Lysine-Specific Histone Demethylases Contribute to Cellular Differentiation and Carcinogenesis
Abstract
:1. Introduction
2. Lysine-Specific Demethylase 1
2.1. KDM1A
2.2. KDM1A and Long Non-Coding RNA
2.3. KDM1A and Cellular Differentiation
2.4. KDM1A and Cancer
2.5. KDM1B
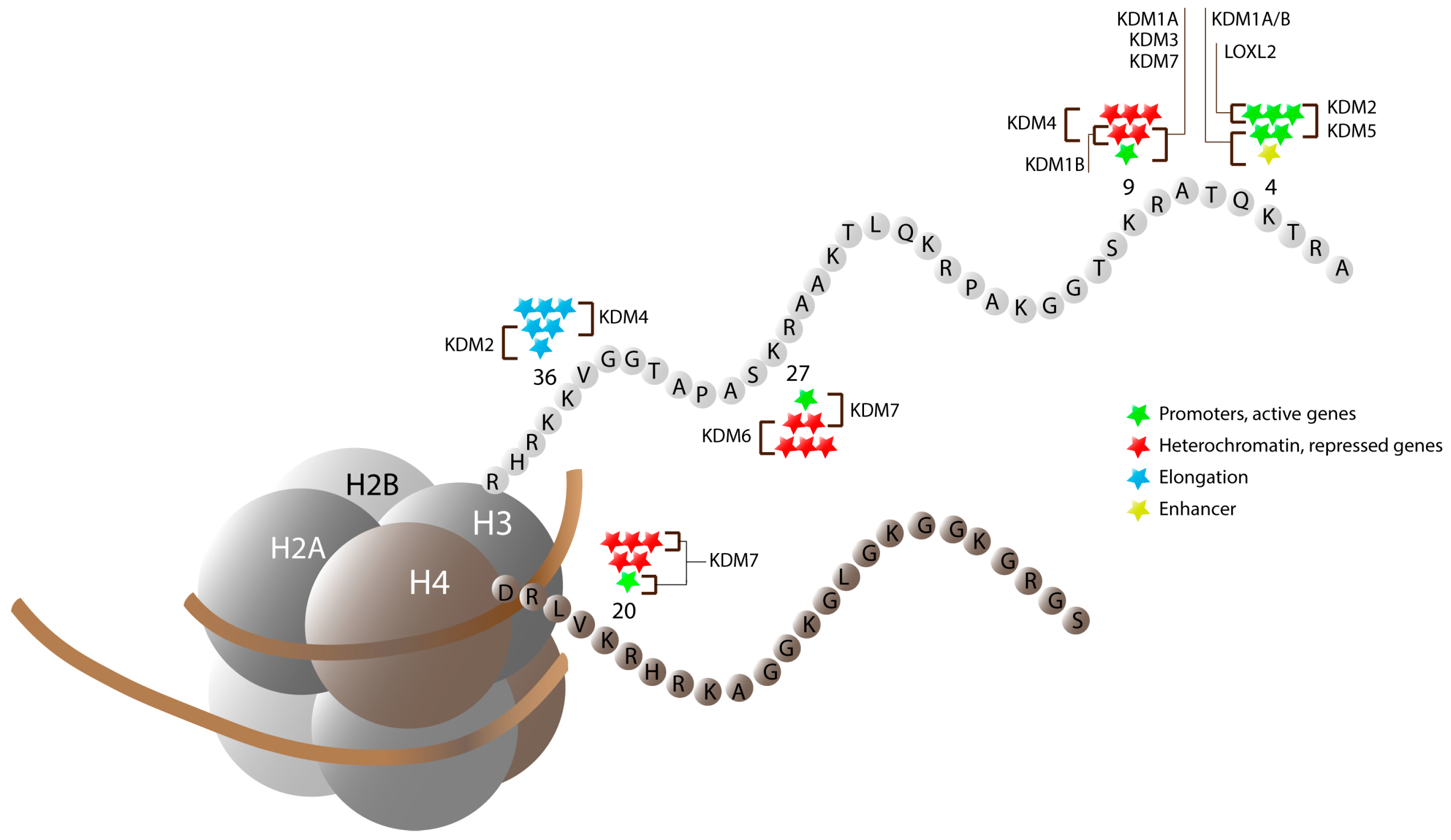
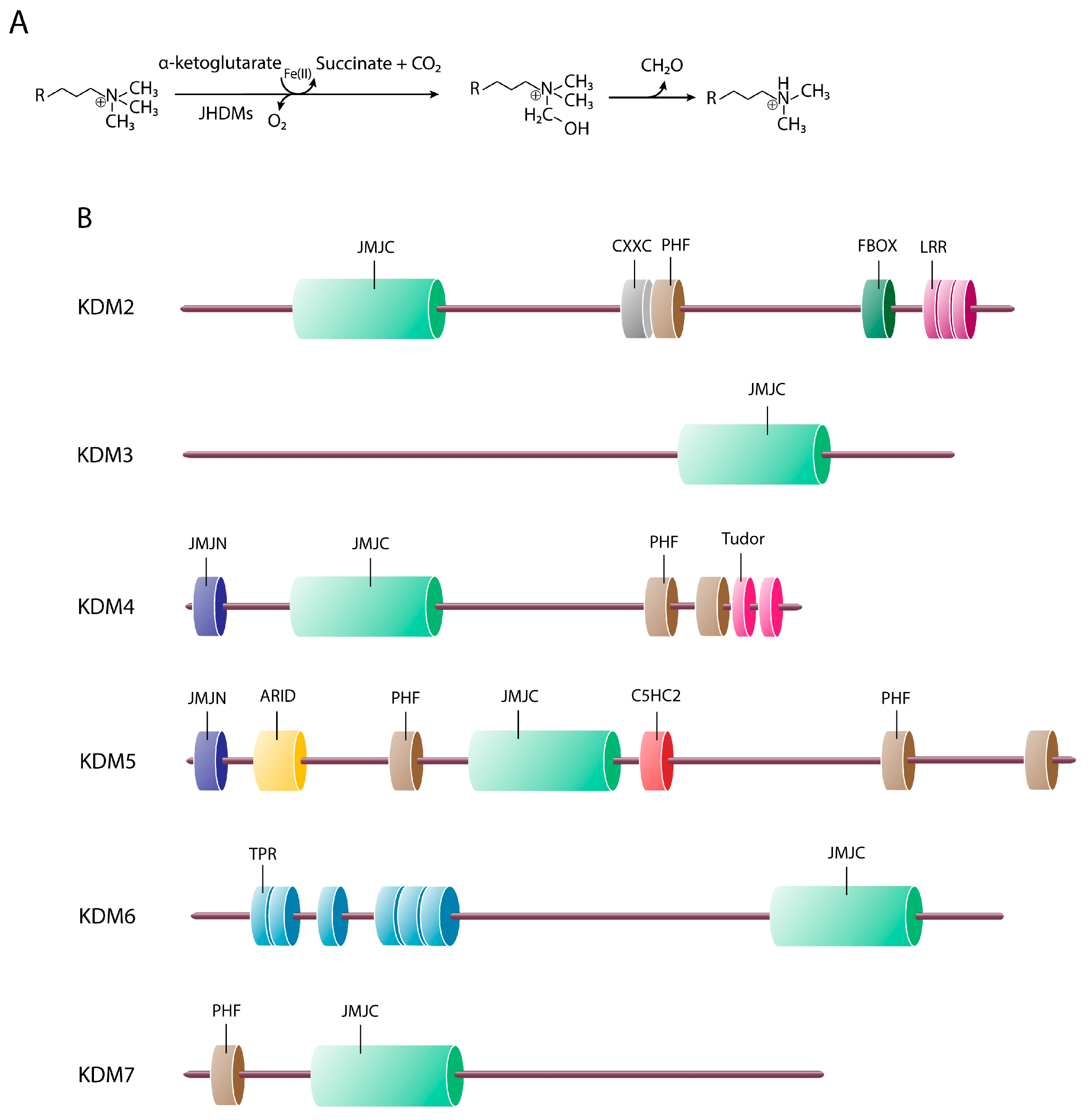
3. Jumonji C Domain-Containing Proteins
3.1. JmjC-Containing Proteins and Cellular Differentiation
3.1.1. KDM2 Cluster (FBXL Subfamily)
3.1.2. KDM3 Cluster (JMJD1 Subfamily)
3.1.3. KDM4 Cluster (JMJD2 Subfamily)
3.1.4. The KDM5 Cluster (JARID1 Subfamily)
3.1.5. KDM6 Cluster (UTX/JMJD3 Subfamily)
3.1.6. The KDM7 Cluster
3.2. JmjC Proteins and Cancer
3.2.1. The KDM2 Cluster (FBXL Subfamily)
3.2.2. The KDM3 Cluster (JMJD1 Subfamily)
3.2.3. The KDM4 Cluster (JMJD2 Subfamily)
3.2.4. The KDM5 Cluster (JARID1 Subfamily)
3.2.5. The KDM6 Cluster (UTX/JMJD3 Subfamily)
3.2.6. The KDM7 Cluster
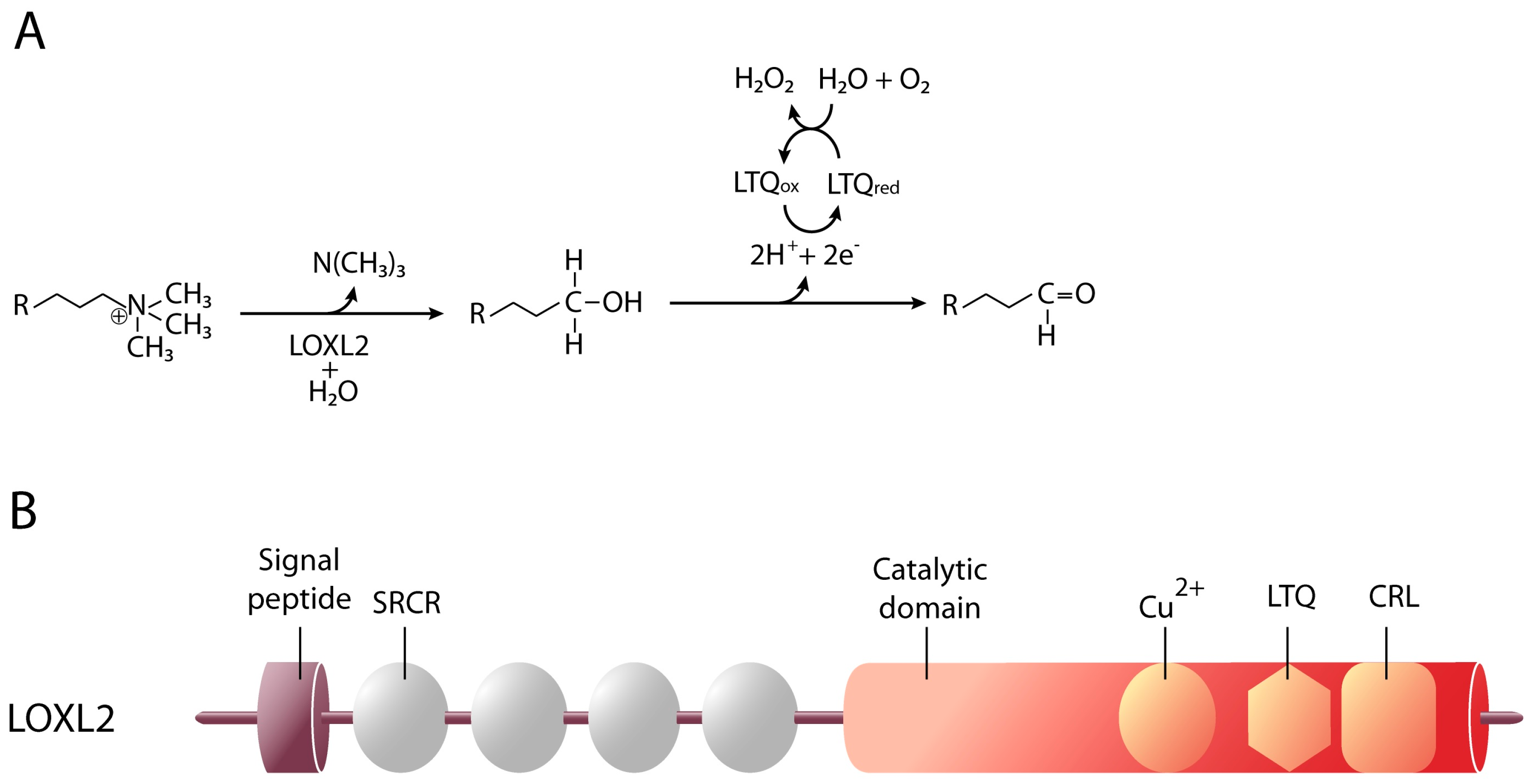
4. Lysyl Oxidase-Like 2
4.1. LOXL2 and Cellular Differentiation
4.2. LOXL2 and Cancer
5. Beyond the Histone Demethylase Function
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wang, Y.; Jia, S. Degrees make all the difference: The multifunctionality of histone H4 lysine 20 methylation. Epigenetics 2009, 4, 273–276. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone h3 lysine 27 methylation in polycomb-group silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef] [PubMed]
- Schotta, G.; Lachner, M.; Sarma, K.; Ebert, A.; Sengupta, R.; Reuter, G.; Reinberg, D.; Jenuwein, T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004, 18, 1251–1262. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, B.E.; Kamal, M.; Lindblad-Toh, K.; Bekiranov, S.; Bailey, D.K.; Huebert, D.J.; McMahon, S.; Karlsson, E.K.; Kulbokas, E.J., 3rd; Gingeras, T.R.; et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 2005, 120, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Harikumar, A.; Meshorer, E. Chromatin remodeling and bivalent histone modifications in embryonic stem cells. EMBO Rep. 2015, 16, 1609–1619. [Google Scholar] [CrossRef] [PubMed]
- Byvoet, P.; Shepherd, G.R.; Hardin, J.M.; Noland, B.J. The distribution and turnover of labeled methyl groups in histone fractions of cultured mammalian cells. Arch. Biochem. Biophys. 1972, 148, 558–567. [Google Scholar] [CrossRef]
- Duerre, J.A.; Lee, C.T. In vivo methylation and turnover of rat brain histones. J. Neurochem. 1974, 23, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Lan, F.; Matson, C.; Mulligan, P.; Whetstine, J.R.; Cole, P.A.; Casero, R.A.; Shi, Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004, 119, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Metzger, E.; Wissmann, M.; Yin, N.; Muller, J.M.; Schneider, R.; Peters, A.H.; Gunther, T.; Buettner, R.; Schule, R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 2005, 437, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Gocke, C.B.; Luo, X.; Borek, D.; Tomchick, D.R.; Machius, M.; Otwinowski, Z.; Yu, H. Structural basis for corest-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol. Cell 2006, 23, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Forneris, F.; Binda, C.; Vanoni, M.A.; Mattevi, A.; Battaglioli, E. Histone demethylation catalysed by LSD1 is a flavin-dependent oxidative process. FEBS Lett. 2005, 579, 2203–2207. [Google Scholar] [CrossRef] [PubMed]
- Karasulu, B.; Patil, M.; Thiel, W. Amine oxidation mediated by lysine-specific demethylase 1: Quantum mechanics/molecular mechanics insights into mechanism and role of lysine 661. J. Am. Chem. Soc. 2013, 135, 13400–13413. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Wang, J.; Hu, Y.; Zhao, H.; Hou, W.; Zhao, H.; Wang, H.; Liao, J.; Xu, X. Modulation of LSD1 phosphorylation by CK2/WIP1 regulates RNF168-dependent 53BP1 recruitment in response to DNA damage. Nucleic Acids Res. 2015, 43, 5936–5947. [Google Scholar] [CrossRef] [PubMed]
- Costa, R.; Arrigoni, G.; Cozza, G.; Lolli, G.; Battistutta, R.; Izpisua Belmonte, J.C.; Pinna, L.A.; Sarno, S. The lysine-specific demethylase 1 is a novel substrate of protein kinase CK2. Biochim. Biophys. Acta 2014, 1844, 722–729. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Xu, G.; Liu, J.; Zhang, N.; Li, L.; Ji, J.; Zhang, J.; Zhang, L.; Wang, G.; Wang, X.; et al. Phosphorylation of LSD1 at Ser112 is crucial for its function in induction of emt and metastasis in breast cancer. Breast Cancer Res. Treat. 2016, 159, 443–456. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wan, X.; Wei, Y.; Liu, X.; Lai, W.; Zhang, L.; Jin, J.; Wu, C.; Shao, Q.; Shao, G.; et al. LSD1-mediated epigenetic modification contributes to ovarian cancer cell migration and invasion. Oncol. Rep. 2016, 35, 3586–3592. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Kim, T.Y.; Kim, M.S.; Kim, M.A.; Park, S.H.; Jang, Y.K. Nuclear import of human histone lysine-specific demethylase LSD1. J. Biochem. 2014, 156, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Metzger, E.; Willmann, D.; McMillan, J.; Forne, I.; Metzger, P.; Gerhardt, S.; Petroll, K.; Von Maessenhausen, A.; Urban, S.; Schott, A.K.; et al. Assembly of methylated KDM1A and CHD1 drives androgen receptor-dependent transcription and translocation. Nat. Struct. Mol. Biol. 2016, 23, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.G.; Wynder, C.; Cooch, N.; Shiekhattar, R. An essential role for corest in nucleosomal histone 3 lysine 4 demethylation. Nature 2005, 437, 432–435. [Google Scholar] [CrossRef] [PubMed]
- Tochio, N.; Umehara, T.; Koshiba, S.; Inoue, M.; Yabuki, T.; Aoki, M.; Seki, E.; Watanabe, S.; Tomo, Y.; Hanada, M.; et al. Solution structure of the swirm domain of human histone demethylase LSD1. Structure 2006, 14, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Shenoy, A.K.; Li, X.; Jin, Y.; Jin, L.; Cai, Q.; Tang, M.; Liu, Y.; Chen, H.; Reisman, D.; et al. MOF acetylates the histone demethylase LSD1 to suppress Epithelial-to-Mesenchymal transition. Cell Rep. 2016, 15, 2665–2678. [Google Scholar] [CrossRef] [PubMed]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563. [Google Scholar] [PubMed]
- Dinger, M.E.; Pang, K.C.; Mercer, T.R.; Mattick, J.S. Differentiating protein-coding and noncoding RNA: Challenges and ambiguities. PLoS Comput. Biol. 2008, 4, e1000176. [Google Scholar] [CrossRef] [PubMed]
- Dinger, M.E.; Amaral, P.P.; Mercer, T.R.; Pang, K.C.; Bruce, S.J.; Gardiner, B.B.; Askarian-Amiri, M.E.; Ru, K.; Solda, G.; Simons, C.; et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008, 18, 1433–1445. [Google Scholar] [CrossRef] [PubMed]
- Loewer, S.; Cabili, M.N.; Guttman, M.; Loh, Y.H.; Thomas, K.; Park, I.H.; Garber, M.; Curran, M.; Onder, T.; Agarwal, S.; et al. Large intergenic non-coding RNA-ROR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 2010, 42, 1113–1117. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.R.; Mondal, T.; Mohammad, F.; Enroth, S.; Redrup, L.; Komorowski, J.; Nagano, T.; Mancini-Dinardo, D.; Kanduri, C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 2008, 32, 232–246. [Google Scholar] [CrossRef] [PubMed]
- Nagano, T.; Mitchell, J.A.; Sanz, L.A.; Pauler, F.M.; Ferguson-Smith, A.C.; Feil, R.; Fraser, P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 2008, 322, 1717–1720. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, E.; Allis, C.D. Rna meets chromatin. Genes Dev. 2005, 19, 1635–1655. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, H. A unified mode of epigenetic gene silencing: RNA meets polycomb group proteins. RNA Biol. 2005, 2, 8–10. [Google Scholar] [CrossRef] [PubMed]
- Vicent, G.P.; Nacht, A.S.; Zaurin, R.; Font-Mateu, J.; Soronellas, D.; Le Dily, F.; Reyes, D.; Beato, M. Unliganded progesterone receptor-mediated targeting of an RNA-containing repressive complex silences a subset of hormone-inducible genes. Genes Dev. 2013, 27, 1179–1197. [Google Scholar] [CrossRef] [PubMed]
- Hirschi, A.; Martin, W.J.; Luka, Z.; Loukachevitch, L.V.; Reiter, N.J. G-quadruplex RNA binding and recognition by the lysine-specific histone demethylase-1 enzyme. RNA 2016, 22, 1250–1260. [Google Scholar] [CrossRef] [PubMed]
- Whyte, W.A.; Bilodeau, S.; Orlando, D.A.; Hoke, H.A.; Frampton, G.M.; Foster, C.T.; Cowley, S.M.; Young, R.A. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 2012, 482, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Fu, X.; Huo, B.; Wang, Y.; Sun, J.; Meng, L.; Hao, T.; Zhao, Z.J.; Hu, X. GATA2 regulates GATA1 expression through LSD1-mediated histone modification. Am. J. Transl. Res. 2016, 8, 2265–2274. [Google Scholar] [PubMed]
- Lopez, C.I.; Saud, K.E.; Aguilar, R.; Berndt, F.A.; Canovas, J.; Montecino, M.; Kukuljan, M. The chromatin modifying complex corest/LSD1 negatively regulates notch pathway during cerebral cortex development. Dev. Neurobiol. 2016, 76, 1360–1373. [Google Scholar] [CrossRef] [PubMed]
- Mulligan, P.; Yang, F.; Di Stefano, L.; Ji, J.Y.; Ouyang, J.; Nishikawa, J.L.; Toiber, D.; Kulkarni, M.; Wang, Q.; Najafi-Shoushtari, S.H.; et al. A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development. Mol. Cell 2011, 42, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Kim, J.; Zhang, R.; Yang, X.; Zhang, Y.; Fang, J.; Chen, Z.; Teng, L.; Chen, X.; Ge, H.; et al. Histone demethylase LSD1 promotes adipocyte differentiation through repressing wnt signaling. Cell Chem. Biol. 2016, 23, 1228–1240. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Ponn, A.; Hu, X.; Law, B.K.; Lu, J. Requirement of the histone demethylase LSD1 in Snai1-mediated transcriptional repression during epithelial-mesenchymal transition. Oncogene 2010, 29, 4896–4904. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Wu, Y.; Li, J.; Dong, C.; Ye, X.; Chi, Y.I.; Evers, B.M.; Zhou, B.P. The snag domain of snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010, 29, 1803–1816. [Google Scholar] [CrossRef] [PubMed]
- Bennesch, M.A.; Segala, G.; Wider, D.; Picard, D. Lsd1 engages a corepressor complex for the activation of the estrogen receptor alpha by estrogen and camp. Nucleic Acids Res. 2016, 44, 8655–8670. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Li, S.; Song, W.; Li, X.; Li, Q.; Zhang, Z.; Han, Y.; Zhang, X.; Miao, S.; Du, R.; et al. Lysine-specific demethylase 1 (LSD1/KDM1A) contributes to colorectal tumorigenesis via activation of the Wnt/β-catenin pathway by down-regulating Dickkopf-1 (DKK1) [corrected]. PLoS ONE 2013, 8, e70077. [Google Scholar] [CrossRef]
- Kahl, P.; Gullotti, L.; Heukamp, L.C.; Wolf, S.; Friedrichs, N.; Vorreuther, R.; Solleder, G.; Bastian, P.J.; Ellinger, J.; Metzger, E.; et al. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res. 2006, 66, 11341–11347. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, V.; Ahmad, S.; Nilsson, E.M.; Helczynski, L.; Kenna, S.; Persson, J.L.; Gudas, L.J.; Mongan, N.P. The lysine specific demethylase-1 (LSD1/KDM1A) regulates VEGF-A expression in prostate cancer. Mol. Oncol. 2013, 7, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, E.C.; Robinson, B.D.; Downes, M.J.; Powell, L.G.; Lee, M.M.; Scherr, D.S.; Gudas, L.J.; Mongan, N.P. Role of androgen receptor and associated lysine-demethylase coregulators, LSD1 and JMJD2A, in localized and advanced human bladder cancer. Mol. Carcinog. 2011, 50, 931–944. [Google Scholar] [CrossRef] [PubMed]
- Lv, T.; Yuan, D.; Miao, X.; Lv, Y.; Zhan, P.; Shen, X.; Song, Y. Over-expression of LSD1 promotes proliferation, migration and invasion in non-small cell lung cancer. PLoS ONE 2012, 7, e35065. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, Y.; Yang, X.H.; Kang, T.; Zhao, Y.; Wang, C.; Evers, B.M.; Zhou, B.P. The deubiquitinase USP28 stabilizes LSD1 and confers stem-cell-like traits to breast cancer cells. Cell Rep. 2013, 5, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Althoff, K.; Beckers, A.; Odersky, A.; Mestdagh, P.; Koster, J.; Bray, I.M.; Bryan, K.; Vandesompele, J.; Speleman, F.; Stallings, R.L.; et al. MiR-137 functions as a tumor suppressor in neuroblastoma by downregulating KDM1A. Int. J. Cancer 2013, 133, 1064–1073. [Google Scholar] [CrossRef] [PubMed]
- Amente, S.; Milazzo, G.; Sorrentino, M.C.; Ambrosio, S.; Di Palo, G.; Lania, L.; Perini, G.; Majello, B. Lysine-specific demethylase (LSD1/KDM1A) and mycn cooperatively repress tumor suppressor genes in neuroblastoma. Oncotarget 2015, 6, 14572–14583. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Zhang, Z.M.; Xia, Y.; Liao, G.Q.; Pan, Y.; Liu, S.; Zhang, Y.; Yan, Z.S. LSD1-mediated epigenetic modification contributes to proliferation and metastasis of colon cancer. Br. J. Cancer 2013, 109, 994–1003. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhu, Y.; Wang, Q.; Hu, H.; Li, Z.; Wang, D.; Zhang, W.; Qi, B.; Ye, J.; Wu, H.; et al. The histone demethylase LSD1 is a novel oncogene and therapeutic target in oral cancer. Cancer Lett. 2016, 374, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.A.; Inamura, K.; Yamauchi, M.; Nishihara, R.; Mima, K.; Sukawa, Y.; Li, T.; Yasunari, M.; Morikawa, T.; Fitzgerald, K.C.; et al. Loss of CDH1 (E-cadherin) expression is associated with infiltrative tumour growth and lymph node metastasis. Br. J. Cancer 2016, 114, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Karytinos, A.; Forneris, F.; Profumo, A.; Ciossani, G.; Battaglioli, E.; Binda, C.; Mattevi, A. A novel mammalian flavin-dependent histone demethylase. J. Biol. Chem. 2009, 284, 17775–17782. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Jiang, J.; Stewart, M.D.; Qi, S.; Yamane, K.; Li, J.; Zhang, Y.; Wong, J. Aof1 is a histone H3K4 demethylase possessing demethylase activity-independent repression function. Cell Res. 2010, 20, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yin, X.; Yang, H.; Xu, Y. Histone demethylase LSD2 acts as an E3 ubiquitin ligase and inhibits cancer cell growth through promoting proteasomal degradation of OGT. Mol. Cell 2015, 58, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Barbera, A.J.; Xu, Y.; Rutenberg, M.; Leonor, T.; Bi, Q.; Lan, F.; Mei, P.; Yuan, G.C.; Lian, C.; et al. Human LSD2/KDM1B/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Mol. Cell 2010, 39, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Chen, F.; Dong, Z.; Hu, D.; Barbera, A.J.; Clark, E.A.; Fang, J.; Yang, Y.; Mei, P.; Rutenberg, M.; et al. LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation. Mol. Cell 2013, 49, 558–570. [Google Scholar] [CrossRef] [PubMed]
- Van Essen, D.; Zhu, Y.; Saccani, S. A feed-forward circuit controlling inducible NF-kappab target gene activation by promoter histone demethylation. Mol. Cell 2010, 39, 750–760. [Google Scholar] [CrossRef] [PubMed]
- Ciccone, D.N.; Su, H.; Hevi, S.; Gay, F.; Lei, H.; Bajko, J.; Xu, G.; Li, E.; Chen, T. Kdm1b is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 2009, 461, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Tsukada, Y.; Fang, J.; Erdjument-Bromage, H.; Warren, M.E.; Borchers, C.H.; Tempst, P.; Zhang, Y. Histone demethylation by a family of jmjc domain-containing proteins. Nature 2006, 439, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zang, J.; Whetstine, J.; Hong, X.; Davrazou, F.; Kutateladze, T.G.; Simpson, M.; Mao, Q.; Pan, C.H.; Dai, S.; et al. Structural insights into histone demethylation by JMJD2 family members. Cell 2006, 125, 691–702. [Google Scholar] [CrossRef] [PubMed]
- Klose, R.J.; Kallin, E.M.; Zhang, Y. JMJC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006, 7, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Iwase, S.; Lan, F.; Bayliss, P.; de la Torre-Ubieta, L.; Huarte, M.; Qi, H.H.; Whetstine, J.R.; Bonni, A.; Roberts, T.M.; Shi, Y. The x-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 2007, 128, 1077–1088. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Hong, T.; Walter, K.L.; Ewalt, M.; Michishita, E.; Hung, T.; Carney, D.; Pena, P.; Lan, F.; Kaadige, M.R.; et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 2006, 442, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Wysocka, J.; Swigut, T.; Xiao, H.; Milne, T.A.; Kwon, S.Y.; Landry, J.; Kauer, M.; Tackett, A.J.; Chait, B.T.; Badenhorst, P.; et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 2006, 442, 86–90. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, L.D.; Regan, L. TPR proteins: The versatile helix. Trends Biochem. Sci. 2003, 28, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Bai, C.; Sen, P.; Hofmann, K.; Ma, L.; Goebl, M.; Harper, J.W.; Elledge, S.J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 1996, 86, 263–274. [Google Scholar] [CrossRef]
- Kobe, B.; Kajava, A.V. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 2001, 11, 725–732. [Google Scholar] [CrossRef]
- Yang, Y.; Hu, L.; Wang, P.; Hou, H.; Lin, Y.; Liu, Y.; Li, Z.; Gong, R.; Feng, X.; Zhou, L.; et al. Structural insights into a dual-specificity histone demethylase CEKDM7A from Caenorhabditis elegans. Cell Res. 2010, 20, 886–898. [Google Scholar] [CrossRef] [PubMed]
- Deng, P.; Chen, Q.M.; Hong, C.; Wang, C.Y. Histone methyltransferases and demethylases: Regulators in balancing osteogenic and adipogenic differentiation of mesenchymal stem cells. Int. J. Oral Sci. 2015, 7, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Rotili, D.; Mai, A. Targeting histone demethylases: A new avenue for the fight against cancer. Genes Cancer 2011, 2, 663–679. [Google Scholar] [CrossRef] [PubMed]
- Frescas, D.; Guardavaccaro, D.; Bassermann, F.; Koyama-Nasu, R.; Pagano, M. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature 2007, 450, 309–313. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Yamaza, T.; Lee, J.S.; Yu, J.; Wang, S.; Fan, G.; Shi, S.; Wang, C.Y. BCOR regulates mesenchymal stem cell function by epigenetic mechanisms. Nat. Cell Biol. 2009, 11, 1002–1009. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Ma, Y.; Ma, P.; Wang, S.; Fan, Z. Demethylation of epiregulin gene by histone demethylase FBXL11 and BCL6 corepressor inhibits osteo/dentinogenic differentiation. Stem Cells 2013, 31, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Dong, R.; Yao, R.; Du, J.; Wang, S.; Fan, Z. Depletion of histone demethylase KDM2A enhanced the adipogenic and chondrogenic differentiation potentials of stem cells from apical papilla. Exp. Cell Res. 2013, 319, 2874–2882. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, T.; Iwasaki, S.; Matsumura, Y.; Kawamura, T.; Tanaka, T.; Abe, Y.; Yamasaki, A.; Tsurutani, Y.; Yoshida, A.; Chikaoka, Y.; et al. The FBXL10/KDM2B scaffolding protein associates with novel polycomb repressive complex-1 to regulate adipogenesis. J. Biol. Chem. 2015, 290, 4163–4177. [Google Scholar] [CrossRef] [PubMed]
- Meshorer, E.; Yellajoshula, D.; George, E.; Scambler, P.J.; Brown, D.T.; Misteli, T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell 2006, 10, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Chambers, I.; Colby, D.; Robertson, M.; Nichols, J.; Lee, S.; Tweedie, S.; Smith, A. Functional expression cloning of nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 2003, 113, 643–655. [Google Scholar] [CrossRef]
- Ivanova, N.; Dobrin, R.; Lu, R.; Kotenko, I.; Levorse, J.; DeCoste, C.; Schafer, X.; Lun, Y.; Lemischka, I.R. Dissecting self-renewal in stem cells with RNA interference. Nature 2006, 442, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Matoba, R.; Niwa, H.; Masui, S.; Ohtsuka, S.; Carter, M.G.; Sharov, A.A.; Ko, M.S. Dissecting OCT3/4-regulated gene networks in embryonic stem cells by expression profiling. PLoS ONE 2006, 1, e26. [Google Scholar] [CrossRef] [PubMed]
- Mitsui, K.; Tokuzawa, Y.; Itoh, H.; Segawa, K.; Murakami, M.; Takahashi, K.; Maruyama, M.; Maeda, M.; Yamanaka, S. The homeoprotein nanog is required for maintenance of pluripotency in mouse epiblast and es cells. Cell 2003, 113, 631–642. [Google Scholar] [CrossRef]
- Okada, Y.; Scott, G.; Ray, M.K.; Mishina, Y.; Zhang, Y. Histone demethylase JHDM2A is critical for TNP1 and PRM1 transcription and spermatogenesis. Nature 2007, 450, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Lockman, K.; Taylor, J.M.; Mack, C.P. The histone demethylase, JMJD1A, interacts with the myocardin factors to regulate SMC differentiation marker gene expression. Circ. Res. 2007, 101, e115–e123. [Google Scholar] [CrossRef] [PubMed]
- Saez, M.A.; Fernandez-Rodriguez, J.; Moutinho, C.; Sanchez-Mut, J.V.; Gomez, A.; Vidal, E.; Petazzi, P.; Szczesna, K.; Lopez-Serra, P.; Lucariello, M.; et al. Mutations in JMJD1C are involved in rett syndrome and intellectual disability. Genet. Med. 2016, 18, 378–385. [Google Scholar] [CrossRef] [PubMed]
- Verrier, L.; Escaffit, F.; Chailleux, C.; Trouche, D.; Vandromme, M. A new isoform of the histone demethylase JMJD2A/KDM4A is required for skeletal muscle differentiation. PLoS Genet. 2011, 7, e1001390. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Fan, Z.; Yu, B.; Chang, J.; Al Hezaimi, K.; Zhou, X.; Park, N.H.; Wang, C.Y. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human mscs. Cell Stem Cell 2012, 11, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Li, X.; Huang, J.X.; Huang, H.Y.; Zhang, Y.Y.; Qian, S.W.; Zhu, H.; Zhang, Y.D.; Liu, Y.; Liu, Y.; et al. Histone demethylase Kdm4b functions as a co-factor of c/ebpbeta to promote mitotic clonal expansion during differentiation of 3t3-l1 preadipocytes. Cell Death Differ. 2012, 19, 1917–1927. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Ward, P.S.; Kapoor, G.S.; Rohle, D.; Turcan, S.; Abdel-Wahab, O.; Edwards, C.R.; Khanin, R.; Figueroa, M.E.; Melnick, A.; et al. Idh mutation impairs histone demethylation and results in a block to cell differentiation. Nature 2012, 483, 474–478. [Google Scholar] [CrossRef] [PubMed]
- Loh, Y.H.; Zhang, W.; Chen, X.; George, J.; Ng, H.H. Jmjd1a and jmjd2c histone H3 lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007, 21, 2545–2557. [Google Scholar] [CrossRef] [PubMed]
- Iwamori, N.; Zhao, M.; Meistrich, M.L.; Matzuk, M.M. The testis-enriched histone demethylase, KDM4D, regulates methylation of histone H3 lysine 9 during spermatogenesis in the mouse but is dispensable for fertility. Biol. Reprod. 2011, 84, 1225–1234. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; van Essen, D.; Saccani, S. Cell-type-specific control of enhancer activity by H3K9 trimethylation. Mol. Cell 2012, 46, 408–423. [Google Scholar] [CrossRef] [PubMed]
- Zoabi, M.; Nadar-Ponniah, P.T.; Khoury-Haddad, H.; Usaj, M.; Budowski-Tal, I.; Haran, T.; Henn, A.; Mandel-Gutfreund, Y.; Ayoub, N. RNA-dependent chromatin localization of KDM4D lysine demethylase promotes H3K9me3 demethylation. Nucleic Acids Res. 2014, 42, 13026–13038. [Google Scholar] [CrossRef] [PubMed]
- Christensen, J.; Agger, K.; Cloos, P.A.; Pasini, D.; Rose, S.; Sennels, L.; Rappsilber, J.; Hansen, K.H.; Salcini, A.E.; Helin, K. Rbp2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell 2007, 128, 1063–1076. [Google Scholar] [CrossRef] [PubMed]
- Benevolenskaya, E.V.; Murray, H.L.; Branton, P.; Young, R.A.; Kaelin, W.G., Jr. Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol. Cell 2005, 18, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Ge, W.; Shi, L.; Zhou, Y.; Liu, Y.; Ma, G.E.; Jiang, Y.; Xu, Y.; Zhang, X.; Feng, H. Inhibition of osteogenic differentiation of human adipose-derived stromal cells by retinoblastoma binding protein 2 repression of RUNX2-activated transcription. Stem Cells 2011, 29, 1112–1125. [Google Scholar] [CrossRef] [PubMed]
- Cloos, P.A.; Christensen, J.; Agger, K.; Helin, K. Erasing the methyl mark: Histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008, 22, 1115–1140. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.; Shaw, A.L.; Madsen, B.; Jensen, K.; Taylor-Papadimitriou, J.; Freemont, P.S. Human PLU-1 has transcriptional repression properties and interacts with the developmental transcription factors BF-1 and PAX9. J. Biol. Chem. 2003, 278, 20507–20513. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.R.; Bartenschlager, H.; Rujirabanjerd, S.; Tzschach, A.; Numann, A.; Janecke, A.R.; Sporle, R.; Stricker, S.; Raynaud, M.; Nelson, J.; et al. A distinctive gene expression fingerprint in mentally retarded male patients reflects disease-causing defects in the histone demethylase KDM5C. Pathogenetics 2010, 3, 2. [Google Scholar] [CrossRef] [PubMed]
- Morales Torres, C.; Laugesen, A.; Helin, K. Utx is required for proper induction of ectoderm and mesoderm during differentiation of embryonic stem cells. PLoS ONE 2013, 8, e60020. [Google Scholar]
- Wang, C.; Lee, J.E.; Cho, Y.W.; Xiao, Y.; Jin, Q.; Liu, C.; Ge, K. Utx regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc. Natl. Acad. Sci. USA 2012, 109, 15324–15329. [Google Scholar] [CrossRef] [PubMed]
- Agger, K.; Cloos, P.A.; Rudkjaer, L.; Williams, K.; Andersen, G.; Christensen, J.; Helin, K. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009, 23, 1171–1176. [Google Scholar] [CrossRef] [PubMed]
- Barradas, M.; Anderton, E.; Acosta, J.C.; Li, S.; Banito, A.; Rodriguez-Niedenfuhr, M.; Maertens, G.; Banck, M.; Zhou, M.M.; Walsh, M.J.; et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4A/ARF by oncogenic ras. Genes Dev. 2009, 23, 1177–1182. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Li, Q.; Ayers, S.; Gu, Y.; Shi, Z.; Zhu, Q.; Chen, Y.; Wang, H.Y.; Wang, R.F. Jmjd3 inhibits reprogramming by upregulating expression of ink4a/arf and targeting phf20 for ubiquitination. Cell 2013, 152, 1037–1050. [Google Scholar] [CrossRef] [PubMed]
- Hemming, S.; Cakouros, D.; Isenmann, S.; Cooper, L.; Menicanin, D.; Zannettino, A.; Gronthos, S. EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells 2014, 32, 802–815. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Yu, B.; Hong, C.; Wang, C.Y. KDM6B epigenetically regulates odontogenic differentiation of dental mesenchymal stem cells. Int. J. Oral Sci. 2013, 5, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Shpargel, K.B.; Sengoku, T.; Yokoyama, S.; Magnuson, T. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 2012, 8, e1002964. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Park, J.W.; Chun, Y.S. Jumonji histone demethylases as emerging therapeutic targets. Pharmacol. Res. 2016, 105, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Ramos, C.; Robert, B. MSH/MSX gene family in neural development. Trends Genet. 2005, 21, 624–632. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Xiang, Y.; Wang, Y.; Li, X.; Xu, L.; Zhu, Z.; Zhang, T.; Zhu, Q.; Zhang, K.; Jing, N.; et al. Dual-specificity histone demethylase KIAA1718 (KDM7A) regulates neural differentiation through FGF4. Cell Res. 2010, 20, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Park, J.W.; Lee, K.H.; Yoon, H.; Shin, D.H.; Ju, U.I.; Seok, S.H.; Lim, S.H.; Lee, Z.H.; Kim, H.H.; et al. Plant homeodomain finger protein 2 promotes bone formation by demethylating and activating runx2 for osteoblast differentiation. Cell Res. 2014, 24, 1231–1249. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Cheung, P.; Kuo, A.J.; Yukl, E.T.; Wilmot, C.M.; Gozani, O.; Patel, D.J. A molecular threading mechanism underlies jumonji lysine demethylase KDM2A regulation of methylated H3K36. Genes Dev. 2014, 28, 1758–1771. [Google Scholar] [CrossRef] [PubMed]
- Frescas, D.; Guardavaccaro, D.; Kuchay, S.M.; Kato, H.; Poleshko, A.; Basrur, V.; Elenitoba-Johnson, K.S.; Katz, R.A.; Pagano, M. KDM2A represses transcription of centromeric satellite repeats and maintains the heterochromatic state. Cell Cycle 2008, 7, 3539–3547. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Jackson, M.W.; Wang, B.; Yang, M.; Chance, M.R.; Miyagi, M.; Gudkov, A.V.; Stark, G.R. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc. Natl. Acad. Sci. USA 2010, 107, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Liu, Y.; Yu, L.; Chen, J.; Hou, J.; Cui, L.; Ma, D.; Lu, W. Histone demethylase KDM2A promotes tumor cell growth and migration in gastric cancer. Tumour Biol. 2015, 36, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, L.; Holowatyj, A.; Jiang, Y.; Yang, Z.Q. Integrated genomic and functional analyses of histone demethylases identify oncogenic KDM2A isoform in breast cancer. Mol. Carcinog. 2016, 55, 977–990. [Google Scholar] [CrossRef] [PubMed]
- Wagner, K.W.; Alam, H.; Dhar, S.S.; Giri, U.; Li, N.; Wei, Y.; Giri, D.; Cascone, T.; Kim, J.H.; Ye, Y.; et al. KDM2A promotes lung tumorigenesis by epigenetically enhancing erk1/2 signaling. J. Clin. Investig. 2013, 123, 5231–5246. [Google Scholar] [CrossRef] [PubMed]
- Rizwani, W.; Schaal, C.; Kunigal, S.; Coppola, D.; Chellappan, S. Mammalian lysine histone demethylase KDM2a regulates E2F1-mediated gene transcription in breast cancer cells. PLoS ONE 2014, 9, e100888. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Minehata, K.; Akagi, K.; Jenkins, N.A.; Copeland, N.G. Tumor suppressor gene identification using retroviral insertional mutagenesis in blm-deficient mice. EMBO J. 2006, 25, 3422–3431. [Google Scholar] [CrossRef] [PubMed]
- Tzatsos, A.; Paskaleva, P.; Ferrari, F.; Deshpande, V.; Stoykova, S.; Contino, G.; Wong, K.K.; Lan, F.; Trojer, P.; Park, P.J.; et al. KDM2B promotes pancreatic cancer via polycomb-dependent and -independent transcriptional programs. J. Clin. Investig. 2013, 123, 727–739. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Nguyen, A.T.; Zhang, Y. Kdm2b/jhdm1b, an H3K36me2-specific demethylase, is required for initiation and maintenance of acute myeloid leukemia. Blood 2011, 117, 3869–3880. [Google Scholar] [CrossRef] [PubMed]
- Gumz, M.L.; Zou, H.; Kreinest, P.A.; Childs, A.C.; Belmonte, L.S.; LeGrand, S.N.; Wu, K.J.; Luxon, B.A.; Sinha, M.; Parker, A.S.; et al. Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin. Cancer Res. 2007, 13, 4740–4749. [Google Scholar] [CrossRef] [PubMed]
- Uemura, M.; Yamamoto, H.; Takemasa, I.; Mimori, K.; Hemmi, H.; Mizushima, T.; Ikeda, M.; Sekimoto, M.; Matsuura, N.; Doki, Y.; et al. Jumonji domain containing 1a is a novel prognostic marker for colorectal cancer: In vivo identification from hypoxic tumor cells. Clin. Cancer Res. 2010, 16, 4636–4646. [Google Scholar] [CrossRef] [PubMed]
- Wade, M.A.; Jones, D.; Wilson, L.; Stockley, J.; Coffey, K.; Robson, C.N.; Gaughan, L. The histone demethylase enzyme KDM3A is a key estrogen receptor regulator in breast cancer. Nucleic Acids Res. 2015, 43, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Krieg, A.J.; Rankin, E.B.; Chan, D.; Razorenova, O.; Fernandez, S.; Giaccia, A.J. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol. Cell. Biol. 2010, 30, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Chen, M.; Eng, R.; DeJong, J.; Sinha, A.U.; Rahnamay, N.F.; Koche, R.; Al-Shahrour, F.; Minehart, J.C.; Chen, C.W.; et al. MLL-AF9- and HOXA9-mediated acute myeloid leukemia stem cell self-renewal requires JMJD1C. J. Clin. Investig. 2016, 126, 997–1011. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.S.; Patchev, V.K.; Obendorf, M. A novel variant of the putative demethylase gene, s-JMJD1C, is a coactivator of the AR. Arch. Biochem. Biophys. 2007, 460, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Cloos, P.A.; Christensen, J.; Agger, K.; Maiolica, A.; Rappsilber, J.; Antal, T.; Hansen, K.H.; Helin, K. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 2006, 442, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Ehrbrecht, A.; Muller, U.; Wolter, M.; Hoischen, A.; Koch, A.; Radlwimmer, B.; Actor, B.; Mincheva, A.; Pietsch, T.; Lichter, P.; et al. Comprehensive genomic analysis of desmoplastic medulloblastomas: Identification of novel amplified genes and separate evaluation of the different histological components. J. Pathol. 2006, 208, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Bollig-Fischer, A.; Kreike, B.; van de Vijver, M.J.; Abrams, J.; Ethier, S.P.; Yang, Z.Q. Genomic amplification and oncogenic properties of the GASC1 histone demethylase gene in breast cancer. Oncogene 2009, 28, 4491–4500. [Google Scholar] [CrossRef] [PubMed]
- Uimonen, K.; Merikallio, H.; Paakko, P.; Harju, T.; Mannermaa, A.; Palvimo, J.; Kosma, V.M.; Soini, Y. GASC1 expression in lung carcinoma is associated with smoking and prognosis of squamous cell carcinoma. Histol. Histopathol. 2014, 29, 797–804. [Google Scholar] [PubMed]
- Yang, Z.Q.; Imoto, I.; Fukuda, Y.; Pimkhaokham, A.; Shimada, Y.; Imamura, M.; Sugano, S.; Nakamura, Y.; Inazawa, J. Identification of a novel gene, GASC1, within an amplicon at 9p23–24 frequently detected in esophageal cancer cell lines. Cancer Res. 2000, 60, 4735–4739. [Google Scholar] [PubMed]
- Hu, C.E.; Liu, Y.C.; Zhang, H.D.; Huang, G.J. JMJD2A predicts prognosis and regulates cell growth in human gastric cancer. Biochem. Biophys. Res. Commun. 2014, 449, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.C.; Lee, C.F.; Li, Y.S.; Chen, Y.R.; Hsiao, P.W.; Chan, M.Y.; Lin, F.M.; Huang, H.D.; Chen, Y.T.; Jeng, Y.M.; et al. Histone demethylase rbp2 promotes lung tumorigenesis and cancer metastasis. Cancer Res. 2013, 73, 4711–4721. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Ge, Z.; Wang, L.; Li, Q.; Wang, N.; Bjorkholm, M.; Jia, J.; Xu, D. The histone demethylase rbp2 is overexpressed in gastric cancer and its inhibition triggers senescence of cancer cells. Gastroenterology 2010, 138, 981–992. [Google Scholar] [CrossRef] [PubMed]
- Roesch, A.; Becker, B.; Meyer, S.; Wild, P.; Hafner, C.; Landthaler, M.; Vogt, T. Retinoblastoma-binding protein 2-homolog 1: A retinoblastoma-binding protein downregulated in malignant melanomas. Mod. Pathol. 2005, 18, 1249–1257. [Google Scholar] [CrossRef] [PubMed]
- Barrett, A.; Madsen, B.; Copier, J.; Lu, P.J.; Cooper, L.; Scibetta, A.G.; Burchell, J.; Taylor-Papadimitriou, J. Plu-1 nuclear protein, which is upregulated in breast cancer, shows restricted expression in normal human adult tissues: A new cancer/testis antigen? Int. J. Cancer 2002, 101, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.J.; Sundquist, K.; Baeckstrom, D.; Poulsom, R.; Hanby, A.; Meier-Ewert, S.; Jones, T.; Mitchell, M.; Pitha-Rowe, P.; Freemont, P.; et al. A novel gene (plu-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J. Biol. Chem. 1999, 274, 15633–15645. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Qi, G.; Tang, F.; Yuan, S.; Wang, Z.; Liang, X.; Li, B.; Yu, S.; Liu, J.; Huang, Q.; et al. Jarid1b promotes metastasis and epithelial-mesenchymal transition via pten/akt signaling in hepatocellular carcinoma cells. Oncotarget 2015, 6, 12723–12739. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Zhu, Z.; Han, G.; Ye, X.; Xu, B.; Peng, Z.; Ma, Y.; Yu, Y.; Lin, H.; Chen, A.P.; et al. Jarid1b is a histone h3 lysine 4 demethylase up-regulated in prostate cancer. Proc. Natl. Acad. Sci. USA 2007, 104, 19226–19231. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Han, S.; Peng, R.; Jiao, C.; Wang, X.; Yang, X.; Yang, R.; Li, X. Depletion of histone demethylase KDM5B inhibits cell proliferation of hepatocellular carcinoma by regulation of cell cycle checkpoint proteins p15 and p27. J. Exp. Clin. Cancer Res. 2016, 35, 37. [Google Scholar] [CrossRef] [PubMed]
- Stein, J.; Majores, M.; Rohde, M.; Lim, S.; Schneider, S.; Krappe, E.; Ellinger, J.; Dietel, M.; Stephan, C.; Jung, K.; et al. KDM5C is overexpressed in prostate cancer and is a prognostic marker for prostate-specific antigen-relapse following radical prostatectomy. Am. J. Pathol. 2014, 184, 2430–2437. [Google Scholar] [CrossRef] [PubMed]
- Rondinelli, B.; Rosano, D.; Antonini, E.; Frenquelli, M.; Montanini, L.; Huang, D.; Segalla, S.; Yoshihara, K.; Amin, S.B.; Lazarevic, D.; et al. Histone demethylase JARID1c inactivation triggers genomic instability in sporadic renal cancer. J. Clin. Investig. 2015, 125, 4625–4637. [Google Scholar] [CrossRef] [PubMed]
- Dalgliesh, G.L.; Furge, K.; Greenman, C.; Chen, L.; Bignell, G.; Butler, A.; Davies, H.; Edkins, S.; Hardy, C.; Latimer, C.; et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 2010, 463, 360–363. [Google Scholar] [CrossRef] [PubMed]
- Gui, Y.; Guo, G.; Huang, Y.; Hu, X.; Tang, A.; Gao, S.; Wu, R.; Chen, C.; Li, X.; Zhou, L.; et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat. Genet. 2011, 43, 875–878. [Google Scholar] [CrossRef] [PubMed]
- Ntziachristos, P.; Tsirigos, A.; Welstead, G.G.; Trimarchi, T.; Bakogianni, S.; Xu, L.; Loizou, E.; Holmfeldt, L.; Strikoudis, A.; King, B.; et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature 2014, 514, 513–517. [Google Scholar] [CrossRef] [PubMed]
- Van Haaften, G.; Dalgliesh, G.L.; Davies, H.; Chen, L.; Bignell, G.; Greenman, C.; Edkins, S.; Hardy, C.; O’Meara, S.; Teague, J.; et al. Somatic mutations of the histone H3K27 demethylase gene utx in human cancer. Nat. Genet. 2009, 41, 521–523. [Google Scholar] [CrossRef] [PubMed]
- Pereira, F.; Barbachano, A.; Silva, J.; Bonilla, F.; Campbell, M.J.; Munoz, A.; Larriba, M.J. KDM6B/JMJD3 histone demethylase is induced by vitamin d and modulates its effects in colon cancer cells. Hum. Mol. Genet. 2011, 20, 4655–4665. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, R.; Nakagawa, S.; Sakamoto, Y.; Karashima, R.; Ida, S.; Imamura, Y.; Ishimoto, T.; Iwagami, S.; Baba, Y.; Miyamoto, Y.; et al. Abstract 5150: JMJD3 suppresses progression of colorectal carcinoma by regulating cell cycle and anti-apoptosis. Cancer Res. 2014, 74, 5150. [Google Scholar] [CrossRef]
- Yamamoto, K.; Tateishi, K.; Kudo, Y.; Sato, T.; Yamamoto, S.; Miyabayashi, K.; Matsusaka, K.; Asaoka, Y.; Ijichi, H.; Hirata, Y.; et al. Loss of histone demethylase KDM6B enhances aggressiveness of pancreatic cancer through downregulation of C/EBPalpha. Carcinogenesis 2014, 35, 2404–2414. [Google Scholar] [CrossRef] [PubMed]
- Anderton, J.A.; Bose, S.; Vockerodt, M.; Vrzalikova, K.; Wei, W.; Kuo, M.; Helin, K.; Christensen, J.; Rowe, M.; Murray, P.G.; et al. The H3K27me3 demethylase, KDM6B, is induced by epstein-barr virus and over-expressed in hodgkin’s lymphoma. Oncogene 2011, 30, 2037–2043. [Google Scholar] [CrossRef] [PubMed]
- Park, W.Y.; Hong, B.J.; Lee, J.; Choi, C.; Kim, M.Y. H3k27 demethylase JMJD3 employs the NF-kappaB and bmp signaling pathways to modulate the tumor microenvironment and promote melanoma progression and metastasis. Cancer Res. 2016, 76, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Perrigue, P.M.; Najbauer, J.; Barciszewski, J. Histone demethylase JMJD3 at the intersection of cellular senescence and cancer. Biochim. Biophys. Acta 2016, 1865, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Ramadoss, S.; Chen, X.; Wang, C.Y. Histone demethylase KDM6B promotes epithelial-mesenchymal transition. J. Biol. Chem. 2012, 287, 44508–44517. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Guo, X.; Wang, Y.; Qiu, W.; Chang, Y.; Zhang, A.; Duan, X. Expression and significance of histone H3K27 demethylases in renal cell carcinoma. BMC Cancer 2012, 12, 470. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Chen, R.; Dimicoli, S.; Bueso-Ramos, C.; Neuberg, D.; Pierce, S.; Wang, H.; Yang, H.; Jia, Y.; Zheng, H.; et al. Global H3K4ME3 genome mapping reveals alterations of innate immunity signaling and overexpression of jmjd3 in human myelodysplastic syndrome CD34+ cells. Leukemia 2013, 27, 2177–2186. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Zhu, Z.; Han, G.; Lin, H.; Xu, L.; Chen, C.D. Jmjd3 is a histone h3k27 demethylase. Cell Res. 2007, 17, 850–857. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Hou, L.; Ding, G.; Li, Y.; Wang, J.; Qian, B.; Sun, J.; Wang, Q. Kdm6b induces epithelial-mesenchymal transition and enhances clear cell renal cell carcinoma metastasis through the activation of slug. Int. J. Clin. Exp. Pathol. 2015, 8, 6334–6344. [Google Scholar] [PubMed]
- Sun, X.; Qiu, J.J.; Zhu, S.; Cao, B.; Sun, L.; Li, S.; Li, P.; Zhang, S.; Dong, S. Oncogenic features of phf8 histone demethylase in esophageal squamous cell carcinoma. PLoS ONE 2013, 8, e77353. [Google Scholar] [CrossRef] [PubMed]
- Bjorkman, M.; Ostling, P.; Harma, V.; Virtanen, J.; Mpindi, J.P.; Rantala, J.; Mirtti, T.; Vesterinen, T.; Lundin, M.; Sankila, A.; et al. Systematic knockdown of epigenetic enzymes identifies a novel histone demethylase PHF8 overexpressed in prostate cancer with an impact on cell proliferation, migration and invasion. Oncogene 2012, 31, 3444–3456. [Google Scholar] [CrossRef] [PubMed]
- Asensio-Juan, E.; Gallego, C.; Martinez-Balbas, M.A. The histone demethylase PHF8 is essential for cytoskeleton dynamics. Nucleic Acids Res. 2012, 40, 9429–9440. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.J.; Dimova, N.V.; Tan, M.K.; Sigoillot, F.D.; King, R.W.; Shi, Y. The G2/M regulator histone demethylase PHF8 is targeted for degradation by the anaphase-promoting complex containing CDC20. Mol. Cell. Biol. 2013, 33, 4166–4180. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.H.; Sarkissian, M.; Hu, G.Q.; Wang, Z.; Bhattacharjee, A.; Gordon, D.B.; Gonzales, M.; Lan, F.; Ongusaha, P.P.; Huarte, M.; et al. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 2010, 466, 503–507. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lin, X.; Wang, S.; Wang, C.; Wang, Q.; Duan, X.; Lu, P.; Wang, Q.; Wang, C.; Liu, X.S.; et al. PHF8 and REST/NRSF co-occupy gene promoters to regulate proximal gene expression. Sci. Rep. 2014, 4, 5008. [Google Scholar] [CrossRef] [PubMed]
- Arteaga, M.F.; Mikesch, J.H.; Qiu, J.; Christensen, J.; Helin, K.; Kogan, S.C.; Dong, S.; So, C.W. The histone demethylase PHF8 governs retinoic acid response in acute promyelocytic leukemia. Cancer Cell 2013, 23, 376–389. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Park, J.W.; Sung, H.S.; Choi, Y.J.; Kim, W.H.; Lee, H.S.; Chung, H.J.; Shin, H.W.; Cho, C.H.; Kim, T.Y.; et al. PHF2 histone demethylase acts as a tumor suppressor in association with p53 in cancer. Oncogene 2015, 34, 2897–2909. [Google Scholar] [CrossRef] [PubMed]
- Williamson, P.R.; Kagan, H.M. Reaction pathway of bovine aortic lysyl oxidase. J. Biol. Chem. 1986, 261, 9477–9482. [Google Scholar] [PubMed]
- Barker, H.E.; Cox, T.R.; Erler, J.T. The rationale for targeting the lox family in cancer. Nat. Rev. Cancer 2012, 12, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.C. Biosynthesis of collagen crosslinks: Increased activity of purified lysyl oxidase with reconstituted collagen fibrils. Proc. Natl. Acad. Sci. USA 1974, 71, 4826–4830. [Google Scholar] [CrossRef] [PubMed]
- Barker, H.E.; Chang, J.; Cox, T.R.; Lang, G.; Bird, D.; Nicolau, M.; Evans, H.R.; Gartland, A.; Erler, J.T. Loxl2-mediated matrix remodeling in metastasis and mammary gland involution. Cancer Res. 2011, 71, 1561–1572. [Google Scholar] [CrossRef] [PubMed]
- Herranz, N.; Dave, N.; Millanes-Romero, A.; Pascual-Reguant, L.; Morey, L.; Diaz, V.M.; Lorenz-Fonfria, V.; Gutierrez-Gallego, R.; Jeronimo, C.; Iturbide, A.; et al. Lysyl oxidase-like 2 (loxl2) oxidizes trimethylated lysine 4 in histone H3. FEBS J. 2016, 283, 4263–4273. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.X.; Mure, M.; Medzihradszky, K.F.; Burlingame, A.L.; Brown, D.E.; Dooley, D.M.; Smith, A.J.; Kagan, H.M.; Klinman, J.P. A crosslinked cofactor in lysyl oxidase: Redox function for amino acid side chains. Science 1996, 273, 1078–1084. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, E.; Tajima, S. Reciprocal regulation of lox and loxl2 expression during cell adhesion and terminal differentiation in epidermal keratinocytes. J. Dermatol. Sci. 2009, 55, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Slaga, T.J.; Budunova, I.V.; Gimenez-Conti, I.B.; Aldaz, C.M. The mouse skin carcinogenesis model. J. Investig. Dermatol. Symp. Proc. 1996, 1, 151–156. [Google Scholar] [PubMed]
- Kim, J.H.; Lee, E.H.; Park, H.J.; Park, E.K.; Kwon, T.G.; Shin, H.I.; Cho, J.Y. The role of lysyl oxidase-like 2 in the odontogenic differentiation of human dental pulp stem cells. Mol. Cells 2013, 35, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Iftikhar, M.; Hurtado, P.; Bais, M.V.; Wigner, N.; Stephens, D.N.; Gerstenfeld, L.C.; Trackman, P.C. Lysyl oxidase-like-2 (loxl2) is a major isoform in chondrocytes and is critically required for differentiation. J. Biol. Chem. 2011, 286, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Iturbide, A.; Pascual-Reguant, L.; Fargas, L.; Cebria, J.P.; Alsina, B.; Garcia de Herreros, A.; Peiro, S. LOXL2 oxidizes methylated TAF10 and controls TFIID-dependent genes during neural progenitor differentiation. Mol. Cell 2015, 58, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Schietke, R.; Warnecke, C.; Wacker, I.; Schodel, J.; Mole, D.R.; Campean, V.; Amann, K.; Goppelt-Struebe, M.; Behrens, J.; Eckardt, K.U.; et al. The lysyl oxidases LOX and LOXL2 are necessary and sufficient to repress E-cadherin in hypoxia: Insights into cellular transformation processes mediated by HIF-1. J. Biol. Chem. 2010, 285, 6658–6669. [Google Scholar] [CrossRef] [PubMed]
- Voloshenyuk, T.G.; Landesman, E.S.; Khoutorova, E.; Hart, A.D.; Gardner, J.D. Induction of cardiac fibroblast lysyl oxidase by TGF-Beta1 requires PI3K/AKT, SMAD3, and MAPK signaling. Cytokine 2011, 55, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Barry-Hamilton, V.; Spangler, R.; Marshall, D.; McCauley, S.; Rodriguez, H.M.; Oyasu, M.; Mikels, A.; Vaysberg, M.; Ghermazien, H.; Wai, C.; et al. Allosteric inhibition of lysyl oxidase-like-2 impedes the development of a pathologic microenvironment. Nat. Med. 2010, 16, 1009–1017. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Ran, Y.L.; Hu, H.; Yu, L.; Liu, Q.; Zhou, Z.; Sun, Y.M.; Sun, L.C.; Pan, J.; Sun, L.X.; et al. Secreted LOXL2 is a novel therapeutic target that promotes gastric cancer metastasis via the SRC/FAK pathway. Carcinogenesis 2009, 30, 1660–1669. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.C.; Tse, A.P.; Huang, Y.P.; Zhu, Y.T.; Chiu, D.K.; Lai, R.K.; Au, S.L.; Kai, A.K.; Lee, J.M.; Wei, L.L.; et al. Lysyl oxidase-like 2 is critical to tumor microenvironment and metastatic niche formation in hepatocellular carcinoma. Hepatology 2014, 60, 1645–1658. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Sengupta, R.; Espejo, A.B.; Lee, M.G.; Dorsey, J.A.; Richter, M.; Opravil, S.; Shiekhattar, R.; Bedford, M.T.; Jenuwein, T.; et al. P53 is regulated by the lysine demethylase lsd1. Nature 2007, 449, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hevi, S.; Kurash, J.K.; Lei, H.; Gay, F.; Bajko, J.; Su, H.; Sun, W.; Chang, H.; Xu, G.; et al. The lysine demethylasE LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 2009, 41, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Sola, S.; Xavier, J.M.; Santos, D.M.; Aranha, M.M.; Morgado, A.L.; Jepsen, K.; Rodrigues, C.M. P53 interaction with JMJD3 results in its nuclear distribution during mouse neural stem cell differentiation. PLoS ONE 2011, 6, e18421. [Google Scholar] [CrossRef] [PubMed]
- Kontaki, H.; Talianidis, I. Lysine methylation regulates E2F1-induced cell death. Mol. Cell 2010, 39, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.S.; Suzuki, T.; Dohmae, N.; Hayami, S.; Unoki, M.; Yoshimatsu, M.; Toyokawa, G.; Takawa, M.; Chen, T.; Kurash, J.K.; et al. Demethylation of Rb regulator MYPT1 by histone demethylase LSD1 promotes cell cycle progression in cancer cells. Cancer Res. 2011, 71, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Hoesel, B.; Schmid, J.A. The complexity of NF-kappaB signaling in inflammation and cancer. Mol. Cancer 2013, 12, 86. [Google Scholar] [CrossRef] [PubMed]
- Farcas, A.M.; Blackledge, N.P.; Sudbery, I.; Long, H.K.; McGouran, J.F.; Rose, N.R.; Lee, S.; Sims, D.; Cerase, A.; Sheahan, T.W.; et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. eLife 2012, 1, e00205. [Google Scholar] [CrossRef] [PubMed]
- Long, H.K.; Blackledge, N.P.; Klose, R.J. Zf-cxxc domain-containing proteins, CpG islands and the chromatin connection. Biochem. Soc. Trans. 2013, 41, 727–740. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Shen, L.; Wan, M.; Taranova, O.; Wu, H.; Zhang, Y. KDM2B maintains murine embryonic stem cell status by recruiting PRC1 complex to cpg islands of developmental genes. Nat. Cell Biol. 2013, 15, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Abe, Y.; Rozqie, R.; Matsumura, Y.; Kawamura, T.; Nakaki, R.; Tsurutani, Y.; Tanimura-Inagaki, K.; Shiono, A.; Magoori, K.; Nakamura, K.; et al. JMJD1A is a signal-sensing scaffold that regulates acute chromatin dynamics via SWI/SNF association for thermogenesis. Nature Commun. 2015, 6, 7052. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.A.; Mohn, S.E.; Weinmann, A.S. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol. Cell 2010, 40, 594–605. [Google Scholar] [CrossRef] [PubMed]

| Names | Cellular Differentiation | Ref. | Putative Cancer Role | Ref. |
|---|---|---|---|---|
| KDM1A | Hematopoiesis, cerebral cortex development, brown adipogenesis, EMT | [24,25,26,27,28,29,30] | Oncogene | [31,32,33,34,35,36,37,38,39,40] |
| KDM1B | Embryo development | [58] | Tumor suppressor | [54] |
| KDM2A | Osteo/dentinogenic differentiation, adipogenesis | [88,89,90] | Pro- and anti-oncogenic functions | [126,127,128,129,130,131,132] |
| KDM2B | Osteogenesis; adipogenesis | [88,91] | Pro- and anti-oncogenic functions | [87,133,134,135] |
| KDM3A | Spermatogenesis; smooth muscle cell differentiation | [97,98] | Oncogene | [136,137,138,139] |
| KDM3B | Leukemogenesis | [98,140] | Tumor suppressor | [139] |
| KDM3C | Neurogenesis? | [99] | Tumor suppressor | [139,140,141] |
| KDM4A | Skeletal muscle cell differentiation | [100] | Oncogene | [142,147] |
| KDM4B | Osteoblast differentiation; adipogenesis | [85,101,102] | Oncogene | [142] |
| KDM4C | Adipogenesis | [103] | Oncogene | [142,143,144,145,146] |
| KDM4D | Spermatogenesis? | [105] | NR | |
| KDM5A | Osteogenesis | [110] | Pro- and anti-oncogenic functions | [136,148,149,150] |
| KDM5B | Neural cell differentiation, craniofacial and development | [111,112] | Oncogene | [151,152,153,154] |
| KDM5C | Brain development | [113] | Oncogene | [156,157] |
| KDM6A | Ectoderm differentiation; cell reprogramming; osteoblast differentiation | [114,115,119] | Tumor suppressor | [158,159,160,161] |
| KDM6B | Neuronal and epidermal differentiation; cell reprogramming; osteogenic and adipogenic differentiation | [101,118,120] | Pro- and anti-oncogenic functions | [116,160,162,163,164,165,166,167,168,169,170,171,172] |
| KDM7A | Neuronal differentiation, craniofacial development | [123] | Oncogene | [173,174,175,176,177,178] |
| KDM7B | Neuronal differentiation | [124] | NR | |
| KDM7C | Osteoblast differentiation; bone formation | [125] | Tumor suppressor | [180] |
| LOXL2 | Keratinogenesis; odontoblast genesis; chondrogenesis; zebrafish development; EMT | [171,173,174,175,176,177] | Oncogene | [166,178,179,180] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verde, G.; Querol-Paños, J.; Cebrià-Costa, J.P.; Pascual-Reguant, L.; Serra-Bardenys, G.; Iturbide, A.; Peiró, S. Lysine-Specific Histone Demethylases Contribute to Cellular Differentiation and Carcinogenesis. Epigenomes 2017, 1, 4. https://doi.org/10.3390/epigenomes1010004
Verde G, Querol-Paños J, Cebrià-Costa JP, Pascual-Reguant L, Serra-Bardenys G, Iturbide A, Peiró S. Lysine-Specific Histone Demethylases Contribute to Cellular Differentiation and Carcinogenesis. Epigenomes. 2017; 1(1):4. https://doi.org/10.3390/epigenomes1010004
Chicago/Turabian StyleVerde, Gaetano, Jessica Querol-Paños, Joan Pau Cebrià-Costa, Laura Pascual-Reguant, Gemma Serra-Bardenys, Ane Iturbide, and Sandra Peiró. 2017. "Lysine-Specific Histone Demethylases Contribute to Cellular Differentiation and Carcinogenesis" Epigenomes 1, no. 1: 4. https://doi.org/10.3390/epigenomes1010004
APA StyleVerde, G., Querol-Paños, J., Cebrià-Costa, J. P., Pascual-Reguant, L., Serra-Bardenys, G., Iturbide, A., & Peiró, S. (2017). Lysine-Specific Histone Demethylases Contribute to Cellular Differentiation and Carcinogenesis. Epigenomes, 1(1), 4. https://doi.org/10.3390/epigenomes1010004





