R. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 2008, 133, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Q.; Yuan, W.; Cao, Z.; Qi, B.; Kumar, S.; Li, Y.; Qian, W. The cytosolic Fe-S cluster assembly component MET18 is required for the full enzymatic activity of ROS1 in active DNA demethylation. Sci. Rep. 2016, 6, 26443. [Google Scholar] [CrossRef] [PubMed]
- Slotkin, R.K.; Vaughn, M.; Borges, F.; Tanurdzic, M.; Becker, J.D.; Feijo, J.A.; Martienssen, R.A. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 2009, 136, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.-K. Active DNA demethylation in plants and animals. Cold Spring Harb. Symp. Quant. Biol. 2012, 77, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Penterman, J.; Zilberman, D.; Huh, J.H.; Ballinger, T.; Henikoff, S.; Fischer, R.L. DNA demethylation in the Arabidopsis genome. Proc. Natl. Acad. Sci. USA 2007, 104, 6752–6757. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 2009, 43, 143–166. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.F.; Ibarra, C.A.; Silva, P.; Zemach, A.; Eshed-Williams, L.; Fischer, R.L.; Zilberman, D. Genome-wide demethylation of Arabidopsis endosperm. Science 2009, 324, 1451–1454. [Google Scholar] [CrossRef] [PubMed]
- Steward, N.; Ito, M.; Yamachuchi, Y.; Koizumi, N.; Sano, H. Periodic DNA methylation in maize nucleosomes and demethylation by environmental stress. J. Biol. Chem. 2002, 277, 37741–37746. [Google Scholar] [CrossRef] [PubMed]
- Kou, H.P.; Li, Y.; Song, X.X.; Ou, X.F.; Xing, S.C.; Ma, J.; Von Wettstein, D.; Liu, B. Heritable alteration in DNA methylation induced by nitrogen-deficiency stress accompanies enhanced tolerance by progenies to the stress in rice (Oryza sativa L.). J. Plant Physiol. 2011, 168, 1685–1693. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Beena, A.S.; Awana, M.; Singh, A. Physiological, biochemical, epigenetic and molecular analyses of wheat (Triticum aestivum) genotypes with contrasting salt tolerance. Front. Plant Sci. 2017, 8, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Jullien, P.E.; Mosquna, A.; Ingouff, M.; Sakata, T.; Ohad, N.; Berger, F. Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis. PLoS Biol. 2008, 6, e194. [Google Scholar] [CrossRef] [PubMed]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; D’Alessio, A.C.; Taranova, O.V.; Hong, K.; Sowers, L.C.; Zhang, Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010, 466, 1129–1133. [Google Scholar] [CrossRef] [PubMed]
- Penn, N.W.; Suwalski, R.; O’Riley, C.; Bojanowski, K.; Yura, R. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem. J. 1972, 126, 781–790. [Google Scholar] [CrossRef] [PubMed]
- Kriaucionis, S.; Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 2009, 324, 929–930. [Google Scholar] [CrossRef] [PubMed]
- Globisch, D.; Munzel, M.; Muller, M.; Michalakis, S.; Wagner, M.; Koch, S.; Brückl, T.; Biel, M.; Carell, T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS ONE 2010, 5, e15367. [Google Scholar] [CrossRef] [PubMed]
- He, Y.F.; Li, B.Z.; Li, Z.; Liu, P.; Wang, Y.; Tang, Q.; Ding, J.; Jia, Y.; Chen, Z.; Li, L.; et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 2011, 333, 1303–1307. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, R.M.; Souza, A.L.; Clish, C.B.; Gehring, M. 5-Hydroxymethylcytosine is not present in appreciable quantities in Arabidopsis DNA. Genes Genome Genet. 2015, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Casadesus, J.; Low, D. Epigenetic gene regulation in the bacterial world. Microbiol. Mol. Biol. Rev. 2006, 70, 830–856. [Google Scholar] [CrossRef] [PubMed]
- Ratel, D.; Ravanat, J.L.; Berger, F.; Wion, D. N6-methyladenine: the other methylated base of DNA. BioEssays 2006, 28, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Blanco, M.A.; Gu, L.; Sendinc, E.; Liu, J.; Aristizabal-Corrales, D.; Hsu, C.H.; Aravind, L.; He, C.; Shi, Y. DNA Methylation on N-adenine in C. elegans. Cell 2015, 161, 868–878. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Huang, H.; Liu, D.; Cheng, Y.; Liu, X.; Zhang, W.; Yin, R.; Zhang, D.; Zhang, P.; Liu, J.; et al. N6-methyladenine DNA modification in Drosophila. Cell 2015, 161, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Luo, G.Z.; Chen, K.; Deng, X.; Yu, M.; Han, D.; Hao, Z.; Liu, J.; Lu, X.; Dore, L.C.; et al. N6-Methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell 2015, 161, 879–892. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Xiong, J.; Yang, Y.; Liu, S.M.; Yuan, B.F.; Feng, Y.Q. Determination of DNA adenine methylation in genomes of mammals and plants by liquid chromatography/mass spectrometry. R. Soc. Chem. Adv. 2015, 5, 64046–64054. [Google Scholar] [CrossRef]
- Bartholomew, B. Regulating the chromatin landscape: Structural and mechanistic perspectives. Ann. Rev. Biochem. 2014, 83, 671–696. [Google Scholar] [CrossRef] [PubMed]
- Spruijt, C.G.; Vermeulen, M. DNA methylation: Old dog, new tricks? Nat. Struct. Mol. Biol. 2014, 21, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Baubec, T.; Colombo, D.F.; Wirbelauer, C.; Schmidt, J.; Burger, L.; Krebs, A.R.; Akalin, A.; Schübeler, D. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 2015, 520, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Bhaumik, S.R.; Smith, E.; Shilatifard, A. Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 2007, 14, 1008–1016. [Google Scholar] [CrossRef] [PubMed]
- Rose, N.; Klose, R.J. Understanding the relationship between DNA methylation and histone lysine methylation. Biochim. Biophys. Acta 2014, 1839, 1362–1372. [Google Scholar] [CrossRef] [PubMed]
- To, T.K.; Kim, J.-M.; Matsui, A.; Kurihara, Y.; Morosawa, T.; Ishida, J.; Tanaka, M.; Endo, T.; Kakutani, T.; Toyoda, T.; et al. Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet. 2011, 7, e1002055. [Google Scholar] [CrossRef] [PubMed]
- Shahbazian, M.D.; Grunstein, M. Functions of site-specific histone acetylation and deacetylation. Ann. Rev. Biochem. 2007, 76, 75–100. [Google Scholar] [CrossRef] [PubMed]
- Saksouk, N.; Simboeck, E.; Déjardin, J. Constitutive heterochromatin formation and transcription in mammals. Epigenet. Chromatin 2015, 8, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Sarnowska, E.; Gratkowska, D.M.; Sacharowski, S.P.; Cwiek, P.; Tohge, T.; Fernie, A.R.; Siedlecki, J.A.; Koncz, C.; Sarnowski, T.J. The role of SWI/SNF chromatin remodeling complexes in hormone crosstalk. Trends Plant Sci. 2016, 21, 594–608. [Google Scholar] [CrossRef] [PubMed]
- Han, S.K.; Sang, Y.; Rodrigues, A.; Wu, M.F.; Rodriguez, P.L.; Wagner, D.; Biol, F. The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell 2012, 24, 4892–48906. [Google Scholar] [CrossRef] [PubMed]
- Benayoun, B.A.; Pollina, E.A.; Brunet, A. Epigenetic regulation of ageing: Linking environmental inputs to genomic stability. Nat. Rev. Mol. Cell Biol. 2015, 16, 593–610. [Google Scholar] [CrossRef] [PubMed]
- Seong, K.H.; Li, D.; Shimizu, H.; Nakamura, R.; Ishii, S. Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell 2011, 145, 1049–1061. [Google Scholar] [CrossRef] [PubMed]
- Zemach, A.; Kim, M.Y.; Silva, P.; Rodrigues, J.A.; Dotson, B.; Brooks, M.D.; Zilberman, D. Local DNA hypomethylation activates genes in rice endosperm. Proc. Natl. Acad. Sci. USA 2010, 107, 18729–18734. [Google Scholar] [CrossRef] [PubMed]
- Zilberman, D.; Gehring, M.; Tran, R.K.; Ballinger, T.; Henikoff, S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2007, 39, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Earley, K.; Lawrence, R.J.; Pontes, O.; Reuther, R.; Enciso, A.J.; Silva, M.; Neves, N.; Gross, M.; Viegas, W.; Pikaard, C.S. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 2006, 20, 1283–1293. [Google Scholar] [CrossRef] [PubMed]
- Rossi, V.; Locatelli, S.; Varotto, S.; Donn, G.; Pirona, R.; Henderson, D.A.; Hartings, H.; Motto, M. Maize histone deacetylase hda101 is involved in plant development, gene transcription, and sequence-specific modulation of histone modification of genes and repeats. Plant Cell 2007, 19, 1145–1162. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.K. Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 2009, 12, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Millar, C.B.; Grunstein, M. Genome-wide patterns of histone modifications in yeast. Nat. Rev. Mol. Cell Biol. 2006, 7, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Shlyueva, D.; Stampfel, G.; Stark, A. Transcriptional enhancers: From properties to genome-wide predictions. Nat. Rev. Genet. 2014, 15, 272–286. [Google Scholar] [CrossRef] [PubMed]
- Sims, R.J., III; Nishioka, K.; Reinberg, D. Histone lysine methylation: A signature for chromatin function. Trends Genet. 2003, 19, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, K.; Ding, Y.; Malkaram, S.; Riethoven, J.M.; Liu, R.; Yang, J.; Laczko, P.; Chen, H.; Xia, Y.; Ladunga, I.; et al. Dynamic changes in genome-wide histone H3 lysine 4 methylation patterns in response to dehydration stress in Arabidopsis thaliana. BMC Plant Biol. 2010, 10, 238. [Google Scholar] [CrossRef] [PubMed]
- Zong, W.; Zhong, X.; You, J.; Xiong, L. Genome-wide profiling of histone H3K4-tri-methylation and gene expression in rice under drought stress. Plant Mol. Biol. 2013, 81, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Widiez, T.; Symeonidi, A.; Luo, C.; Lam, E.; Lawton, M.; Rensing, S.A. The chromatin landscape of the moss Physcomitrella patens and its dynamics during development and drought stress. Plant J. 2014, 79, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.; Muller, A.; Napoli, C.A.; Selinger, D.A.; Pikaard, C.S.; Richards, E.J.; Bender, J.; Mount, D.W.; Jorgensen, R.A. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002, 30, 5036–5055. [Google Scholar] [CrossRef] [PubMed]
- Pontvianne, F.; Blevins, T.; Pikaard, C.S. Arabidopsis histone lysine methyltransferases. Adv. Bot. Res. 2010, 53, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Sridhar, V.V.; Zhu, J.; Kapoor, A.; Zhu, J.-K. Distinctive core histone post-translational modification patterns in Arabidopsis thaliana. PLoS ONE 2007, 2, e1210. [Google Scholar] [CrossRef] [PubMed]
- Camporeale, G.; Oommen, A.M.; Griffin, J.B.; Sarath, G.; Zempleni, J. K12-biotinylated histone H4 marks heterochromatin in human lymphoblastoma cells. J. Nutr. Biochem. 2007, 18, 760–768. [Google Scholar] [CrossRef] [PubMed]
- Sani, E.; Herzyk, P.; Perrella, G.; Colot, V.; Amtmann, A. Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol. 2013, 14, R59. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Casas-Mollano, J.A.; Xu, J.; Riethoven, J.-J.M.; Zhang, C.; Cerutti, H. Osmotic stress induces phosphorylation of histone H3 at threonine 3 in pericentromeric regions of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2015, 112, 8487–8492. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Ding, Y.; Sun, X.; Xie, S.; Wang, D.; Liu, X.; Su, L.; Wei, W.; Pan, L.; Zhou, D.X. Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis. J. Exp. Bot. 2016, 67, 1703–1713. [Google Scholar] [CrossRef] [PubMed]
- Borsani, O.; Zhu, J.; Verslues, P.E.; Sunkar, R.; Zhu, J.K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 2005, 123, 1279–1291. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Liu, X.; Thorn, G.; Duan, J.; Tian, L. Biochemical and biophysical research communications expression analysis of histone acetyltransferases in rice under drought stress. Biochem. Biophys. Res. Commun. 2014, 443, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Eichten, S.R.; Hermanson, P.J.; Zaunbrecher, V.M.; Song, J.; Wendt, J.; Rosenbaum, H.; Madzima, T.H.; Sloan, A.E.; Huang, J.; et al. Genetic perturbation of the maize methylome. Plant Cell 2014, 26, 4602–4616. [Google Scholar] [CrossRef] [PubMed]
- Su, L.-C.; Deng, B.; Liu, S.; Li, L.-M.; Hu, B.; Zhong, Y.-T.; Li, L. Isolation and characterization of an osmotic stress and ABA induced histone deacetylase in Arachis hypogaea. Front. Plant Sci. 2015, 6, 512. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Luo, M.; Wang, Y.; Wu, K. Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 2010, 61, 3345–3353. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Liu, X.; Singh, P.; Cui, Y.; Zimmerli, L.; Wu, K. Chromatin modifications and remodeling in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 129–136. [Google Scholar] [CrossRef] [PubMed]
- González, R.M.; Ricardi, M.M.; Iusem, N.D. Epigenetic marks in an adaptive water stress-responsive gene in tomato roots under normal and drought conditions. Epigenetics 2013, 8, 864–872. [Google Scholar] [CrossRef] [PubMed]
- Bocchini, M.; Bartucca, M.L.; Ciancaleoni, S.; Mimmo, T.; Cesco, S.; Pii, Y.; Albertini, E.; Del Buono, D. Iron deficiency in barley plants: Phytosiderophore release, iron translocation, and DNA methylation. Front. Plant Sci. 2015, 6, 514. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-M.; Sasaki, T.; Ueda, M.; Sako, K.; Seki, M. Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 2015, 6, 114. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; How-Kit, A.; Stammitti, L.; Teyssier, E.; Rolin, D.; Mortain-Bertrand, A.; Halle, S.; Liu, M.; Kong, J.; Wu, C.; et al. A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc. Natl. Acad. Sci. USA 2015, 112, 10804–10809. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, P.H.; Burhans, W.C.; Curcio, M.J. Retrotransposition is associated with genome instability during chronological aging. Proc. Natl. Acad. Sci. USA 2011, 108, 20376–20381. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Gaubert, H.; Bucher, E.; Mirouze, M.; Vaillant, I.; Paszkowski, J. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 2011, 472, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Kaldis, A.; Tsementzi, D.; Tanriverdi, O.; Vlachonasios, K.E. Arabidopsis thaliana transcriptional co-activators ADA2 band SGF29a are implicated in salt stress responses. Planta 2011, 233, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Ji, D.; Li, S.; Wang, P.; Li, Q.; Xiang, F. The dynamic changes of DNA methylation and histone modifications of salt responsive transcription factor genes in soybean. PLoS ONE 2012, 7, e41274. [Google Scholar] [CrossRef] [PubMed]
- Pandey, G.; Sharma, N.; Sahu, P.P.; Prasad, M. Chromatin-based epigenetic regulation of plant abiotic stress response. Curr. Genom. 2016, 17, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S. Epigenetic memory of stress responses in plants. J. Phytochem. Biochem. 2018, 2, e102. [Google Scholar]
- Verhoeven, K.J.F.; VanGurp, T.P. Transgenerational effects of stress exposure on offspring phenotypes in apomictic dandelion. PLoS ONE 2012, 7, e38605. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, M.; Wang, G.; Meyers, B.C.; Fellowship, D.Y. Plants regenerated from tissue culture contain stable epigenome changes in rice. eLife 2013, 2, e00354. [Google Scholar]
- Luna, E.; Bruce, T.J.A.; Roberts, M.R.; Flors, V.; Ton, J. Next-generation systemic acquired resistance. Plant Physiol. 2012, 158, 844–853. [Google Scholar] [CrossRef] [PubMed]
- Luna, E.; Ton, J. The epigenetic machinery controlling transgenerational systemic acquired resistance. Plant Signal. Behav. 2012, 7, 615–618. [Google Scholar] [CrossRef] [PubMed]
- Becker, C.; Hagmann, J.; Müller, J.; Koenig, D.; Stegle, O.; Borgwardt, K.; Weigel, D. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 2011, 480, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.J.; Schultz, M.D.; Lewsey, M.G.; O’Malley, R.C.; Urich, M.A.; Libiger, O.; Schork, N.J.; Ecker, J.R. Transgenerational epigenetic instability is a source of novel methylation variants. Science 2011, 334, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Havecker, E.R.; Wallbridge, L.M.; Fedito, P.; Hardcastle, T.J.; Baulcombe, D.C. Metastable differentially methylated regions within Arabidopsis inbred populations are associated with modified expression of non-coding transcripts. PLoS ONE 2012, 7, e45242. [Google Scholar] [CrossRef] [PubMed]
- Ni, Z.; Kim, E.D.; Ha, M.; Lackey, E.; Liu, J.; Zhang, Y.; Sun, Q.; Chen, Z.J. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 2009, 457, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Shivaprasad, P.V.; Dunn, R.M.; Santos, B.A.; Bassett, A.; Baulcombe, D.C. Extraordinary transgressive phenotypes of hybrid tomato are influenced by epigenetics and small silencing RNAs. EMBO J. 2012, 31, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Chen, L.; Xia, H.; Wei, H.; Lou, Q.; Li, M.; Li, T.; Luo, L. Transgenerational epimutations induced by multi-generation drought imposition mediate rice plant’s adaptation to drought condition. Sci. Rep. 2017, 7, 39843. [Google Scholar] [CrossRef] [PubMed]
- Lämke, J.; Bäurle, I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 2017, 18, e124. [Google Scholar] [CrossRef] [PubMed]
- Richards, E.J. Inherited epigenetic variation—Revisiting soft inheritance. Nat. Rev. Genet. 2006, 7, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Hauben, M.; Haesendonckx, B.; Standaert, E.; Van Der Kelen, K.; Azmi, A.; Akpo, H.; Van Breusegem, F.; Guisez, Y.; Bots, M.; Lambert, B.; et al. Energy use efficiency is characterized by an epigenetic component that can be directed. Proc. Natl. Acad. Sci. USA 2009, 106, 20109–20114. [Google Scholar] [CrossRef] [PubMed]
- Samota, M.K.; Sasi, M.; Awana, M.; Yadav, O.P.; Amitha Mithra, S.V.; Tyagi, A.; Kumar, S.; Singh, A. Elicitor-induced biochemical and molecular manifestations to improve drought tolerance in rice (Oryza sativa L.) through seed-priming. Front. Plant Sci. 2017, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Buck-Koehntop, B.A.; Defossez, P.A. On how mammalian transcription factors recognize methylated DNA. Epigenetics 2013, 8, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Tao, Y.; Gao, X.; Zhang, L.; Li, X.; Zou, W.; Ruan, K.; Wang, F.; Xu, G.; Hu, R. A CRISPR-based approach for targeted DNA demethylation. Cell Discov. 2016, 2, 1600. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Arul, L.; Talwar, D.; Raina, S.K. PCR amplification of minimal gene expression cassette: An alternative, low cost and easy approach to ‘clean DNA’ transformation. Curr. Sci. 2006, 91, 930–934. [Google Scholar]
- Kumar, S.; Chandra, A.; Pandey, K.C. Bacillus thuringiensis (Bt) transgenic crop: An environment friendly insect-pest management strategy. J. Environ. Biol. 2008, 29, 641–653. [Google Scholar] [PubMed]
- Stricker, S.H.; Köferle, A.; Beck, S. From profiles to function in epigenomics. Nat. Rev. Genet. 2017, 18, 51–66. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, S. Epigenomics of Plant Responses to Environmental Stress. Epigenomes 2018, 2, 6. https://doi.org/10.3390/epigenomes2010006
Kumar S. Epigenomics of Plant Responses to Environmental Stress. Epigenomes. 2018; 2(1):6. https://doi.org/10.3390/epigenomes2010006
Chicago/Turabian StyleKumar, Suresh. 2018. "Epigenomics of Plant Responses to Environmental Stress" Epigenomes 2, no. 1: 6. https://doi.org/10.3390/epigenomes2010006
APA StyleKumar, S. (2018). Epigenomics of Plant Responses to Environmental Stress. Epigenomes, 2(1), 6. https://doi.org/10.3390/epigenomes2010006




