Comparative Transcriptome Analysis Reveals Epithelial Growth Factor Receptor (EGFR) Pathway and Secreted C-Type Lectins as Essential Drivers of Leg Regeneration in Periplaneta americana
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Histological Analysis
2.3. Collection of Biological Material
2.4. RNA Extraction and Sequencing
2.5. Quality Control and Read Mapping
2.6. Differential Expression Analysis and Functional Enrichment
2.7. Real-Time Quantitative PCR
2.8. Double-Stranded RNA (dsRNA) Treatment
3. Results and Discussion
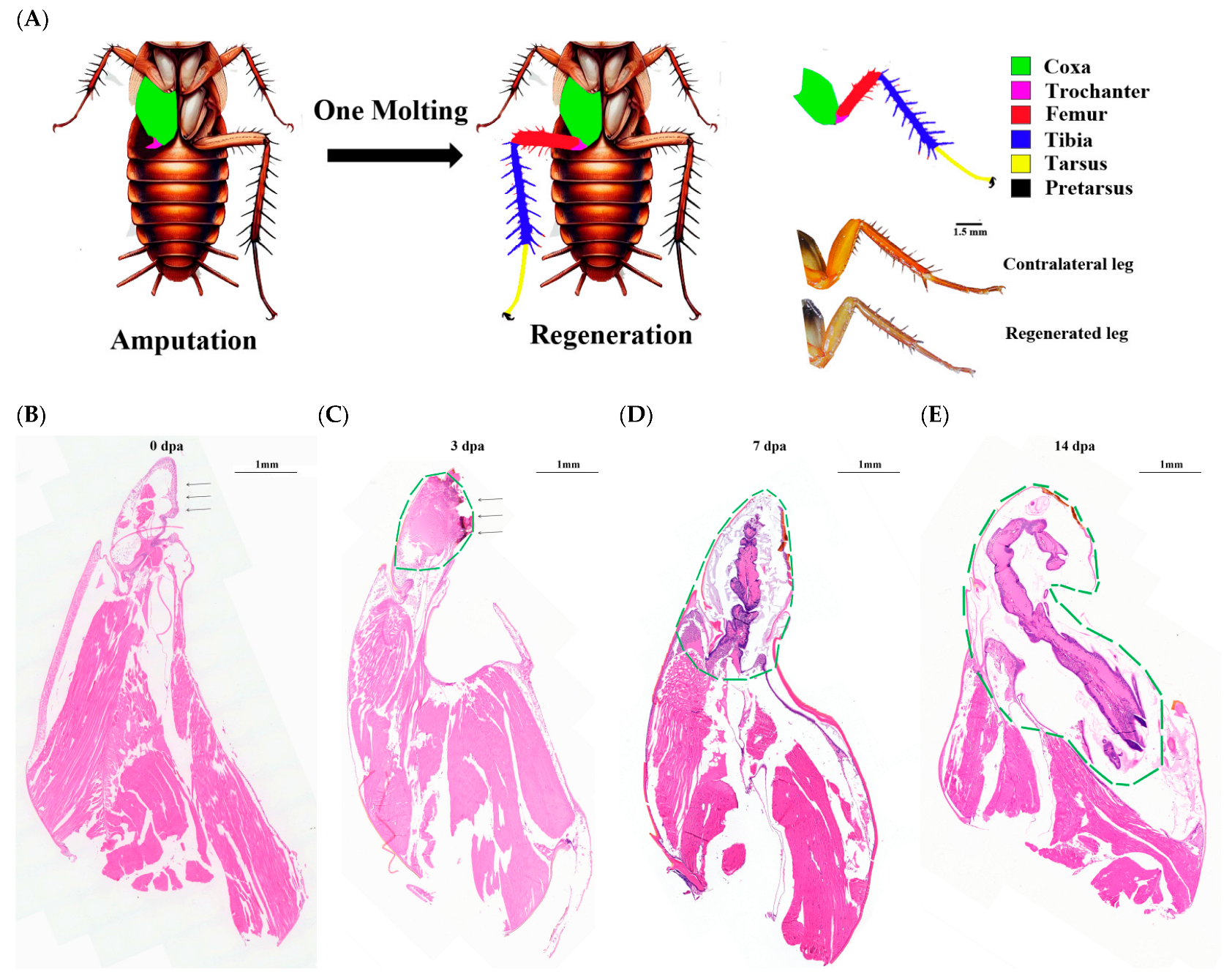
3.1. Morphological Profiles and Histological Analysis of Regeneration Legs
3.2. Analyses of Sequencing Quality
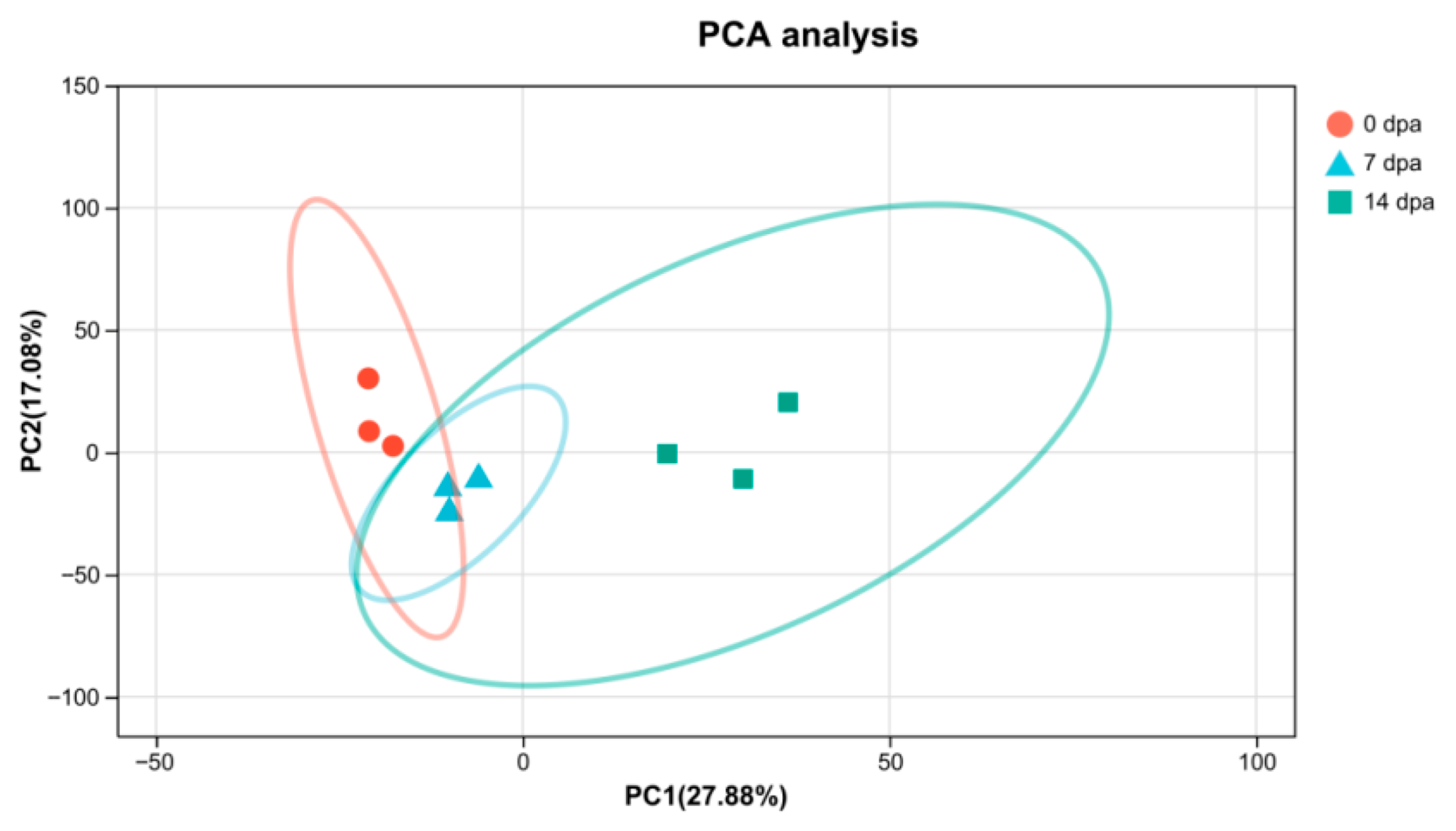
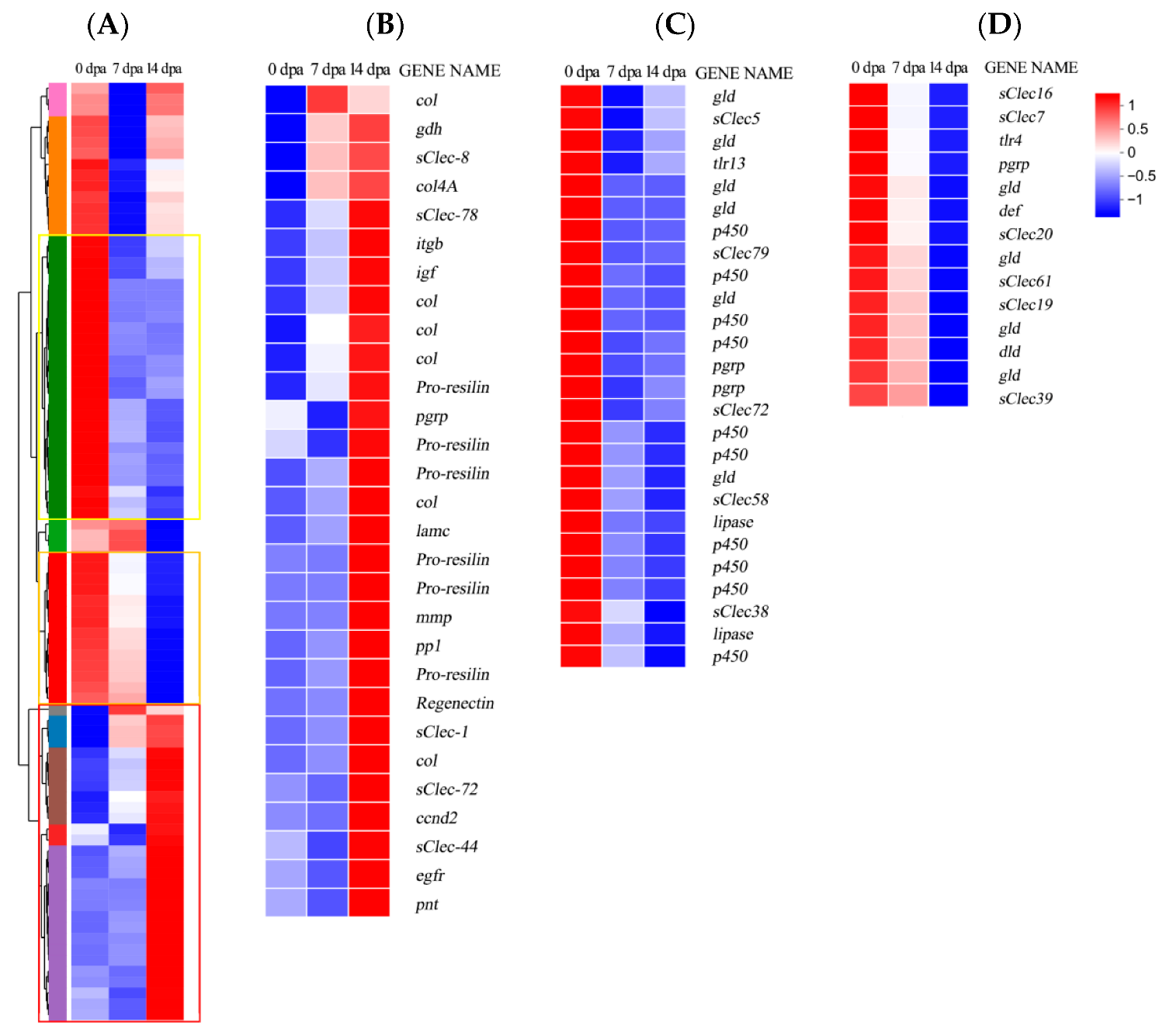
3.3. Differential Expression of Genes
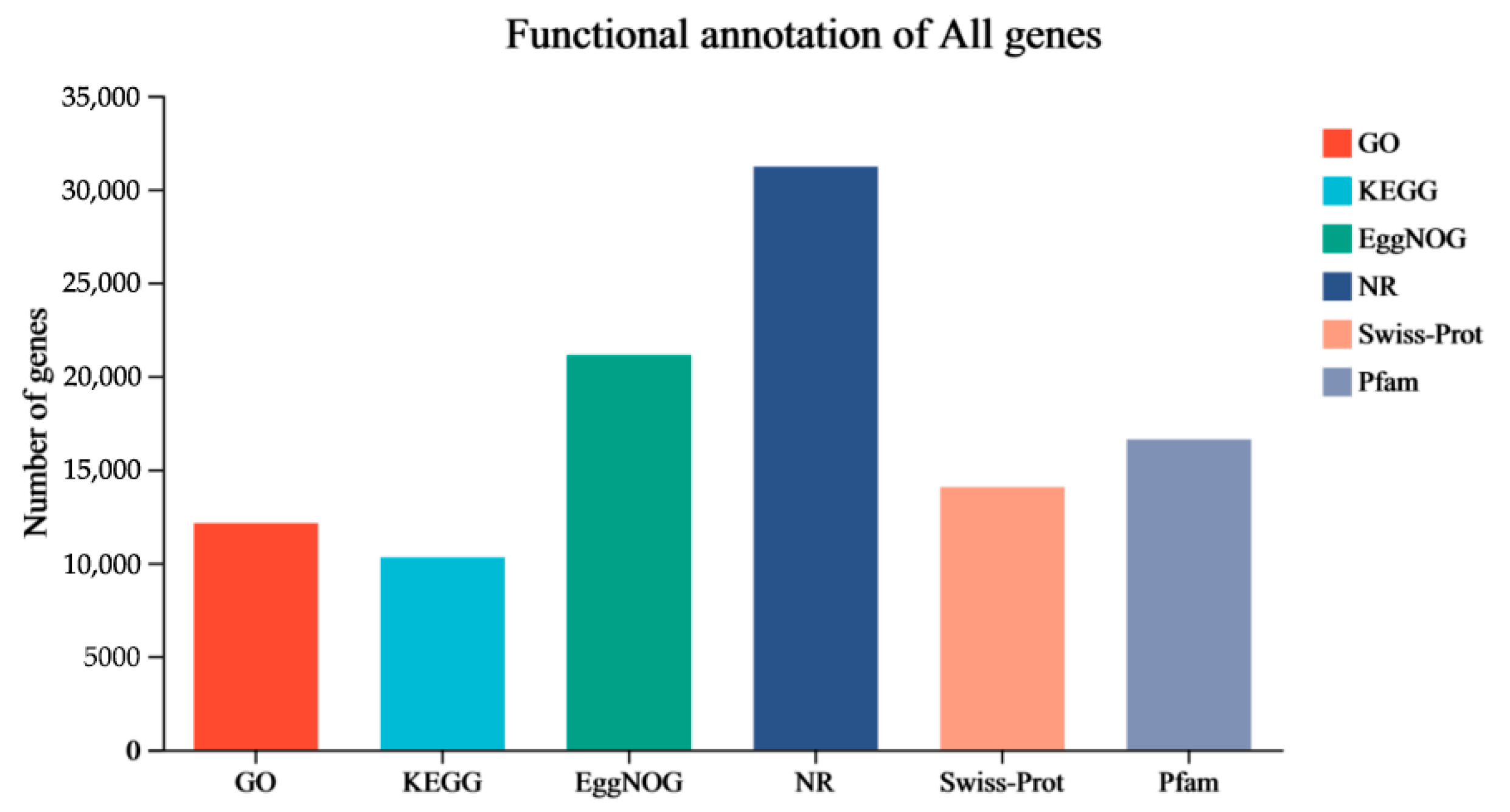
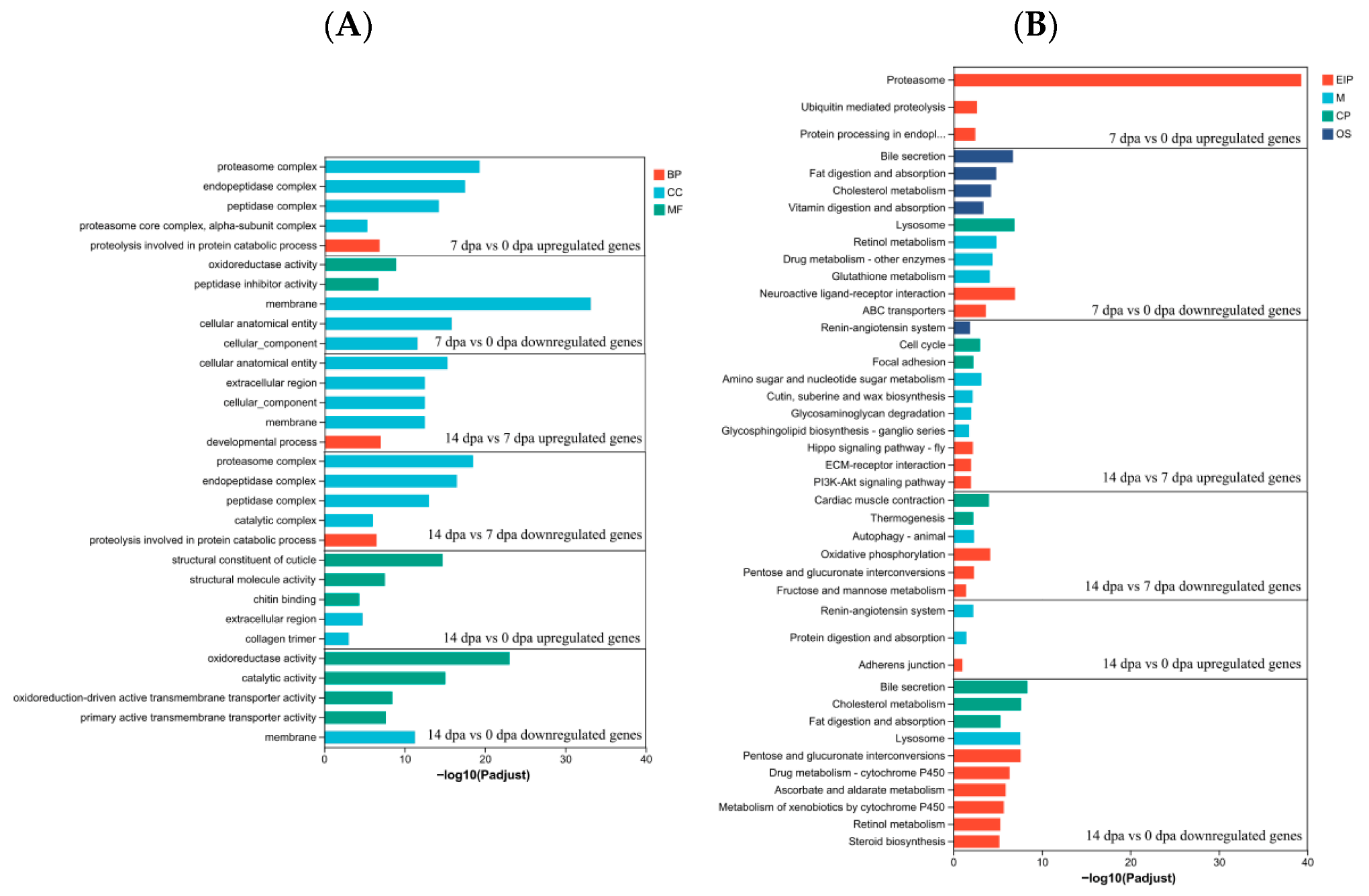
3.4. Functional Annotation and Pathway Assignment DEGs
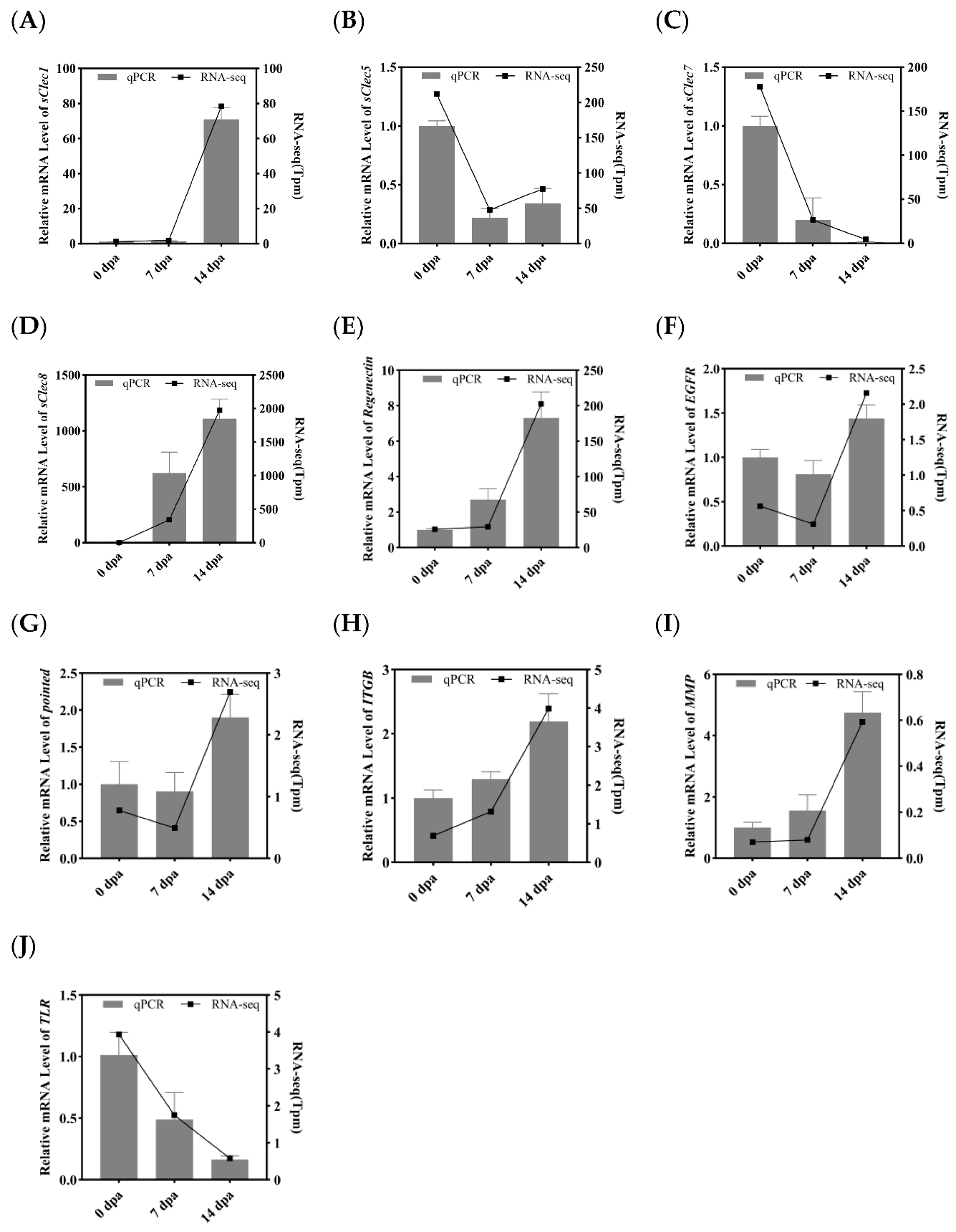
3.5. Validation of Differentially Expressed Transcripts by qRT-PCR
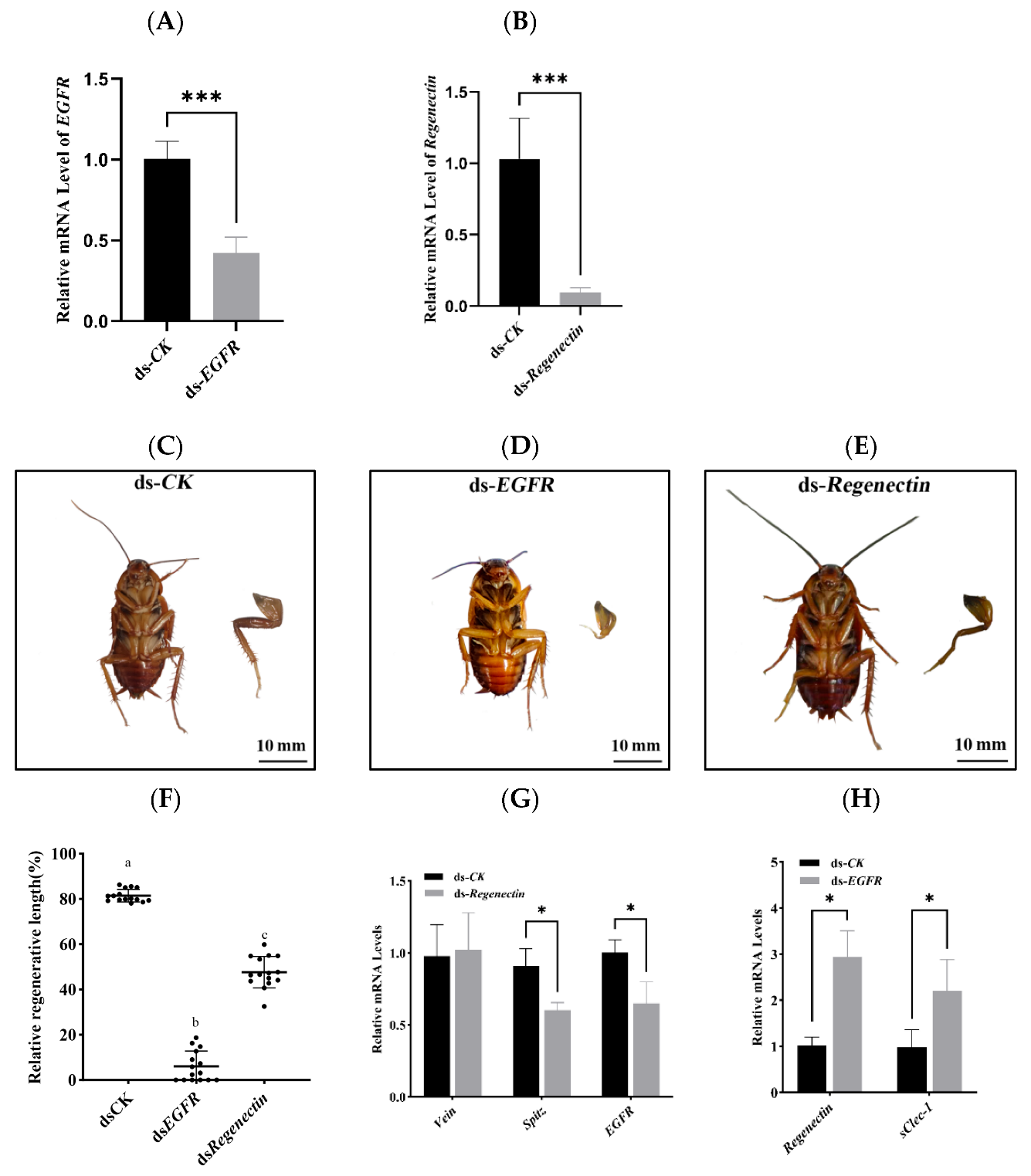
3.6. Functional Analysis of EGFR and Regenectin During Leg Regeneration
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Goss, R.J. Principles of Regeneration; Elsevier: Amsterdam, The Netherlands, 1969; ISBN 978-1-4832-3250-8. [Google Scholar]
- Simon, A.; Tanaka, E.M. Limb Regeneration. WIREs Dev. Biol. 2013, 2, 291–300. [Google Scholar] [CrossRef]
- Poss, K.D. Advances in Understanding Tissue Regenerative Capacity and Mechanisms in Animals. Nat. Rev. Genet. 2010, 11, 710–722. [Google Scholar] [CrossRef]
- Tanaka, E.M. The Molecular and Cellular Choreography of Appendage Regeneration. Cell 2016, 165, 1598–1608. [Google Scholar] [CrossRef] [PubMed]
- Joven, A.; Elewa, A.; Simon, A. Model Systems for Regeneration: Salamanders. Development 2019, 146, dev167700. [Google Scholar] [CrossRef]
- Goldman, J.A.; Poss, K.D. Gene Regulatory Programmes of Tissue Regeneration. Nat. Rev. Genet. 2020, 21, 511–525. [Google Scholar] [CrossRef]
- Zhong, J.; Jing, A.; Zheng, S.; Li, S.; Zhang, X.; Ren, C. Physiological and Molecular Mechanisms of Insect Appendage Regeneration. Cell Regen. 2023, 12, 9. [Google Scholar] [CrossRef]
- Alwes, F.; Enjolras, C.; Averof, M. Live Imaging Reveals the Progenitors and Cell Dynamics of Limb Regeneration. eLife 2016, 5, e19766. [Google Scholar] [CrossRef]
- Wang, J.; Chen, X.; Hou, X.; Wang, J.; Yue, W.; Huang, S.; Xu, G.; Yan, J.; Lu, G.; Hofreiter, M.; et al. “Omics” Data Unveil Early Molecular Response Underlying Limb Regeneration in the Chinese Mitten Crab, Eriocheir sinensis. Sci. Adv. 2022, 8, eabl4642. [Google Scholar] [CrossRef] [PubMed]
- Seifert, A.W.; Monaghan, J.R.; Smith, M.D.; Pasch, B.; Stier, A.C.; Michonneau, F.; Maden, M. The Influence of Fundamental Traits on Mechanisms Controlling Appendage Regeneration. Biol. Rev. 2012, 87, 330–345. [Google Scholar] [CrossRef]
- Mito, T.; Inoue, Y.; Kimura, S.; Miyawaki, K.; Niwa, N.; Shinmyo, Y.; Ohuchi, H.; Noji, S. Involvement of Hedgehog, Wingless, and Dpp in the Initiation of Proximodistal Axis Formation during the Regeneration of Insect Legs, a Verification of the Modified Boundary Model. Mech. Dev. 2002, 114, 27–35. [Google Scholar] [CrossRef]
- Tanaka, E.M.; Reddien, P.W. The Cellular Basis for Animal Regeneration. Dev. Cell 2011, 21, 172–185. [Google Scholar] [CrossRef]
- Shibata, E.; Yokota, Y.; Horita, N.; Kudo, A.; Abe, G.; Kawakami, K.; Kawakami, A. Fgf Signalling Controls Diverse Aspects of Fin Regeneration. Development 2016, 143, 2920–2929. [Google Scholar] [CrossRef]
- Koriyama, Y.; Homma, K.; Sugitani, K.; Higuchi, Y.; Matsukawa, T.; Murayama, D.; Kato, S. Upregulation of IGF-I in the Goldfish Retinal Ganglion Cells during the Early Stage of Optic Nerve Regeneration. Neurochem. Int. 2007, 50, 749–756. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Mito, T.; Miyawaki, K.; Ohuchi, H.; Noji, S. EGFR Signaling Is Required for Re-Establishing the Proximodistal Axis during Distal Leg Regeneration in the Cricket Gryllus bimaculatus Nymph. Dev. Biol. 2008, 319, 46–55. [Google Scholar] [CrossRef]
- Hamada, Y.; Bando, T.; Nakamura, T.; Ishimaru, Y.; Mito, T.; Noji, S.; Tomioka, K.; Ohuchi, H. Leg regeneration is epigenetically regulated by histone H3K27 methylation in the cricket Gryllus bimaculatus. Development 2015, 142, 2916–2927. [Google Scholar] [CrossRef] [PubMed]
- Bando, T.; Ishimaru, Y.; Kida, T.; Hamada, Y.; Matsuoka, Y.; Nakamura, T.; Ohuchi, H.; Noji, S.; Mito, T. Analysis of RNA-Seq Data Reveals Involvement of JAK/STAT Signalling during Leg Regeneration in the Cricket Gryllus bimaculatus. Development 2013, 140, 959–964. [Google Scholar] [CrossRef]
- Zhou, H.; Wang, W.; Yan, S.; Zhang, J.; Wang, D.; Shen, J. JAK/STAT Signaling Regulates the Harmonia axyridis Leg Regeneration by Coordinating Cell Proliferation. Dev. Biol. 2022, 483, 98–106. [Google Scholar] [CrossRef]
- Zhou, H.; Ma, Z.; Wang, Z.; Yan, S.; Wang, D.; Shen, J. Hedgehog Signaling Regulates Regenerative Patterning and Growth in Harmonia axyridis Leg. Cell. Mol. Life Sci. 2021, 78, 2185–2197. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhu, S.; Jia, Q.; Yuan, D.; Ren, C.; Li, K.; Liu, S.; Cui, Y.; Zhao, H.; Cao, Y.; et al. The Genomic and Functional Landscapes of Developmental Plasticity in the American Cockroach. Nat. Commun. 2018, 9, 1008. [Google Scholar] [CrossRef]
- Liu, Y.; Dai, X.; Hu, H.; Zhou, J.; Peng, Y.; Yuan, H.; Song, Q. Isolation and Purification of an Oligopeptide from Periplaneta Americana and Its Mechanism of Promoted Wound Healing. Int. J. Pept. Res. Ther. 2023, 29, 65. [Google Scholar] [CrossRef]
- Zhang, X.-S.; Wei, L.; Zhang, W.; Zhang, F.-X.; Li, L.; Li, L.; Wen, Y.; Zhang, J.-H.; Liu, S.; Yuan, D.; et al. ERK-Activated CK-2 Triggers Blastema Formation during Appendage Regeneration. Sci. Adv. 2024, 10, eadk8331. [Google Scholar] [CrossRef]
- Ren, C.; Wen, Y.; Zheng, S.; Zhao, Z.; Li, E.Y.; Zhao, C.; Liao, M.; Li, L.; Zhang, X.; Liu, S.; et al. Two Transcriptional Cascades Orchestrate Cockroach Leg Regeneration. Cell Rep. 2024, 43, 113889. [Google Scholar] [CrossRef] [PubMed]
- Nachtrab, G.; Poss, K.D. Toward a Blueprint for Regeneration. Development 2012, 139, 2639–2642. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nowotarski, S.H.; Sánchez Alvarado, A. Widening Perspectives on Regenerative Processes through Growth. npj Regen. Med. 2016, 1, 16015. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Godwin, J.; Kuraitis, D.; Rosenthal, N. Extracellular Matrix Considerations for Scar-Free Repair and Regeneration: Insights from Regenerative Diversity among Vertebrates. Int. J. Biochem. Cell Biol. 2014, 56, 47–55. [Google Scholar] [CrossRef]
- Singh, D.; Rai, V.; K Agrawal, D. Regulation of Collagen I and Collagen III in Tissue Injury and Regeneration. Cardiol. Cardiovasc. Med. 2023, 7, 5–16. [Google Scholar] [CrossRef]
- Sharma, S.; Rai, V.K.; Narang, R.K.; Markandeywar, T.S. Collagen-Based Formulations for Wound Healing: A Literature Review. Life Sci. 2022, 290, 120096. [Google Scholar] [CrossRef]
- Weis-Fogh, T. Molecular Interpretation of the Elasticity of Resilin, a Rubber-like Protein. J. Mol. Biol. 1961, 3, 648–667. [Google Scholar] [CrossRef]
- Michels, J.; Appel, E.; Gorb, S.N. Functional Diversity of Resilin in Arthropoda. Beilstein J. Nanotechnol. 2016, 7, 1241–1259. [Google Scholar] [CrossRef] [PubMed]
- Mott, J.D.; Werb, Z. Regulation of Matrix Biology by Matrix Metalloproteinases. Curr. Opin. Cell Biol. 2004, 16, 558–564. [Google Scholar] [CrossRef] [PubMed]
- Nanba, D.; Toki, F.; Asakawa, K.; Matsumura, H.; Shiraishi, K.; Sayama, K.; Matsuzaki, K.; Toki, H.; Nishimura, E.K. EGFR-Mediated Epidermal Stem Cell Motility Drives Skin Regeneration through COL17A1 Proteolysis. J. Cell Biol. 2021, 220, e202012073. [Google Scholar] [CrossRef]
- Zelensky, A.N.; Gready, J.E. The C-type Lectin-like Domain Superfamily. FEBS J. 2005, 272, 6179–6217. [Google Scholar] [CrossRef]
- Yue, R.; Shen, B.; Morrison, S.J. Clec11a/Osteolectin Is an Osteogenic Growth Factor That Promotes the Maintenance of the Adult Skeleton. eLife 2016, 5, e18782. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ji, Y.; Feng, F.; Liu, Z.; Qian, L.; Shen, H.; Lao, L. C-Type Lectin Domain-Containing Protein CLEC3A Regulates Proliferation, Regeneration and Maintenance of Nucleus Pulposus Cells. Cell. Mol. Life Sci. 2022, 79, 435. [Google Scholar] [CrossRef]
- Yun, C.-H. Relationship between Tissue Regeneration and Immune Modulation. Tissue Eng. Regen. Med. 2023, 20, 325–327. [Google Scholar] [CrossRef]
- Pasten, C.; Ortiz-Pineda, P.A.; García-Arrarás, J.E. Ubiquitin–Proteasome System Components Are Upregulated during Intestinal Regeneration. Genesis 2012, 50, 350–365. [Google Scholar] [CrossRef]
- Zhao, J.; Zhai, B.; Gygi, S.P.; Goldberg, A.L. mTOR Inhibition Activates Overall Protein Degradation by the Ubiquitin Proteasome System as Well as by Autophagy. Proc. Natl. Acad. Sci. USA 2015, 112, 15790–15797. [Google Scholar] [CrossRef]
- Meyer-Schwesinger, C. The Ubiquitin–Proteasome System in Kidney Physiology and Disease. Nat. Rev. Nephrol. 2019, 15, 393–411. [Google Scholar] [CrossRef]
- Zhong, Z.; Jiao, Z.; Yu, F.-X. The Hippo Signaling Pathway in Development and Regeneration. Cell Rep. 2024, 43, 113926. [Google Scholar] [CrossRef]
- Riley, S.E.; Feng, Y.; Hansen, C.G. Hippo-Yap/Taz Signalling in Zebrafish Regeneration. npj Regen. Med. 2022, 7, 9. [Google Scholar] [CrossRef]
- Su, M.; Soomro, S.H.; Jie, J.; Fu, H. Effects of the Extracellular Matrix on Myelin Development and Regeneration in the Central Nervous System. Tissue Cell 2021, 69, 101444. [Google Scholar] [CrossRef]
- Wang, S.; Fu, L.; Wang, B.; Cai, Y.; Jiang, J.; Shi, Y.-B. Thyroid Hormone Receptor- and Stage-Dependent Transcriptome Changes Affect the Initial Period of Xenopus Tropicalis Tail Regeneration. BMC Genom. 2024, 25, 1260. [Google Scholar] [CrossRef] [PubMed]
- Avila-Martinez, N.; Gansevoort, M.; Verbakel, J.; Jayaprakash, H.; Araujo, I.M.; Vitorino, M.; Tiscornia, G.; Van Kuppevelt, T.H.; Daamen, W.F. Matrisomal Components Involved in Regenerative Wound Healing in Axolotl and Acomys: Implications for Biomaterial Development. Biomater. Sci. 2023, 11, 6060–6081. [Google Scholar] [CrossRef]
- Reddy, P.C.; Gungi, A.; Unni, M. Cellular and Molecular Mechanisms of Hydra Regeneration. In Evo-Devo: Non-Model Species in Cell and Developmental Biology; Tworzydlo, W., Bilinski, S.M., Eds.; Results and Problems in Cell Differentiation; Springer International Publishing: Cham, Switzerland, 2019; Volume 68, pp. 259–290. ISBN 978-3-030-23458-4. [Google Scholar]
- Haddadi, N.; Lin, Y.; Travis, G.; Simpson, A.M.; Nassif, N.T.; McGowan, E.M. PTEN/PTENP1: ‘Regulating the Regulator of RTK-Dependent PI3K/Akt Signalling’, New Targets for Cancer Therapy. Mol. Cancer 2018, 17, 37. [Google Scholar] [CrossRef] [PubMed]
- Peterson, C.; Chandler, H.L. Insulin Facilitates Corneal Wound Healing in the Diabetic Environment through the RTK-PI3K/Akt/mTOR Axis in Vitro. Mol. Cell. Endocrinol. 2022, 548, 111611. [Google Scholar] [CrossRef]
- Tito, C.; Masciarelli, S.; Colotti, G.; Fazi, F. EGF Receptor in Organ Development, Tissue Homeostasis and Regeneration. J. Biomed. Sci. 2025, 32, 24. [Google Scholar] [CrossRef] [PubMed]
- Raja, K.K.B.; Yeung, K.; Shim, Y.-K.; Mardon, G. Integrative Genomic Analyses Reveal Putative Cell Type-Specific Targets of the Drosophila Ets Transcription Factor Pointed. BMC Genom. 2024, 25, 103. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhou, C.; Chen, Y.; Ma, W.; Cheng, Y.; Chen, J.; Bai, Y.; Luo, W.; Li, N.; Du, E.; et al. Egfr Signaling Promotes Juvenile Hormone Biosynthesis in the German Cockroach. BMC Biol. 2022, 20, 278. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Liu, Z.; Zhang, J.; Zhang, J.; Wu, Q.; Kang, A.; Wu, Y.; Zhou, Y.; Sun, Y. A Novel C-Type Lectin 4E from Cromileptes Altivelis (CaCTL4E) Participates in Antibacterial Innate Immunity. Aquaculture 2025, 601, 742282. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, H.; Huang, Y.; Yang, Y.; Yekefenhazi, D.; Zou, W.; Han, F. Characterization and Immune Functions of LcβLectin from Large Yellow Croaker (Larimichthys crocea): A Potential Antiviral Defense Molecule. Int. J. Mol. Sci. 2025, 26, 3251. [Google Scholar] [CrossRef]
- Chen, P.; Ai, H.; Liu, Z.; Li, C.; Li, B. The Dual Functions of a Newly Identified C-Type Lectin (TcCTL17) in the Immunity and Development of Tribolium castaneum. Bull. Entomol. Res. 2025, 115, 251–264. [Google Scholar] [CrossRef]
- Zhang, Y.; Ai, H.; Wang, Y.; Zhang, P.; Du, L.; Wang, J.; Wang, S.; Gao, H.; Li, B. A Pattern Recognition Receptor C-type Lectin TcCTL14 Contributes to Immune Response and Development in the Red Flour Beetle, Tribolium castaneum. Insect Sci. 2023, 30, 1363–1377. [Google Scholar] [CrossRef]
- Arai, T.; Kawasaki, K.; Kubo, T.; Natori, S. Cloning of cDNA for Regenectin, a Humoral C-Type Lectin of Periplaneta Americana, and Expression of the Regenectin Gene during Leg Regeneration. Insect Biochem. Mol. Biol. 1998, 28, 987–994. [Google Scholar] [CrossRef]
- Rosenkranz, A.A.; Slastnikova, T.A. Epidermal Growth Factor Receptor: Key to Selective Intracellular Delivery. Biochem. Mosc. 2020, 85, 967–993. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Teng, Y.; Xie, K.; He, X.; Chen, G.; Zhang, K.; Liao, Q.; Zhang, J.; Zhou, X.; Zhu, Y.; et al. REG3A Secreted by Peritumoral Acinar Cells Enhances Pancreatic Ductal Adenocarcinoma Progression via Activation of EGFR Signaling. Cell Commun. Signal. 2025, 23, 96. [Google Scholar] [CrossRef]
- Wang, J.-X.; Cao, B.; Ma, N.; Wu, K.-Y.; Chen, W.-B.; Wu, W.; Dong, X.; Liu, C.-F.; Gao, Y.-F.; Diao, T.-Y.; et al. Collectin-11 Promotes Cancer Cell Proliferation and Tumor Growth. JCI Insight 2023, 8, e159452. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Sun, N.; Wu, X.; Wu, J.; Xian, S.; Wang, D.; Pei, Y. Comparative Transcriptome Analysis Reveals Epithelial Growth Factor Receptor (EGFR) Pathway and Secreted C-Type Lectins as Essential Drivers of Leg Regeneration in Periplaneta americana. Insects 2025, 16, 934. https://doi.org/10.3390/insects16090934
Liu X, Sun N, Wu X, Wu J, Xian S, Wang D, Pei Y. Comparative Transcriptome Analysis Reveals Epithelial Growth Factor Receptor (EGFR) Pathway and Secreted C-Type Lectins as Essential Drivers of Leg Regeneration in Periplaneta americana. Insects. 2025; 16(9):934. https://doi.org/10.3390/insects16090934
Chicago/Turabian StyleLiu, Xiaoxuan, Nan Sun, Xiaojuan Wu, Jiajia Wu, Shuqi Xian, Dayong Wang, and Yechun Pei. 2025. "Comparative Transcriptome Analysis Reveals Epithelial Growth Factor Receptor (EGFR) Pathway and Secreted C-Type Lectins as Essential Drivers of Leg Regeneration in Periplaneta americana" Insects 16, no. 9: 934. https://doi.org/10.3390/insects16090934
APA StyleLiu, X., Sun, N., Wu, X., Wu, J., Xian, S., Wang, D., & Pei, Y. (2025). Comparative Transcriptome Analysis Reveals Epithelial Growth Factor Receptor (EGFR) Pathway and Secreted C-Type Lectins as Essential Drivers of Leg Regeneration in Periplaneta americana. Insects, 16(9), 934. https://doi.org/10.3390/insects16090934







