The Pea (Pisum sativum L.) Rogue Paramutation is Accompanied by Alterations in the Methylation Pattern of Specific Genomic Sequences
Abstract
:1. Introduction
2. Results
2.1. Genetic Similarity between Rogue and Non-Rogue Epigenotypes
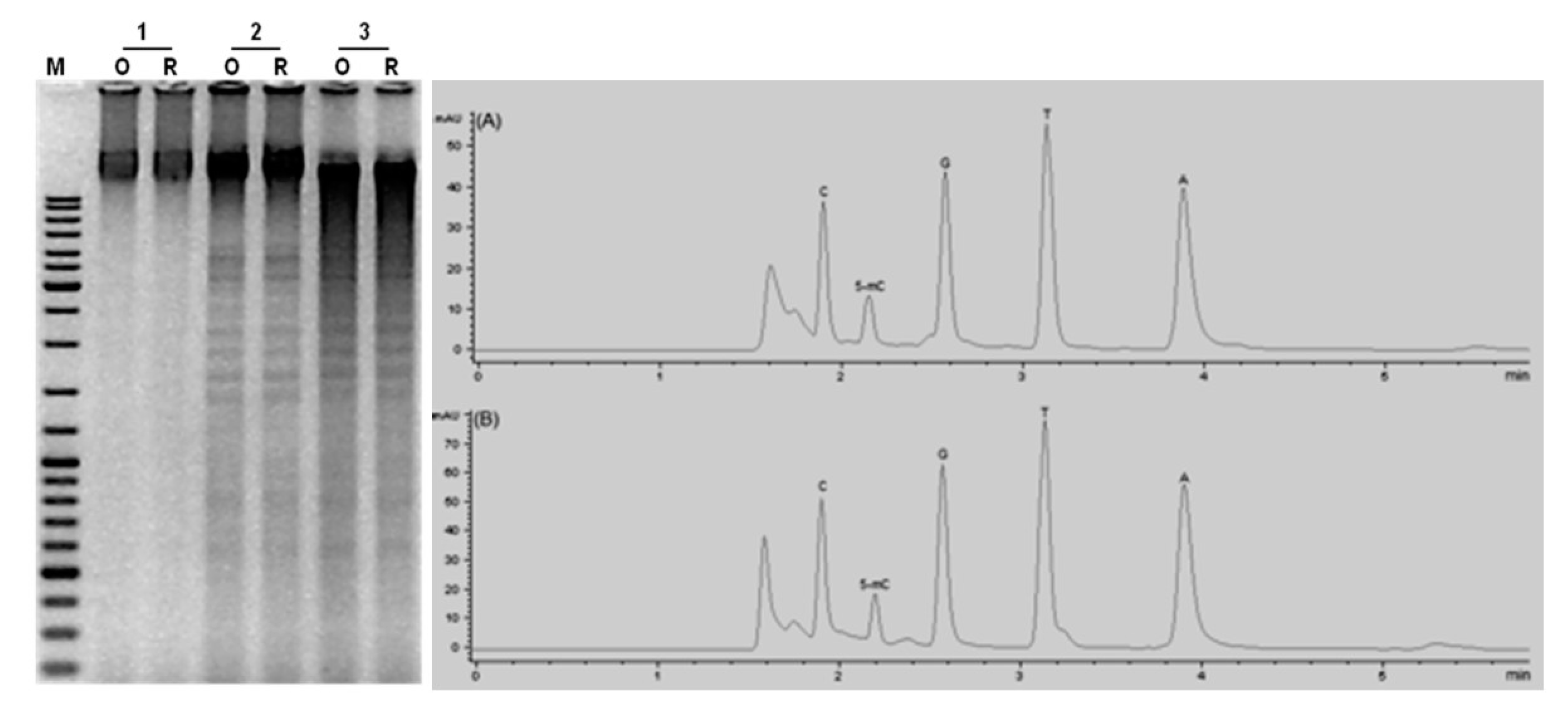
2.2. Genome-Wide Methylation Analysis
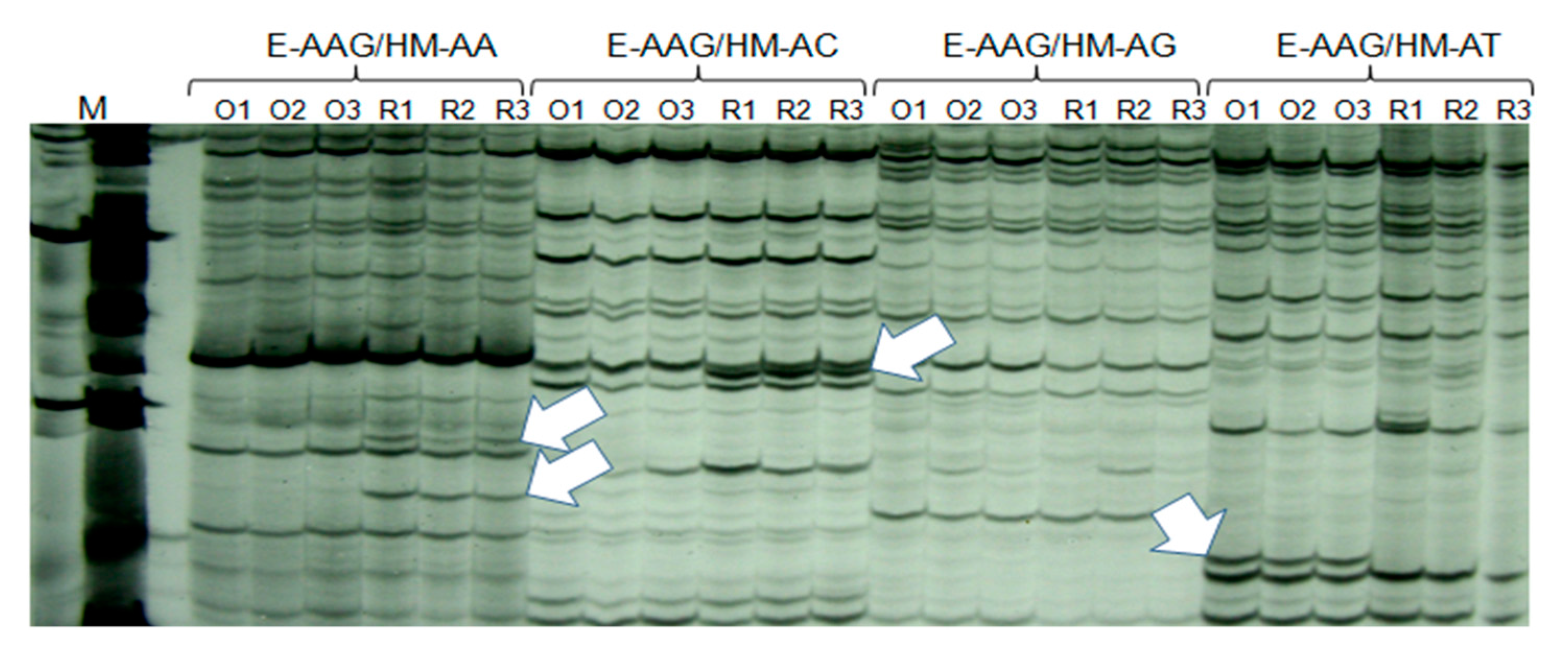
2.3. MS-AFLP Analysis
2.4. Differential Methylation in Pollen DNA
2.5. Expression of the Differentially Methylated Sequences
3. Discussion
4. Material and Methods
4.1. Plant Material
4.2. DNA Extraction
4.3. RAPD and SSR Analyses
4.4. HpaII and MspI Restriction Analysis of Genomic DNA
4.5. HPLC Analysis
4.6. MS-AFLPs Analysis
4.7. Confirmation of the Differential Methylation
4.8. Meiotic Inheritance of Differential Methylation
4.9. RNA Extraction and cDNA Synthesis
4.10. RT-qPCR
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bateson, W.; Pellew, C. On the genetics of “rogues” among culinary peas (Pisum sativum L.). J. Genet. 1915, 13–36. [Google Scholar] [CrossRef]
- Bateson, W.; Pellew, C. The genetics of “rogues” among culinary peas (Pisum sativum). Proc. R. Soc. Lond. 1920, 91, 186–195. [Google Scholar] [CrossRef]
- Brink, R.A. Paramutation at the R Locus in Maize. Cold Spring Harb. Symp. Quant. Biol. 1958, 23, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Rassoulzadegan, M.; Grandjean, V.; Gounon, P.; Vincent, S.; Gillot, I.; Cuzin, F. RNA-mediated non-Mendelian inheritance of an epigenetic change in the mouse. Nature 2006, 441, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Arteaga-Vazquez, M.A.; Chandler, V.L. Paramutation in maize: RNA mediated trans-generational gene silencing. Curr. Opin. Genet. Dev. 2010, 20, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Hollick, J.B. Paramutation and development. Annu. Rev. Cell. Dev. Biol. 2010, 26, 557–579. [Google Scholar] [CrossRef] [PubMed]
- Erhard, K.F.; Hollick, J.B. Paramutation: A process for acquiring trans-generational regulatory states. Curr. Opin. Plant Biol. 2011, 14, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Pilu, R. Paramutation: Just a curiosity or fine tuning of gene expression in the next generation? Curr. Genom. 2011, 12, 298–306. [Google Scholar] [CrossRef]
- Alleman, M.; Sidorenko, L.; McGinnis, K.; Seshadri, V.; Dorweiler, J.E.; White, J.; Sikkink, K.; Chandler, V.L. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 2006, 442, 295–298. [Google Scholar] [CrossRef] [PubMed]
- Sidorenko, L.; Dorweiler, J.E.; Cigan, A.M.; Arteaga-Vazquez, M.; Vyas, M.; Kermicle, J.; Jurcin, D.; Brzeski, J.; Cai, Y.; Chandler, V.L. A dominant mutation in mediator of paramutation2, one of three second-largest subunits of a plant-specific RNA polymerase, disrupts multiple siRNA silencing processes. PLoS Genet. 2009, 5, e1000725. [Google Scholar] [CrossRef] [PubMed]
- Hale, C.J.; Stonaker, J.L.; Gross, S.M.; Hollick, J.B. A novel Snf2 protein maintains trans-generational regulatory states established by paramutation in maize. PLoS Biol. 2007, 5, e275. [Google Scholar] [CrossRef] [PubMed]
- Barbour, J.-E.R.; Liao, I.T.; Stonaker, J.L.; Lim, J.P.; Lee, C.C.; Parkinson, S.E.; Kermicle, J.; Simon, S.; Meyers, B.C.; Williams-Carrier, R.; et al. Required to maintain repression2 is a novel protein that facilitates locus-specific paramutation in maize. Plant Cell. 2012, 24, 1761–1775. [Google Scholar] [CrossRef] [PubMed]
- Erhard, K.F., Jr.; Stonaker, J.L.; Parkinson, S.E.; Lim, J.P.; Hale, C.J.; Hollick, J.B. RNA polymerase IV functions in paramutation in Zea mays. Science 2009, 323, 1201–1205. [Google Scholar] [CrossRef]
- Stonaker, J.L.; Lim, J.P.; Erhard, K.F., Jr.; Hollick, J.B. Diversity of Pol IV function is defined by mutations at the maize rmr7 locus. PLoS Genet. 2009, 5, e1000706. [Google Scholar] [CrossRef] [PubMed]
- Gouil, Q.; Novák, O.; Baulcombe, D.C. SLTAB2 is the paramutated SULFUREA locus in tomato. J. Exp. Bot. 2016, 67, 2655–2664. [Google Scholar] [CrossRef]
- Meyer, P.; Heidmann, I.; Niedenhof, I. Differences in DNA-methylation are associated with a paramutation phenomenon in transgenic petunia. Plant J. 1993, 4, 89–100. [Google Scholar] [CrossRef]
- Das, O.P.; Messing, J. Variegated phenotype and developmental methylation changes of a maize allele originating from epimutation. Genetics 1994, 136, 1121–1141. [Google Scholar] [PubMed]
- Sidorenko, L.V.; Peterson, T. Transgene-Induced Silencing Identifies sequences involved in the establishment of paramutation of the maize p1 gene. Plant Cell. 2001, 13, 319–335. [Google Scholar] [CrossRef] [PubMed]
- Walker, E.L.; Panavas, T. Structural features and methylation patterns associated with paramutation at the r1 locus in Zea mays. Genetics 2001, 159, 1201–1215. [Google Scholar] [PubMed]
- Haring, M.; Bader, R.; Louwers, M.; Schwabe, A.; van Driel, R.; Stam, M. The role of DNA methylation, nucleosome occupancy and histone modifications in paramutation. Plant J. 2010, 63, 366–378. [Google Scholar] [CrossRef] [PubMed]
- Belele, C.L.; Sidorenko, L.; Stam, M.; Bader, R.; Arteaga-Vazquez, M.A.; Chandler, V.L. Specific tandem repeats are sufficient for paramutation-induced trans-generational silencing. PLoS Genet. 2013, 9, e1003773. [Google Scholar] [CrossRef] [PubMed]
- Brotherton, W., Jr. Further studies on the inheritance of rogue types in garden peas (Pisum sativum). J. Agric. Res. 1923, 24, 815–852. [Google Scholar]
- Brotherton, W., Jr. Gamete production in certain crosses with rogues in peas. J. Agric. Res. 1924, 28, 815–852. [Google Scholar]
- Bunten, I. A preliminary report on the chromosome complement of “rabbit eared rogues” in culinary peas (Pisum sativum L.). Am. J. Bot. 1930, 17, 139–142. [Google Scholar] [CrossRef]
- Pyke, K.A.; Hedley, C.L. Aspects of the rogue phenotype of peas. Plant Sci. Lett. 1984, 35, 87–90. [Google Scholar] [CrossRef]
- Pereira, G.; Marques, C.; Ribeiro, R.; Formiga, S.; Dâmaso, M.; Tavares Sousa, M.; Farinhó, M.; Leitão, J.M. Identification of DNA markers linked to an induced mutated gene conferring resistance to powdery mildew in pea (Pisum sativum L.). Euphytica 2010, 171, 327–335. [Google Scholar] [CrossRef]
- Loridon, K.; McPhee, K.; Morin, J.; Dubreuil, P.; Pilet-Nayel, M.L.; Aubert, G.; Rameau, C.; Baranger, A.; Coyne, C.; Lejeune-Hènaut, I.; et al. Microsatellite marker polymorphism and mapping in pea (Pisum sativum L.). Theor. Appl. Genet. 2005, 11, 1022–1103. [Google Scholar] [CrossRef] [PubMed]
- Elisiário, P.J.; Justo, E.M.; Leitão, J.M. Identification of mandarin hybrids by isozyme and RAPD analysis. Sci. Hort. 1999, 81, 287–300. [Google Scholar] [CrossRef]
- Carlier, J.D.; Sousa, N.H.; Santo, T.E.; d’Eeckenbrugge, G.C.; Leitão, J.M. A genetic map of pineapple (Ananas comosus (L.) Merr.) including SCAR, CAPS, SSR and EST-SSR markers. Mol. Breed. 2012, 29, 245–260. [Google Scholar] [CrossRef]
- Cervera, M.T.; Ruiz-Garcia, L.; Martinez-Zapater, J. Analysis of DNA methylation in Arabidopsis thaliana based on methylation-sensitive AFLP markers. Mol. Gen. Genom. 2002, 268, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Reyna-López, G.E.; Simpson, J.; Ruiz-Herrera, J. Differences in DNA methylation pattern are detectable during the dimorphic transition of fungi by amplification of restriction polymorphisms. Mol. Gen. Genet. 1997, 253, 703–710. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.Z.; Xu, C.G.; Saghai-Maroof, M.A.; Zhang, Q. Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Mol. Gen. Genet. 1999, 261, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; van de Lee, T.; Hornes, M.; Frijters, A.; Pot, J.; Peleman, J.; Kuiper, M. AFLP: A new technique for DNA fingerprinting. Nucl. Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef] [PubMed]
- Baurens, F.-C.; Nicolleau, J.; Legavre, T.; Verdeil, J.-L.; Monteuuis, O. Genomic DNA methylation of juvenile and mature Acacia mangium micropropagated in vitro with reference to leaf morphology as a phase change marker. Tree Physiol. 2004, 24, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Farinhó, M.; Carlier, J.; Svetleva, D.; Coelho, P.; Monteiro, A.; Leitão, J. Mapping of a locus for adult plant resistance to downy mildew in broccoli (Brassica oleracea convar. italica). Theor. Appl. Genet. 2004, 109, 1392–1398. [Google Scholar] [CrossRef] [PubMed]
- Farinhó, M.; Coelho, P.; Monteiro, A.; Leitão, J. SCAR and CAPS markers flanking the Brassica oleracea L. Pp523 downy mildew resistance locus demarcate a genomic region syntenic to the top arm end of Arabidopsis thaliana L. chromosome 1. Euphytica 2007, 157, 215–221. [Google Scholar] [CrossRef]
- Available online: www.ncbi.nlm.nih.gov (accessed on 3 February 2017).
- Available online: www.jcvi.org/cgi-bin/medicago/overview.cgi (accessed on 3 February 2017).
- Available online: www.coolseasonfoodlegume.org (accessed on 3 February 2017).

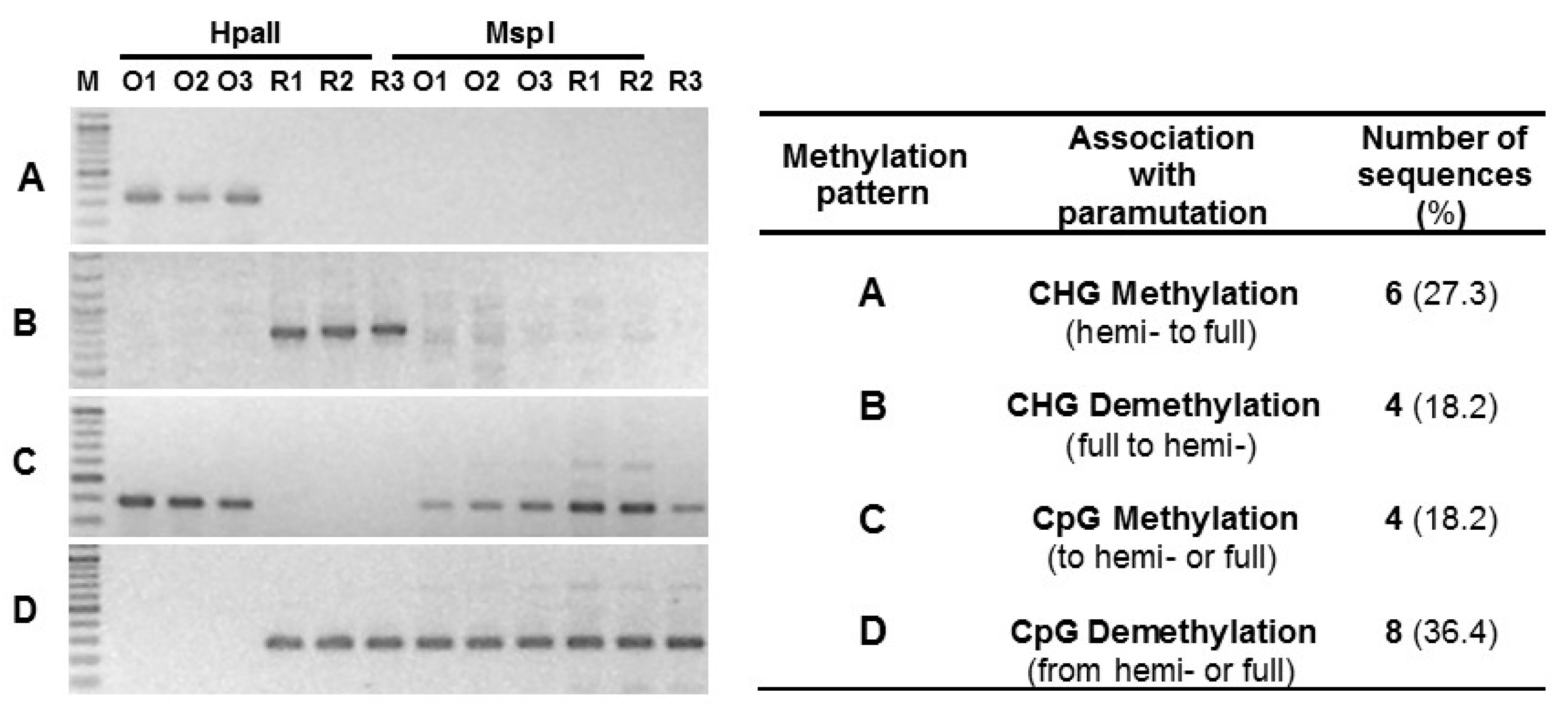
| CHG Methylation | CHG Demethylation | CpG Methylation | CpG Demethylation | |
|---|---|---|---|---|
| Maintained in pollen DNA | ACA/AG_560_ O AGC/AT_466_O ** | ACA/AG_735_R ACT/AC_451_R AGG/AG_705_R | AAG/AG_366_O AGC/AT_134_O | AAG/AC_613_R ACA/AA_749_R AGC/AA_202_R AGG/AC_384_R TA/CA_260_R |
| Altered in pollen DNA | AAC/AA_300_O ACG/CC_81_O AGG/AT_139_O AGG/AT_302_O | ACT/CA_584_R | ACT/AG_449_O ACA/CT_546_O | AAC/AA_174_R AAG/AA_197_R AAG/AA_325_R |
| Sequence | Methylation Pattern (Leaves) * | Methylation Pattern (Pollen) * |
|---|---|---|
| AAC/AA_174_R | D | Non-methylated |
| AAC/AA_300_O | A | C |
| AAG/AA_197_R | D | Fully CHG methylated |
| AAG/AA_325_R | D | Non-methylated |
| AAG/AC_613_R | D | D |
| AAG/AG_366_O | C | C |
| ACA/AA_749_R | D | D |
| ACA/AG_560_O | A | A |
| ACA/AG_735_R | B | B |
| ACA/CT_546_O | C | Non-methylated |
| ACG/CC_81_O | A | C |
| ACT/AC_451_R | B | B |
| ACT/AG_449_O | C | D |
| ACT/CA_584_R | B | D |
| AGC/AA_202_R | D | D |
| AGC/AT_134_O | C | C |
| AGC/AT_466_O | C | C |
| AGG/AC_384_R | D | D |
| AGG/AG_705_R | B | B |
| AGG/AT_139_O | A | C |
| AGG/AT_302_O | A | C |
| TA/CA_260_R | D | D |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santo, T.E.; Pereira, R.J.; Leitão, J.M. The Pea (Pisum sativum L.) Rogue Paramutation is Accompanied by Alterations in the Methylation Pattern of Specific Genomic Sequences. Epigenomes 2017, 1, 6. https://doi.org/10.3390/epigenomes1010006
Santo TE, Pereira RJ, Leitão JM. The Pea (Pisum sativum L.) Rogue Paramutation is Accompanied by Alterations in the Methylation Pattern of Specific Genomic Sequences. Epigenomes. 2017; 1(1):6. https://doi.org/10.3390/epigenomes1010006
Chicago/Turabian StyleSanto, Tatiana E., Ricardo J. Pereira, and José M. Leitão. 2017. "The Pea (Pisum sativum L.) Rogue Paramutation is Accompanied by Alterations in the Methylation Pattern of Specific Genomic Sequences" Epigenomes 1, no. 1: 6. https://doi.org/10.3390/epigenomes1010006
APA StyleSanto, T. E., Pereira, R. J., & Leitão, J. M. (2017). The Pea (Pisum sativum L.) Rogue Paramutation is Accompanied by Alterations in the Methylation Pattern of Specific Genomic Sequences. Epigenomes, 1(1), 6. https://doi.org/10.3390/epigenomes1010006






