Development of a Diagnostic Marker for Phlebotomus papatasi to Initiate a Potential Vector Surveillance Program in North America
Abstract
1. Introduction
2. Materials and Methods
2.1. Sand Fly Collection and Storage
2.2. Taxonomic Identification
2.3. Genomic DNA Extraction and Sample Preparation
2.4. Primer Design
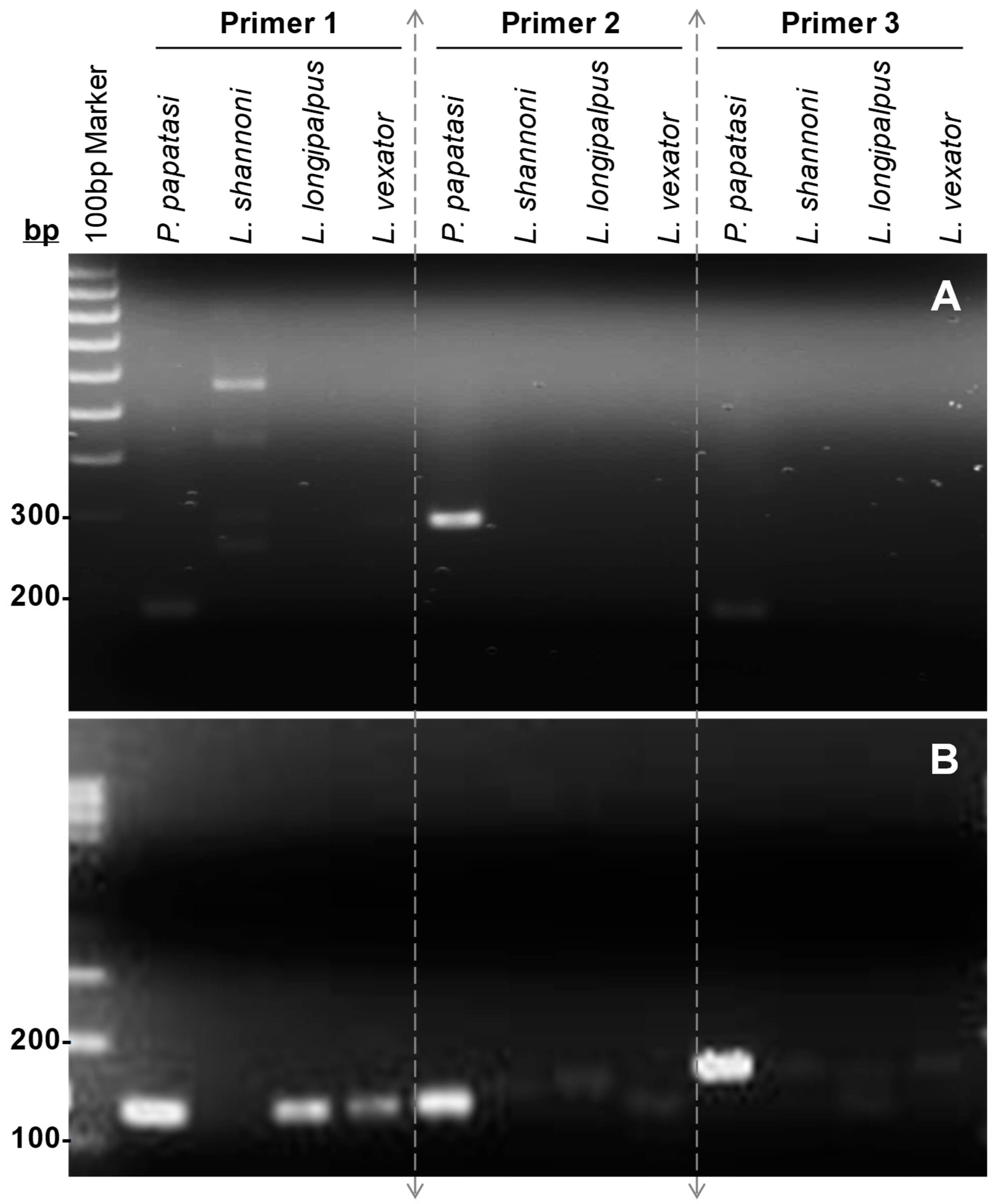
2.5. PCR Amplification
2.6. In Silico Analysis of the Specificity of the Selected Diagnostic Marker
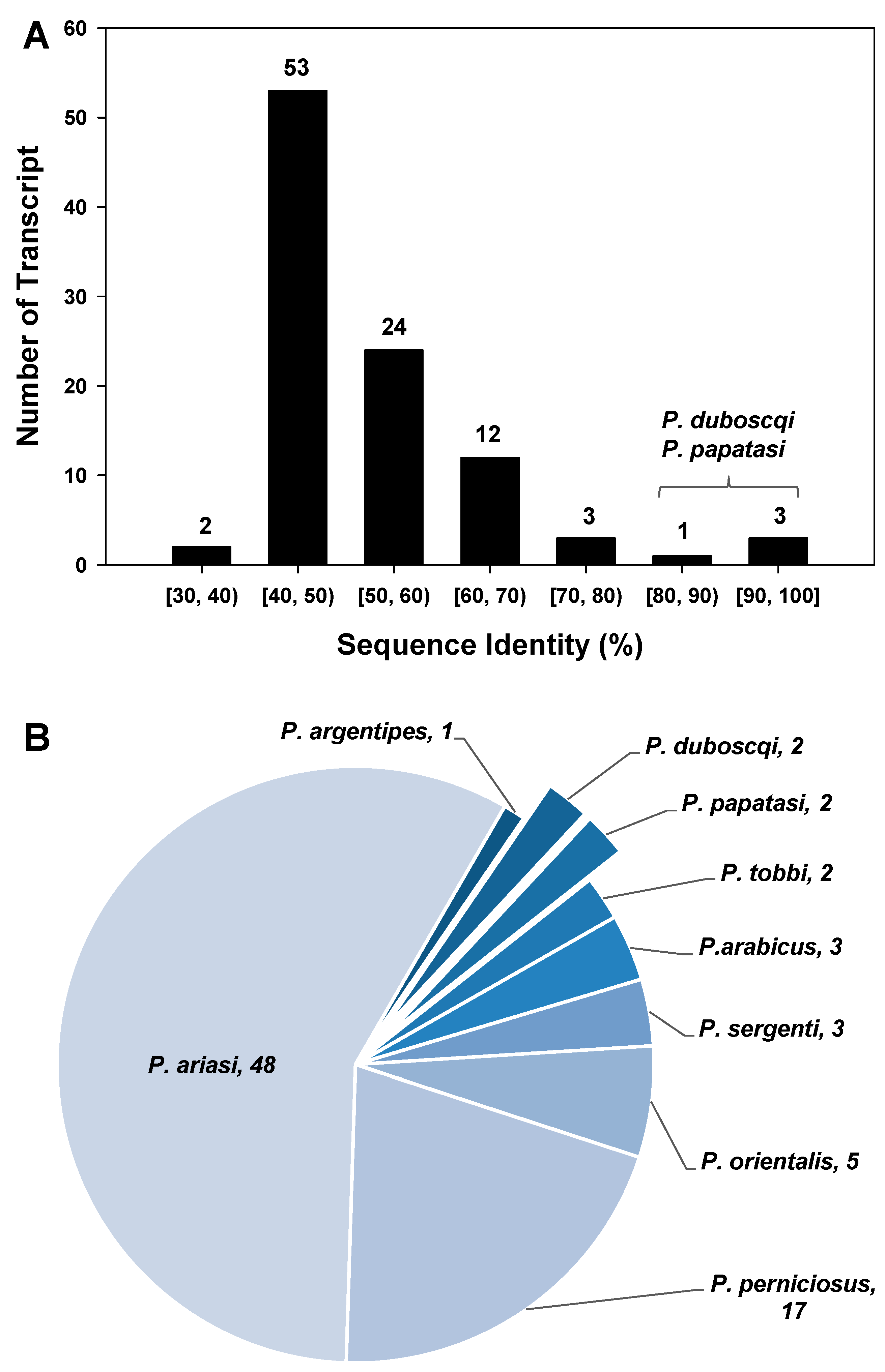
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alvar, J.; Velez, I.D.; Bern, C.; Herrero, M.; Desjeux, P.; Cano, J.; Jannin, J.; den Boer, M.; WHO Leishmaniasis Control Team. Leishmaniasis worldwide and global estimates of its incidence. PLoS ONE 2012, 7, e35671. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Leishmaniasis: Fact Sheet. Available online: http://www.who.int/news-room/fact-sheets/detail/leishmaniasis (accessed on 14 September 2018).
- Maroli, M.; Feliciangeli, M.D.; Bichaud, L.; Charrel, R.N.; Gradoni, L. Phlebotomine sandflies and the spreading of leishmaniases and other diseases of public health concern. Med. Vet. Entomol. 2013, 27, 123–147. [Google Scholar] [CrossRef] [PubMed]
- Molyneux, D.H.; Ashford, R.W. The Biology of Trypanosoma and Leishmania, Parasites of Man and Domestic Animals; Taylor & Francis Ltd.: London, UK, 1983; ISBN 0850662427. [Google Scholar]
- Young, D.G.; Duran, M.A. Guide to the Identification and Geographic Distribution of Lutzomyia Sand Flies in Mexico, the West. Indies, Central and South. America (Diptera: Psychodidae); Walter Reed Army Inst of Research: Washington, DC, USA, 1994. [Google Scholar]
- Ferro, C.; Cardenas, E.; Corredor, D.; Morales, A.; Munstermann, L.E. Life cycle and fecundity analysis of Lutzomyia shannoni (Dyar) (Diptera: Psychodidae). Mem. Inst. Oswaldo Cruz 1998, 93, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Lawyer, P.G.; Young, D.G. Experimental transmission of Leishmania mexicana to hamsters by bites of phlebotomine sand flies (Diptera: Psychodidae) from the United States. J. Med. Entomol. 1987, 24, 458–462. [Google Scholar] [CrossRef] [PubMed]
- Minter, L.M. Mesoscale Spatial and Temporal Distribution of Lutzomyia spp. (Diptera: Psychodidae) in Deciduous Habitats of the Eastern United States; University of Kentucky: Lexington, KY, USA, 2010. [Google Scholar]
- McIlwee, B.E.; Weis, S.E.; Hosler, G.A. Incidence of endemic human cutaneous leishmaniasis in the United States. JAMA Dermatol. 2018, 154, 1032–1039. [Google Scholar] [CrossRef] [PubMed]
- Kipp, E.J.; Mariscal, J.; Armijos, R.X.; Weigel, M.; Waldrup, K. Genetic evidence of enzootic leishmaniasis in a stray canine and Texas mouse from sites in west and central Texas. Mem. Inst. Oswaldo Cruz 2016, 111, 652–654. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.F.; Bradley, K.K.; Wright, J.H.; Glowicz, J. Case report: Emergence of autochthonous cutaneous leishmaniasis in northeastern Texas and southeastern Oklahoma. Am. J. Trop. Med. Hyg. 2013, 88, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Douvoyiannis, M.; Khromachou, T.; Byers, N.; Hargreaves, J.; Murray, H.W. Cutaneous leishmaniasis in North Dakota. Clin. Infect. Dis. 2014, 59, e73–e75. [Google Scholar] [CrossRef] [PubMed]
- Aspock, H.; Gerersdorfer, T.; Formayer, H.; Walochnik, J. Sandflies and sandfly-borne infections of humans in Central Europe in the light of climate change. Wien. Klin. Wochenschr. 2008, 120, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Vaselek, S.; Ayhan, N.; Oguz, G.; Erisoz Kasap, O.; Savic, S.; Di Muccio, T.; Gradoni, L.; Ozbel, Y.; Alten, B.; Petric, D. Sand fly and Leishmania spp. survey in Vojvodina (Serbia): First detection of Leishmania infantum DNA in sand flies and the first record of Phlebotomus (Transphlebotomus) mascittii Grassi, 1908. Parasites Vectors 2017, 10, 444. [Google Scholar] [CrossRef] [PubMed]
- Poeppl, W.; Obwaller, A.G.; Weiler, M.; Burgmann, H.; Mooseder, G.; Lorentz, S.; Rauchenwald, F.; Aspock, H.; Walochnik, J.; Naucke, T.J. Emergence of sandflies (Phlebotominae) in Austria, a Central European country. Parasitol. Res. 2013, 112, 4231–4237. [Google Scholar] [CrossRef] [PubMed]
- Ready, P.D. Managing the spread of canine leishmaniosis in Europe. Vet. Rec. 2017, 180, 44–46. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, C.; Wang, O.; Strutz, S.E.; Gonzalez-Salazar, C.; Sanchez-Cordero, V.; Sarkar, S. Climate change and risk of leishmaniasis in North America: Predictions from ecological niche models of vector and reservoir species. PLoS Negl. Trop. Dis. 2010, 4, e585. [Google Scholar] [CrossRef] [PubMed]
- Moo-Llanes, D.A.; Pech-May, A.; Ibarra-Cerdena, C.N.; Rebollar-Tellez, E.A.; Ramsey, J.M. Inferring distributional shifts of epidemiologically important North and Central American sandflies from Pleistocene to future scenarios. Med. Vet. Entomol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Trájer, A.J.; Bede-Fazekas, Á.; Hufnagel, L.; Horváth, L.; Bobvos, J. The effect of climate change on the potential distribution of the European Phlebotomus species. Appl. Ecol. Environ. Res. 2013, 11, 189–208. [Google Scholar] [CrossRef]
- Cross, E.R.; Hyams, K.C. The potential effect of global warming on the geographic and seasonal distribution of Phlebotomus papatasi in southwest Asia. Environ. Health Perspect. 1996, 104, 724–727. [Google Scholar] [CrossRef] [PubMed]
- Bates, P.A. Transmission of Leishmania metacyclic promastigotes by phlebotomine sand flies. Int. J. Parasitol. 2007, 37, 1097–1106. [Google Scholar] [CrossRef] [PubMed]
- Haddad, N.; Saliba, H.; Altawil, A.; Villinsky, J.; Al-Nahhas, S. Cutaneous leishmaniasis in the central provinces of Hama and Edlib in Syria: Vector identification and parasite typing. Parasites Vectors 2015, 8, 524. [Google Scholar] [CrossRef] [PubMed]
- Orshan, L.; Elbaz, S.; Ben-Ari, Y.; Akad, F.; Afik, O.; Ben-Avi, I.; Dias, D.; Ish-Shalom, D.; Studentsky, L.; Zonstein, I. Distribution and dispersal of Phlebotomus papatasi (Diptera: Psychodidae) in a zoonotic cutaneous leishmaniasis focus, the Northern Negev, Israel. PLoS Negl. Trop. Dis. 2016, 10, e0004819. [Google Scholar] [CrossRef] [PubMed]
- Ali, R.M.; Loutfy, N.F.; Awad, O.M.; Suliman, N.K. Bionomics of phlebotomine sandfly species in west Alexandria, Egypt. J. Entomol. Zool. Stud. 2016, 4, 349–353. [Google Scholar]
- Haouas, N.; Amer, O.; Alshammri, F.F.; Al-Shammari, S.; Remadi, L.; Ashankyty, I. Cutaneous leishmaniasis in northwestern Saudi Arabia: Identification of sand fly fauna and parasites. Parasites Vectors 2017, 10, 544. [Google Scholar] [CrossRef] [PubMed]
- Ozbel, Y.; Karakus, M.; Arserim, S.K.; Kalkan, S.O.; Toz, S. Molecular detection and identification of Leishmania spp. in naturally infected Phlebotomus tobbi and Sergentomyia dentata in a focus of human and canine leishmaniasis in western Turkey. Acta Trop. 2016, 155, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Zivdari, M.; Hejazi, S.H.; Mirhendi, S.H.; Jafari, R.; Rastegar, H.A.; Abtahi, S.M. Molecular identification of leishmania parasites in sand flies (Diptera, Psychodidae) of an endemic foci in Poldokhtar, Iran. Adv. Biomed. Res. 2018, 7, 124. [Google Scholar] [CrossRef] [PubMed]
- Vahabi, A.; Rassi, Y.; Oshaghi, M.; Sayyadi, M.; Rafizadeh, S. Detection of Leishmania major DNA within wild caught Phlebotomus papatasi and species composition of sand flies in endemic focus of cutaneous leishmaniasis, in western Iran. J. Parasit. Dis. 2016, 40, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Rafizadeh, S.; Saraei, M.; Abaei, M.R.; Oshaghi, M.A.; Mohebali, M.; Peymani, A.; Naserpour-Farivar, T.; Bakhshi, H.; Rassi, Y. Molecular detection of Leishmania major and L. turanica in Phlebotomus papatasi and first natural infection of P. salehi to L. major in North-East of Iran. J. Arthropod. Borne Dis. 2016, 10, 141–147. [Google Scholar] [PubMed]
- Kovacic, B.W. The Presence of Populations of Phlebotominae at Fort Campbell, Kentucky; University of Kentucky: Lexington, KY, USA, 2007. [Google Scholar]
- Aronson, N. Leishmaniasis in relation to service in Iraq/Afghanistan, U.S. armed forces, 2001–2006. Med. Surv. Mon. Rep. 2007, 14, 2–5. [Google Scholar]
- Minter, L.; Kovacic, B.; Claborn, D.M.; Lawyer, P.; Florin, D.; Brown, G. New state records for Lutzomyia shannoni and Lutzomyia vexator. J. Med. Entomol. 2009, 46, 965–968. [Google Scholar] [CrossRef] [PubMed]
- Maroli, M.; Fausto, A.M.; Sabatinelli, G.; Majori, G. Phlebotomines (Diptera, Psychodidae) from Burkina Faso. A note on the sandfly species collected in domestic resting sites. Ann. Parasitol. Hum. Comp. 1986, 61, 683–688. [Google Scholar] [CrossRef] [PubMed]
- Young, D.; Perkins, P. Phlebotomine sand flies of North America (Diptera: Psychodidae) [Lutzomyia]. Mosquito News (USA) 1984, 44, 263–304. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Minter, L.M.; Yu, T.; Florin, D.A.; Nukmal, N.; Brown, G.C.; Zhou, X. Molecular identification of sand flies (Diptera: Psychodidae) in Eastern North America by using PCR-RFLP. J. Med. Entomol. 2013, 50, 920–924. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.S.; Kato, H.; Fukushige, M.; Wagatsuma, Y.; Itoh, M. Application of RFLP-PCR-based identification for sand fly surveillance in an area endemic for kala-azar in Mymensingh, Bangladesh. J. Parasitol. Res. 2012, 2012, 467821. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Kato, H.; Cáceres, A.G.; Gomez, E.A.; Velez, L.; Mimori, T.; Zhang, F.; Iwata, H.; Korenaga, M.; Sakurai, T. Genotyping of sand fly species in Peruvian Andes where leishmaniasis is endemic. Acta Trop. 2012, 121, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Latrofa, M.S.; Annoscia, G.; Dantas-Torres, F.; Traversa, D.; Otranto, D. Towards a rapid molecular identification of the common phlebotomine sand flies in the Mediterranean region. Vet. Parasitol. 2012, 184, 267–270. [Google Scholar] [CrossRef] [PubMed]
- Dokianakis, E.; Tsirigotakis, N.; Christodoulou, V.; Poulakakis, N.; Antoniou, M. DNA sequencing confirms PCR-RFLP identification of wild caught Larroussius sand flies from Crete and Cyprus. Acta Trop. 2016, 164, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Al-Dakhil, A.A.; Al-Ajmi, R.A.; Siddiqi, N.J.; Ayaad, T.H. Molecular typing of phlebotomine sand flies in al-madinah and asir regions, Saudi Arabia using PCR–RFLP of 18S ribosomal RNA gene. Saudi J. Biol. Sci. 2017, 24, 1697–1703. [Google Scholar] [CrossRef] [PubMed]
- Nzelu, C.O.; Caceres, A.G.; Arrunategui-Jimenez, M.J.; Lanas-Rosas, M.F.; Yanez-Trujillano, H.H.; Luna-Caipo, D.V.; Holguin-Mauricci, C.E.; Katakura, K.; Hashiguchi, Y.; Kato, H. DNA barcoding for identification of sand fly species (Diptera: Psychodidae) from leishmaniasis-endemic areas of Peru. Acta Trop. 2015, 145, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, M.A.C.; Vivero, R.J.; Vélez, I.D.; Porter, C.H.; Uribe, S. DNA barcoding for the identification of sand fly species (Diptera, Psychodidae, Phlebotominae) in Colombia. PLoS ONE 2014, 9, e85496. [Google Scholar] [CrossRef]
- De Souza Pinto, I.; das Chagas, B.D.; Rodrigues, A.A.F.; Ferreira, A.L.; Rezende, H.R.; Bruno, R.V.; Falqueto, A.; Andrade-Filho, J.D.; Galati, E.A.B.; Shimabukuro, P.H.F. DNA barcoding of Neotropical sand flies (Diptera, Psychodidae, Phlebotominae): Species identification and discovery within Brazil. PLoS ONE 2015, 10, e0140636. [Google Scholar] [CrossRef]
- Nzelu, C.O.; Kato, H.; Puplampu, N.; Desewu, K.; Odoom, S.; Wilson, M.D.; Sakurai, T.; Katakura, K.; Boakye, D.A. First detection of Leishmania tropica DNA and Trypanosoma species in Sergentomyia sand flies (Diptera: Psychodidae) from an outbreak area of cutaneous leishmaniasis in Ghana. PLoS Negl. Trop. Dis. 2014, 8, e2630. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.P.; Srinivasan, R.; Jambulingam, P. DNA barcoding for identification of sand flies (Diptera: Psychodidae) in India. Mol. Ecol. Resour. 2012, 12, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, V.; Halada, P.; Hlavackova, K.; Dokianakis, E.; Antoniou, M.; Volf, P. Identification of phlebotomine sand flies (Diptera: Psychodidae) by matrix-assisted laser desorption/ionization time of flight mass spectrometry. Parasites Vectors 2014, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Lafri, I.; Almeras, L.; Bitam, I.; Caputo, A.; Yssouf, A.; Forestier, C.L.; Izri, A.; Raoult, D.; Parola, P. Identification of Algerian field-caught Phlebotomine sand fly vectors by MALDI-TOF MS. PLoS Negl. Trop. Dis. 2016, 10, e0004351. [Google Scholar] [CrossRef] [PubMed]
- Halada, P.; Hlavackova, K.; Dvorak, V.; Volf, P. Identification of immature stages of phlebotomine sand flies using MALDI-TOF MS and mapping of mass spectra during sand fly life cycle. Insect Biochem. Mol. Biol. 2018, 93, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Rosetto, M.; Belardinelli, M.; Fausto, A.M.; Marchini, D.; Bongiorno, G.; Maroli, M.; Mazzini, M. A mammalian-like lipase gene is expressed in the female reproductive accessory glands of the sand fly Phlebotomus papatasi (Diptera, Psychodidae). Insect Mol. Biol. 2003, 12, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, J.G.; Belkaid, Y.; Rowton, E.; Ribeiro, J.M. The salivary apyrase of the blood-sucking sand fly Phlebotomus papatasi belongs to the novel Cimex family of apyrases. J. Exp. Biol. 2001, 204, 229–237. [Google Scholar] [PubMed]
- Hamasaki, R.; Kato, H.; Terayama, Y.; Iwata, H.; Valenzuela, J.G. Functional characterization of a salivary apyrase from the sand fly, Phlebotomus duboscqi, a vector of Leishmania major. J. Insect Physiol. 2009, 55, 1044–1049. [Google Scholar] [CrossRef] [PubMed]
- Gebre-Michael, T.; Pratlong, F.; Lane, R.P. Phlebotomus (Phlebotomus) duboscqi (Diptera: Phlebotominae), naturally infected with Leishmania major in southern Ethiopia. Trans. R. Soc. Trop. Med. Hyg. 1993, 87, 10–11. [Google Scholar] [CrossRef]


| Gene | Accession Number | Primer | Sequence (5′-3′) | Amplicion Length (bp) |
|---|---|---|---|---|
| Mammalian-like lipse | AY179968 | LIP1F | CTGCGAGGCCAACGTGGACA | 132 |
| LIP1R | GCGCAGAGGTCACAGAGGTCG | |||
| LIP2F | CCGGCCACTACGGTGTTGAGG | 135 | ||
| LIP2R | CCAGTTCCGACGATCGATTT | |||
| LIP3F | ACGTCACACTCTTCCCCAAC | 172 | ||
| LIP3R | TAGTCGCACTTGGCCTTCTT | |||
| Salivary apyrase | AF261768 | APY1F | ACAAAGGACGAGGAGCTGAA | 178 |
| APY1R | GTTGCCCATTCTGCCTTAAA | |||
| APY2F | TGGCACGAAGCTGTTAATTG | 228 | ||
| APY1R | GTCA TCAC TATC GGGG AGGA | |||
| APY3F | ACCA ATGC AGAC CTCA TTCC | 174 | ||
| APY3R | TTTG ATCC AGAG GGAG TTGC |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Merchant, A.; Yu, T.; Shi, J.; Zhou, X. Development of a Diagnostic Marker for Phlebotomus papatasi to Initiate a Potential Vector Surveillance Program in North America. Insects 2018, 9, 162. https://doi.org/10.3390/insects9040162
Merchant A, Yu T, Shi J, Zhou X. Development of a Diagnostic Marker for Phlebotomus papatasi to Initiate a Potential Vector Surveillance Program in North America. Insects. 2018; 9(4):162. https://doi.org/10.3390/insects9040162
Chicago/Turabian StyleMerchant, Austin, Tian Yu, Jizhe Shi, and Xuguo Zhou. 2018. "Development of a Diagnostic Marker for Phlebotomus papatasi to Initiate a Potential Vector Surveillance Program in North America" Insects 9, no. 4: 162. https://doi.org/10.3390/insects9040162
APA StyleMerchant, A., Yu, T., Shi, J., & Zhou, X. (2018). Development of a Diagnostic Marker for Phlebotomus papatasi to Initiate a Potential Vector Surveillance Program in North America. Insects, 9(4), 162. https://doi.org/10.3390/insects9040162






