Functional Divergence of Two General Odorant-Binding Proteins to Sex Pheromones and Host Plant Volatiles in Adoxophyes orana (Lepidoptera: Tortricidae)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing
2.2. RNA Extraction, cDNA Synthesis, and qRT-PCR
2.3. Signal Peptides Prediction, Sequence Alignment and Phylogenetic Analyses
2.4. Prokaryotic Expression and Purification of rAoraGOBPs
2.5. Fluorescence Competitive Binding Assay
2.6. Homology Modeling, Molecular Docking, and Molecular Dynamic Simulation
2.7. Statistic Analysis
3. Results
3.1. Sequence Analysis of AoraGOBP1 and AoraGOBP2
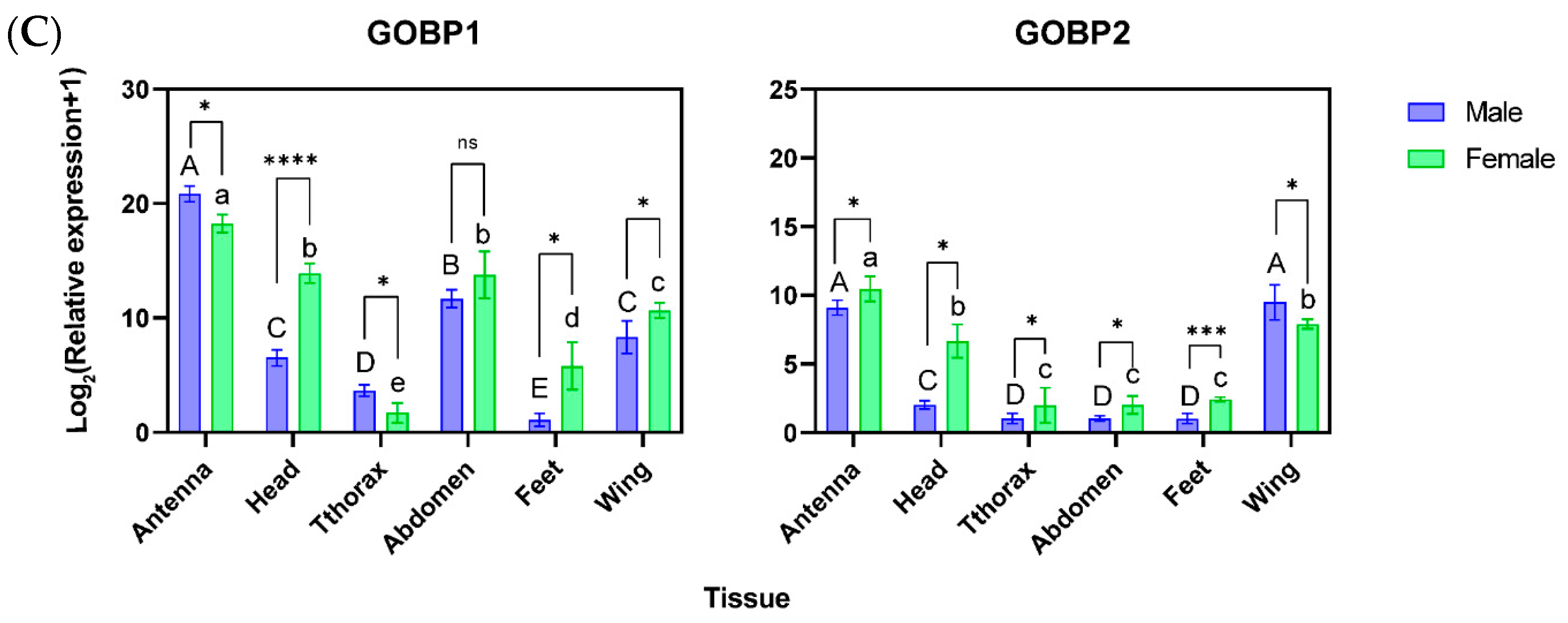
3.2. Spatial–Temporal Patterns of OBPs in A. orona
3.3. Expression and Purification of rAoraGOBPs
3.4. Ligand Binding of rAoraGOBPs
3.5. Protein Structure Modeling and Molecular Docking
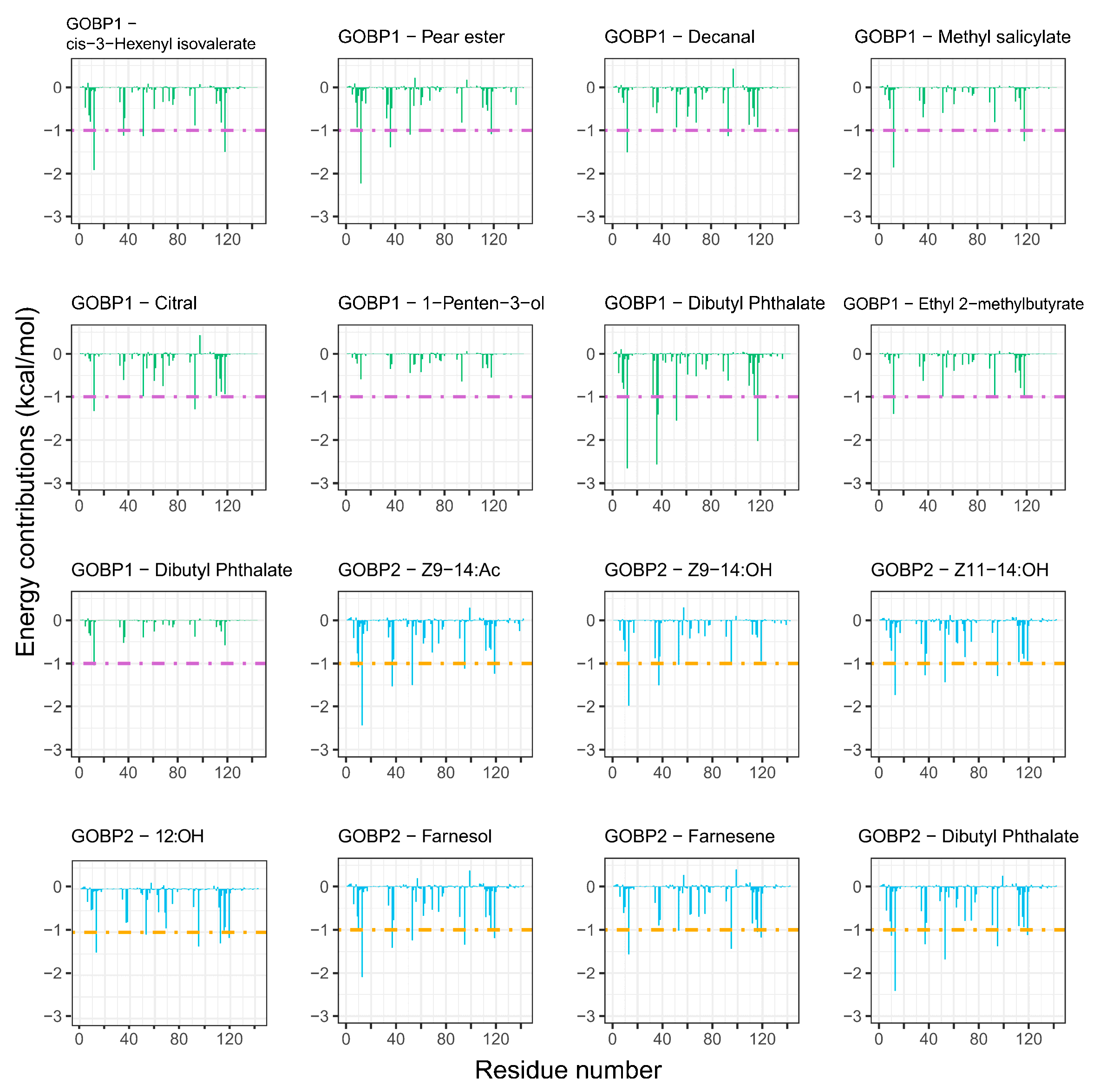
3.6. Per-Residue Free Energy Decomposition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| OBP | Odorant-binding protein |
| CSP | Chemosensory protein |
| OR | Odorant receptor |
| IR | Ionotropic receptor |
| GR | Gustory receptor |
| SNMP | Sensory neuron membrane proteins |
| RT-qPCR | Real-time quantitative polymeric chain reaction |
| One-way ANOVA | One-way analysis of variance |
| SDS-PAGE | Sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| MD simulation | Molecular dynamics simulation |
References
- Pelosi, P.; Iovinella, I.; Zhu, J.; Wang, G.; Dani, F.R. Beyond Chemoreception: Diverse Tasks of Soluble Olfactory Proteins in Insects. Biol. Rev. 2018, 93, 184–200. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Turlings, T.C.J. Plant Volatiles as Mate-Finding Cues for Insects. Trends Plant Sci. 2018, 23, 100–111. [Google Scholar] [CrossRef]
- Gadenne, C.; Barrozo, R.B.; Anton, S. Plasticity in Insect Olfaction: To Smell or Not to Smell? Annu. Rev. Entomol. 2016, 61, 317–333. [Google Scholar] [CrossRef]
- Anton, S.; Cortesero, A.-M. Plasticity in Chemical Host Plant Recognition in Herbivorous Insects and Its Implication for Pest Control. Biology 2022, 11, 1842. [Google Scholar] [CrossRef]
- Zhou, S.; Jander, G. Molecular Ecology of Plant Volatiles in Interactions with Insect Herbivores. J. Exp. Bot. 2022, 73, 449–462. [Google Scholar] [CrossRef]
- Piñero, J.C.; Dorn, S. Synergism between Aromatic Compounds and Green Leaf Volatiles Derived from the Host Plant Underlies Female Attraction in the Oriental Fruit Moth. Entomol. Exp. Appl. 2007, 125, 185–194. [Google Scholar] [CrossRef]
- Varela, N.; Avilla, J.; Anton, S.; Gemeno, C. Synergism of Pheromone and Host-plant Volatile Blends in the Attraction of Grapholita Molesta Males. Entomol. Exp. Appl. 2011, 141, 114–122. [Google Scholar] [CrossRef]
- Piñero, J.C.; Dorn, S. Response of Female Oriental Fruit Moth to Volatiles from Apple and Peach Trees at Three Phenological Stages. Entomol. Exp. Appl. 2009, 131, 67–74. [Google Scholar] [CrossRef]
- Yan, X.; Ma, L.; Li, X.; Chang, L.; Liu, Q.; Song, C.; Zhao, J.; Qie, X.; Deng, C.; Wang, C.; et al. Identification and Evaluation of Cruciferous Plant Volatiles Attractive to Plutella xylostella L. (Lepidoptera: Plutellidae). Pest. Manag. Sci. 2023, 79, 5270–5282. [Google Scholar] [CrossRef] [PubMed]
- Leal, W.S. Odorant Reception in Insects: Roles of Receptors, Binding Proteins, and Degrading Enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef]
- Krieger, J.; Breer, H. Olfactory Reception in Invertebrates. Science 1999, 286, 720–723. [Google Scholar] [CrossRef]
- Jing, D.; Zhang, T.; Bai, S.; Prabu, S.; He, K.; Dewer, Y.; Wang, Z. GOBP1 Plays a Key Role in Sex Pheromones and Plant Volatiles Recognition in Yellow Peach Moth, Conogethes punctiferalis (Lepidoptera: Crambidae). Insects 2019, 10, 302. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Q.; Yan, Q.; Li, L.L.; Xu, J.W.; Mang, D.; Wang, X.L.; Hoh, H.H.; Ye, J.; Ju, Q.; Ma, Y.; et al. Different Binding Properties of Two General-Odorant Binding Proteins in Athetis lepigone with Sex Pheromones, Host Plant Volatiles and Insecticides. Pestic. Biochem. Phys. 2020, 164, 173–182. [Google Scholar] [CrossRef]
- Li, L.L.; Xu, B.Q.; Li, C.Q.; Li, B.L.; Chen, X.L.; Li, G.W. Different Binding Affinities of Three General Odorant-Binding Proteins in Grapholita funebrana (Treitscheke) (Lepidoptera: Tortricidae) to Sex Pheromones, Host Plant Volatiles, and Insecticides. J. Econ. Entomol. 2022, 115, 1129–1145. [Google Scholar] [CrossRef]
- Yang, H.H.; Li, S.-P.; Yin, M.Z.; Zhu, X.Y.; Li, J.B.; Zhang, Y.N.; Li, X.M. Functional Differentiation of Two General Odorant-Binding Proteins to Sex Pheromones in Spodoptera frugiperda. Pestic. Biochem. Phys. 2023, 191, 105348. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Li, R.; Cheng, S.; Zhou, T.; Liu, J. The Mythimna separata General Odorant Binding Protein 2 (MsepGOBP2) Is Involved in the Larval Detection of the Sex Pheromone (Z)-11-Hexadecenal. Pest. Manag. Sci. 2023, 79, 2005–2026. [Google Scholar] [CrossRef] [PubMed]
- Milonas, P.G.; Savopoulou-Soultani, M. Seasonal Abundance and Population Dynamics of Adoxophyes orana (Lepidoptera: Tortricidae) in Northern Greece. Int. J. Pest. Manag. 2006, 52, 45–51. [Google Scholar] [CrossRef]
- Park, H.; Park, I.J.; Lee, S.Y.; Han, K.S.; Yang, C.Y.; Boo, K.S.; Park, K.T.; Lee, J.; Cho, S. Molecular Identification of Adoxophyes orana Complex (Lepidoptera: Tortricidae) in Korea and Japan. J. Asia-Pac. Entomol. 2008, 11, 49–52. [Google Scholar] [CrossRef]
- Li, G.; Wang, H.; Yang, W.; Chen, X.; Li, B.; Chen, Y. Influence of Host Plants on the Development, Survivorship, and Fecundity of the Summer Fruit Tortrix Moth, Adoxophyes orana (Lepidoptera: Tortricidae). Entomol. Res. 2021, 51, 499–508. [Google Scholar] [CrossRef]
- Sun, L.; Liu, Y.; Zhang, H.; Yan, W.; Yue, Q.; Qiu, G. Molecular Characterization of the Ryanodine Receptor from Adoxophyes orana and Its Response to Lethal and Sublethal Doses of Chlorantraniliprole. J. Integr. Agric. 2021, 20, 1585–1595. [Google Scholar] [CrossRef]
- Tsai, C.L.; Shih, L.C.; Yeh, W.B.; Byun, B.K.; Jinbo, U.; Ning, F.Y.; Sung, I.H. Genetic Differentiation and Species Diversification of the Adoxophyes orana Complex (Lepidoptera: Tortricidae) in East Asia. J. Econ. Entomol. 2023, 116, 1885–1893. [Google Scholar] [CrossRef]
- Minks, A.K.; Voerman, S. Sex Pheromones of the Summerfruit Tortrix Moth, Adoxophyes orana: Trapping Performance in the Field. Entomol. Exp. Appl. 1973, 16, 541–549. [Google Scholar] [CrossRef]
- Den Otter, C.J.; Schuil, H.A.; Oosten, A.S. Reception of Host-Plant Ddors and Female Sex Pheromone in Adoxophyes orana (Lepidoptera: Tortricidae): Electrophysiology and Morphology. Entomol. Exp. Appl. 1978, 24, 570–578. [Google Scholar] [CrossRef]
- Yang, C.Y.; Han, K.S.; Boo, K.S. Sex Pheromones and Reproductive Isolation of Three Species in Genus Adoxophyes. J. Chem. Ecol. 2009, 35, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Teufel, F.; Almagro-Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 Predicts All Five Types of Signal Peptides Using Protein Language Models. Nat. Biotech. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent Updates to the Phylogenetic Tree Display and Annotation Tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein Folding Accessible to All. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Kim, G.; Lee, S.; Levy Karin, E.; Kim, H.; Moriwaki, Y.; Ovchinnikov, S.; Steinegger, M.; Mirdita, M. Easy and Accurate Protein Structure Prediction Using ColabFold. Nat. Protoc. 2025, 20, 620–642. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 Update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, X.; Gan, J.; Chen, S.; Xiao, Z.X.; Cao, Y. CB-Dock2: Improved Protein–Ligand Blind Docking by Integrating Cavity Detection, Docking and Homologous Template Fitting. Nucleic Acids Res. 2022, 50, W159–W164. [Google Scholar] [CrossRef] [PubMed]
- The PyMOL Molecular Graphics System, Version 2.5.0, Schrödinger, LLC. Available online: https://github.com/schrodinger/pymol-open-source (accessed on 21 June 2025).
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple Ligand–Protein Interaction Diagrams for Drug Discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Showalter, S.A.; Brüschweiler, R. Validation of Molecular Dynamics Simulations of Biomolecules Using NMR Spin Relaxation as Benchmarks: Application to the AMBER99SB Force Field. J. Chem. Theory Comput. 2007, 3, 961–975. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef]
- Ma, Y.; Li, Y.; Wei, Z.Q.; Hou, J.H.; Si, Y.X.; Zhang, J.; Dong, S.L.; Yan, Q. Identification and Functional Characterization of General Odorant Binding Proteins in Orthaga achatina. Insects 2023, 14, 216. [Google Scholar] [CrossRef]
- Li, Y.; Song, W.; Wang, S.; Miao, W.; Liu, Z.; Wu, F.; Wang, J.; Sheng, S. Binding Characteristics and Structural Dynamics of Two General Odorant-Binding Proteins with Plant Volatiles in the Olfactory Recognition of Glyphodes pyloalis. Insect Biochem. Mol. Biol. 2024, 173, 104177. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Fu, X.B.; Cui, H.C.; Zhao, L.; Yu, J.Z.; Li, H.L. Functional Characteristics, Electrophysiological and Antennal Immunolocalization of General Odorant-Binding Protein 2 in Tea Geometrid, Ectropis obliqua. Int. J. Mol. Sci. 2018, 19, 875. [Google Scholar] [CrossRef]
- Liu, X.L.; Wu, Z.R.; Liao, W.; Zhang, X.Q.; Pei, Y.W.; Liu, M. The Binding Affinity of Two General Odorant Binding Proteins in Spodoptera Frugiperda to General Volatiles and Insecticides. Int. J. Biol. Macromol. 2023, 252, 126338. [Google Scholar] [CrossRef]
- Liu, N.Y.; Yang, K.; Liu, Y.; Xu, W.; Anderson, A.; Dong, S.L. Two General-Odorant Binding Proteins in Spodoptera litura Are Differentially Tuned to Sex Pheromones and Plant Odorants. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2015, 180, 23–31. [Google Scholar] [CrossRef]
- Yin, N.N.; Yang, A.J.; Wu, C.; Xiao, H.Y.; Guo, Y.R.; Liu, N.Y. Genome-Wide Analysis of Odorant-Binding Proteins in Papilio xuthus with Focus on the Perception of Two PxutGOBPs to Host Odorants and Insecticides. J. Agric. Food. Chem. 2022, 70, 10747–10761. [Google Scholar] [CrossRef]
- Huang, G.Z.; Liu, J.T.; Zhou, J.J.; Wang, Q.; Dong, J.Z.; Zhang, Y.J.; Li, X.C.; Li, J.; Gu, S.H. Expressional and Functional Comparisons of Two General Odorant Binding Proteins in Agrotis ipsilon. Insect Biochem. Mol. Biol. 2018, 98, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.Y.; Yang, F.; Yang, K.; He, P.; Niu, X.H.; Xu, W.; Anderson, A.; Dong, S.-L. Two Subclasses of Odorant-binding Proteins inSpodoptera exigua Display Structural Conservation and Functional Divergence. Insect Mol. Biol. 2015, 24, 167–182. [Google Scholar] [CrossRef]
- Li, G.; Chen, X.; Li, B.; Zhang, G.; Li, Y.; Wu, J. Binding Properties of General Odorant Binding Proteins from the Oriental Fruit Moth, Grapholita Molesta (Busck) (Lepidoptera: Tortricidae). PLoS ONE 2016, 11, e0155096. [Google Scholar] [CrossRef] [PubMed]
- Binder, B.F.; Robbins, J.C.; Wilson, R.L. Chemically Mediated Ovipositional Behaviors of the European Corn Borer, Ostrinia nubilalis (Lepidoptera: Pyralidae). J. Chem. Ecol. 1995, 21, 1315–1327. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liu, M.; Lv, C.; Tian, Z.; Li, R.; Li, Y.; Zhang, Y.; Liu, J. Identifying the Key Role of Plutella xylostella General Odorant Binding Protein 2 in Perceiving a Larval Attractant, (E,E)-2,6-Farnesol. J. Agric. Food Chem. 2024, 72, 5690–5698. [Google Scholar] [CrossRef]
- Bradley, S.J.; Suckling, D.M. Factors Influencing Codling Moth Larval Response to α-Farnesene. Entomolo. Exp. Appl. 1995, 75, 221–227. [Google Scholar] [CrossRef]
- Hern, A.; Dorn, S. Sexual Dimorphism in the Olfactory Orientation of Adult Cydia pomonella in Response to α-Farnesene. Entomol. Exp. Appl. 1999, 92, 63–72. [Google Scholar] [CrossRef]





| Ligand Name | CAS Number | Ki (μM) | Ligand Name | CAS Number | Ki (μM) | ||
|---|---|---|---|---|---|---|---|
| GOBP1 | GOBP2 | GOBP1 | GOBP2 | ||||
| Z9-14:Ac | 35153-15-2 | 6.46 ± 0.18 | 1-hexanol | 111-27-3 | - | - | |
| Z11-14:Ac | 20711-10-8 | - | - | Benzyl alcohol | 100-51-6 | - | - |
| Z9-14:OH | 35153-15-2 | - | 1.49 ± 0.01 | (Z)-3-hexen-1-ol | 928-96-1 | - | - |
| Z11-14:OH | 34010-15-6 | - | 1.24 ± 0.02 | Farnesol | 4602-84-0 | 1.13 ± 0.02 | |
| E9-14:Ac | - | - | Nerolidol | 3790-78-1 | - | - | |
| 12:OH | 88170-32-5 | - | 1.74 ± 0.03 | 1-Penten-3-ol | 616-25-1 | 10.19 ± 0.14 | |
| Butyl butanoate | 109-21-7 | - | - | Linalool | 78-70-6 | - | - |
| Methyl salicylate | 119-36-8 | 4.41 ± 0.20 | - | Heptan-1-ol | 111-70-6 | - | - |
| ethyl acetate | 141-78-6 | - | - | Decan-1-ol | 112-30-1 | - | - |
| Ethyl valerate | 539-82-2 | - | - | Cineole | 470-82-6 | - | - |
| Dibutyl phthalate | 84-74-2 | 10.27 ± 0.07 | 5.33 ± 0.04 | (Z,E)-phytol | 7541-49-3 | - | - |
| Ethyl isovalerate | 108-64-5 | - | - | Isooctyl alcohol | 26952-21-6 | - | - |
| cis-3-Hexenyl butyrate | 16491-36-4 | - | - | Farnesene | 502-61-4 | 6.96 ± 0.32 | |
| Methyl oleate | 112-62-9 | - | - | Camphene | 5794-04-7 | - | - |
| Butyl acetate | 123-86-4 | - | - | (Z)-β-ocimene | 13877-91-3 | - | - |
| Ethyl Butyrate | 105-54-4 | - | - | α-Pinene | 7785-70-8 | - | - |
| Pear ester | 3025-30-7 | 5.08 ± 0.13 | - | β-Caryophyllene | 87-44-5 | - | - |
| Ethyl Tiglate | 5837-78-5 | - | - | 3-carene | 4497-92-1 | - | - |
| Isoamyl acetate | 123-92-2 | - | - | 2-methylprop-1-enylbenzene | 768-49-0 | - | - |
| Methyl palmitate | 112-39-0 | - | - | α-phellandrene | 99-83-2 | - | - |
| Butyl propanoate | 590-01-2 | - | - | 3,7-Dimethyl-2,6-octadienenitrile | 5146-66-7 | - | - |
| (Z)-3-Hexen-1-yl 3-methylbutanoate | 35154-45-1 | 6.21 ± 0.18 | - | Benzonitrile | 100-47-0 | - | - |
| cis-3-hexenyl acetate | 1708-82-3 | - | - | ||||
| Ethyl hexanoate | 123-66-0 | - | - | ||||
| Ethyl 2-methylbutanoate | 7452-79-1 | 11.78 ± 0.08 | - | ||||
| Heptanal | 111-71-7 | - | - | ||||
| Hexanal | 66-25-1 | - | - | ||||
| Decanal | 112-31-2 | 0.78 ± 0.15 | - | ||||
| Benzaldehyde | 100-52-7 | - | - | ||||
| Trans-2-Hexenal | 6728-26-3 | - | - | ||||
| Citral | 5392-40-5 | 11.70 ± 0.44 | - | ||||
| Nonanal | 124-19-6 | - | - | ||||
| Isobutyraldehyde | 78-84-2 | 11.88 ± 0.41 | - | ||||
| Complex | Electrostatic Energy (ΔGele) | van der Waals Energy (ΔGvdw) | Polar Solvation Energy (ΔGPB) | Nonpolar Solvation Energy (ΔGSA) | Total Binding Energy (ΔGbind) |
|---|---|---|---|---|---|
| AoraGOBP1–cis-3-Hexenyl isovalerate | −1.89 ± 0.01 | −31.21 ± 0.01 | 8.96 ± 0.01 | −4.49 ± 0.00 | −28.62 ± 0.02 |
| AoraGOBP1–Pear ester | −1.12 ± 0.01 | −35.63 ± 0.01 | 10.48 ± 0.01 | −5.34 ± 0.00 | −31.61 ± 0.02 |
| AoraGOBP1–Decanal | −2.62 ± 0.01 | −28.95 ± 0.01 | 8.40 ± 0.01 | −4.37 ± 0.00 | −27.54 ± 0.02 |
| AoraGOBP1–Methyl salicylate | −1.71 ± 0.01 | −21.97 ± 0.01 | 9.75 ± 0.01 | −3.36 ± 0.00 | −17.29 ± 0.01 |
| AoraGOBP1–Citral | −0.70 ± 0.02 | −26.81 ± 0.01 | 7.15 ± 0.02 | −4.10 ± 0.00 | −24.46 ± 0.01 |
| AoraGOBP1–1-Penten-3-ol | −3.00 ± 0.02 | −13.32 ± 0.01 | 6.33 ± 0.02 | −3.99 ± 0.00 | −12.34 ± 0.02 |
| AoraGOBP1–Dibutyl phthalate | −6.42 ± 0.02 | −44.93 ± 0.02 | 18.69 ± 0.02 | −6.04 ± 0.00 | −24.46 ± 0.01 |
| AoraGOBP1– Ethyl 2-methylbutanoate | −1.57 ± 0.01 | −21.82 ± 0.01 | 6.75 ± 0.02 | −3.34 ± 0.00 | −19.97 ± 0.01 |
| AoraGOBP1–Isobutyraldehyde | −1.13 ± 0.01 | −12.28 ± 0.01 | 4.20 ± 0.01 | −1.99 ± 0.00 | −11.20 ± 0.01 |
| AoraGOBP2–Z9-14:Ac | −2.31 ± 0.02 | −46.26 ± 0.02 | 12.07 ± 0.01 | −6.69 ± 0.00 | −43.18 ± 0.02 |
| AoraGOBP2–Z9-14:OH | −4.96 ± 0.02 | −38.65 ± 0.02 | 10.02 ± 0.01 | −5.75 ± 0.00 | −39.34 ± 0.02 |
| AoraGOBP2–Z11-14:OH | −7.86 ± 0.04 | −38.41 ± 0.02 | 12.94 ± 0.03 | −5.87 ± 0.00 | −39.20 ± 0.02 |
| AoraGOBP2–12:OH | −11.39 ± 0.03 | −34.23 ± 0.02 | 14.49 ± 0.02 | −5.39 ± 0.00 | −36.53 ± 0.02 |
| AoraGOBP2–Farnesol | −0.82 ± 0.01 | −36.78 ± 0.02 | 7.41 ± 0.01 | −5.30 ± 0.00 | −35.49 ± 0.02 |
| AoraGOBP2–Farnesene | −3.56 ± 0.02 | −39.35 ± 0.02 | 10.99 ± 0.02 | −5.57 ± 0.00 | −37.49 ± 0.02 |
| AoraGOBP2–Dibutyl phthalate | −7.13 ± 0.02 | −44.73 ± 0.04 | 19.34 ± 0.01 | −6.41 ± 0.01 | −38.94 ± 0.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, S.; Liu, Y.; Chen, X.; Luo, K.; Zhao, J.; Li, G.; Li, B. Functional Divergence of Two General Odorant-Binding Proteins to Sex Pheromones and Host Plant Volatiles in Adoxophyes orana (Lepidoptera: Tortricidae). Insects 2025, 16, 880. https://doi.org/10.3390/insects16090880
Ren S, Liu Y, Chen X, Luo K, Zhao J, Li G, Li B. Functional Divergence of Two General Odorant-Binding Proteins to Sex Pheromones and Host Plant Volatiles in Adoxophyes orana (Lepidoptera: Tortricidae). Insects. 2025; 16(9):880. https://doi.org/10.3390/insects16090880
Chicago/Turabian StyleRen, Shaoqiu, Yuhan Liu, Xiulin Chen, Kun Luo, Jirong Zhao, Guangwei Li, and Boliao Li. 2025. "Functional Divergence of Two General Odorant-Binding Proteins to Sex Pheromones and Host Plant Volatiles in Adoxophyes orana (Lepidoptera: Tortricidae)" Insects 16, no. 9: 880. https://doi.org/10.3390/insects16090880
APA StyleRen, S., Liu, Y., Chen, X., Luo, K., Zhao, J., Li, G., & Li, B. (2025). Functional Divergence of Two General Odorant-Binding Proteins to Sex Pheromones and Host Plant Volatiles in Adoxophyes orana (Lepidoptera: Tortricidae). Insects, 16(9), 880. https://doi.org/10.3390/insects16090880






