Simple Summary
Fruit flies in the genus Zeugodacus are destructive agricultural pests of Asia and expanding to other regions such as sub-Saharan Africa. Growth in international trade and travel increases the inadvertent introduction of pests such as fruit flies, especially through their immature life stages. Immature stages make it challenging to diagnose the organism to the species level using morphology-based taxonomy. In contrast, molecular approaches, particularly COI-based real-time PCR assays, provide swift and precise species identification independent of their sex, life stages, and intactness of the specimen. In the present study, we developed the Zeugodacus cucurbitae-specific real-time PCR, with high sensitivity and specificity, to meet international accreditation standards (ISO 17025) and obtained approval to use it through International Accreditation New Zealand (IANZ—ISO 17025).
Abstract
Fruit flies that belong to the genus Zeugodacus (Diptera: Tephritidae) pose significant threats as invasive pests of agricultural crops in Asia and sub-Saharan Africa. The intensification of transboundary trade in fresh horticultural produce has increased the risk of introducing invasive species such as fruit flies, more so through the inadvertent transport of their immature developmental stages. Such immature stages of fruit flies belonging to the Tephritidae family are frequently intercepted at the international borders worldwide and are unable to be identified to the species level using morphological characteristics. Molecular identification using mitochondrial Cytochrome Oxidase I (COI) gene has proven to be quite useful, as they are not constrained by developmental stages, sex, or colour morphs of the pest species in question. Also, real-time PCR-based species-specific assays offer quicker turnaround time since they do not require any post-PCR procedures. This study evaluated the utility of a real-time PCR assay based on the COI gene region to identify Zeugodacus cucurbitae from other Tephritid species. The developed real-time PCR assay provides a swift and precise way of discriminating between these highly invasive pest species during an interception event for rapid decision making. High specificity, having no cross-reactions with closely related Tephritids, and sensitivity of the developed assay will be extremely useful in discriminating Z. cucurbitae from other closely related fruit fly species. Z. cucurbitae-specific real-time PCR developed in this study is appropriate for organizations that carry out routine diagnostics to facilitate fresh produce imports and exports. Our assay is fully optimized for rapid deployment at international borders, offering reliable detection of the target species regardless of developmental stage, sex, or geographic origins.
1. Introduction
Zeugodacus cucurbitae Coquillet, commonly known as melon fly and native to India is a very serious pest of cucurbit crops [1]. It was first described by Coquillet in 1899 from specimens collected in the Hawaiian Islands and classified it under the genus Bactrocera. It is therefore sometimes cited as Bactrocera (Zeugodacus) cucurbitae and is said to be widely distributed in Asia and Africa. It belongs to the Tephritidae family, a diverse collection of true fruit flies that are known for their invasiveness and crop damage [2]. Specifically, fruit flies belonging to the genus Zeugodacus (Diptera: Tephritidae) are among the world’s major invasive pests [1] and are highly polyphagous, and this behaviour intensifies their pest status, as it can cause significant economic losses [3].
The female Z. cucurbitae lay her eggs beneath the epidermis of host fruits and vegetables, and the larvae develop inside the fruit, leading to tissue damage and making the produce unmarketable [4]. They can infest more than 125 plant species, primarily within the Cucurbitaceae family, including various commercially cultivated crops [5,6,7,8,9,10]. Amongst the reported host plants, bitter gourd (Momordica charantia), muskmelon (Cucumis melo), snap melon (Cucumis melo var. momordica), and snake gourd (Trichosanthes anguina and T. cucumeria) are the most preferred [11]. Recently, records from India indicate the expansion of the host range to include tomatoes [12].
A recent investigation into the macrogeographic population dynamics reveals that Z. cucurbitae is prevalent across the African continent, the islands of the Indian Ocean, Asia, New Guinea, the Mariana Islands, and Hawaii [10,13]. They successfully invaded Africa in the 20th century, starting in West Africa and progressively spreading eastward, with Mozambique being the most recently documented invasion [7]. Z. cucurbitae remain active throughout the year on various hosts, with their spread largely aided by trade, favourable climate conditions, and relaxed biosecurity measures in specific regions.
Z. cucurbitae have been encountered at New Zealand’s borders from Asian countries as eggs or larvae and can cause delays in the clearance of fresh produce, as there is currently no rapid and precise test or assay available to identify this pest species. The early detection of exotic insect pest populations, followed by rapid management responses, has resulted in successful control and eradication of numerous species known to inflict ecological or economic damage [14]. A significant challenge is the interception of a large percentage of immature invertebrates (such as eggs, larvae, or pupae) and their body parts that are collected and can be challenging or impossible to identify to the species level using morphological methods. This calls for a method of identification by which a rapid and accurate sex and developmental stage non-limiting inventory of species is achieved, helping classification and management efforts. Such a method would also help in quick and accurate discrimination of native and exotic invasive species at the port of entry which is important from the viewpoint of biosecurity and quarantine.
Molecular diagnostics based on various mitochondrial and nuclear markers is a powerful diagnostic tool [15,16,17,18], as this method is independent of completeness of the specimen, life stages, colour morphs, and sex [19]. DNA-based molecular markers are essential for classifying living organisms, advancing from visible traits to concealed genetic attributes [20]. Mitochondrial markers, particularly the cytochrome c oxidase subunit I (COI) gene, have become immensely popular due to their high copy number, rapid evolutionary rate, and lack of introns, rendering them an excellent diagnostic marker among the closely related species [15,21,22,23]. Moreover, the COI gene of Z. cucurbitae is distinct enough from the other closely related fruit fly species, supporting the design of species-specific assays based on these nucleotide differences. However, PCR-based methods such as PCR-RFLP [24] and barcoding [15] are more time-consuming, as they require post-PCR procedures such as gel electrophoresis and sequencing, respectively. On the other hand, species identification is possible in 2–3 hours’ time with species-specific real-time PCR-based assays, without post-PCR procedures [25]. Such assays have been extensively employed in routine diagnostics around the world for various biosecurity applications, including the identification of invasive insects [26].
In the present study, we developed and tested the Z. cucurbitae-specific real-time PCR assay and further validated and accredited it to the ISO 17025 standard by extensive testing to demonstrate the specificity and sensitivity of this developed method. Here we present the development and validation results for both the singleplex and duplex real-time PCR assay (with 18S rRNA as an internal control) for Z. cucurbitae, including their sensitivity, specificity, and use of a blind panel test.
2. Materials and Methods
2.1. Sample Collection and Identification
Specimens of Z. cucurbitae at various life stages were obtained from the insect collection resource at the Plant Health and Environment Laboratory (PHEL)—Ministry for Primary Industries (MPI), Auckland, New Zealand, which includes specimens intercepted at our borders and collected from various countries. Additionally, the non-target fruit fly species were also sourced, either as previously extracted genomic DNA or as intact specimens/body parts of the relevant species available in the insect collection at PHEL-MPI. All these specimens were identified by experts at PHEL-MPI either through traditional [27] or integrated taxonomy.
2.2. DNA Extraction Method from Fruit Fly Species
Total DNA from each fruit fly specimen was extracted employing the DNeasy Blood and Tissue kit (Qiagen, Valencia, CA, USA). A portion of an adult leg, a piece of larva/pupae, or eggs were utilized for genomic DNA extraction. Specimens were incubated for an hour at 56 °C in 180 µL of ATL buffer and 20 µL proteinase K, and then the extraction was performed according to the manufacturer’s instructions. Amplification competency of all DNA samples was confirmed by real-time PCR amplification of a commercially available 18S rRNA as an internal control (Applied Biosystems; Foster City, MA, USA). A detailed list of species and specimens deployed in our study is provided in Table 1.

Table 1.
List of specimens employed in the present study.
2.3. PCR Amplification and Sequencing of Fruit Flies
All target species were identified to species either by morphology or by DNA barcoding using LCO 1490 and HCO 2198 primers [28] prior to any further experiments. The PCR reaction contained GoTaq Green master mix (Promega, Madison, WI, USA), 250 nM each of the primers, 0.5 mg/mL BSA, and was conducted using the following PCR cycling parameters: initial denaturation at 94 °C for 5 min; 40 cycles of 94 °C for 15 s, 50 °C for 30 s, 72 °C for 45 s; final extension at 72 °C for 7 min. The amplified products were resolved on 1.0% agarose gel, stained with SYBR™ Safe DNA Gel Stain (Invitrogen, Waltham, MA, USA), and visualized in a gel documentation system (UVP, Ulm, Germany). PCR products were sequenced with the same primers used for PCR. Sanger sequencing of PCR products was performed by EcoGene® (EcoGene, Auckland, New Zealand). All sequences generated from this study have been submitted to NCBI-GenBank under the accession number PV731299–PV731305.
2.4. Sequence Analyses
COI sequences generated from this study were assembled (de novo) and aligned (MUSCLE alignment) using Geneious Prime 2021.1.1 software [29]. Homology search for Z. cucurbitae sequences was carried out using the BLAST programme in NCBI-GenBank (http://www.ncbi.nlm.nih.gov, last accessed on 16 July 2025) and the Barcode of Life Data Systems V4 (BOLD—https://boldsystems.org/, last accessed on 16 July 2025). Furthermore, the sequence variations in the Z. cucurbitae COI gene region were determined with other existing sequences in both NCBI-GenBank and BOLD database using the Geneious Prime 2021.1.1 software [29]. COI sequences belonging to various closely related species within Tephritidae were systematically aligned and analyzed to delineate suitable regions for the design of suitable primers and probes (Figure 1).
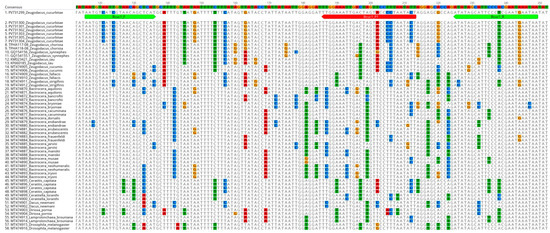
Figure 1.
Sequence alignment of Z. cucurbitae with phylogenetically related non-target species. The sequences PV731299 to PV731304 represent Z. cucurbitae COI sequences obtained from the present study, while the remaining sequences were sourced from the NCBI-GenBank and BOLD databases. Bcuc1_F, Bcuc1_R, and Bcuc1_P1 were the forward primer, reverse primer (both are highlighted in green), and probe (highlighted in red), respectively, developed in this study.
2.5. Z. cucurbitae Real-Time PCR Assay Design
The primer and probe sequences were selected based on the alignment of the partial COI gene sequences of Z. cucurbitae, and the non-target species, including Z. chorista, Z. synnephes, Z. cucumis, and Z. tau. The primers and probes were designed manually, and their secondary structures, along with other thermodynamic characteristics, were analyzed using Geneious Prime 2021.1.1 software and OligoAnalyzer from Integrated DNA Technologies (https://sg.idtdna.com/calc/analyzer, last accessed on 16 July 2025). Such designed primers and probes were further evaluated in Geneious Prime® 2021.1.1 by employing the function for testing specific primers against the chosen sequence or the sequence alignment. We carefully designed primers and probes by ensuring the complete in-silico inclusivity of all known haplotypes of Z. cucurbitae so far and maximized in silico exclusivity against the closely related species. The primers and probe employed in this study are provided in Table 2.

Table 2.
Primer and probe sequences used in this study.
2.6. Real-Time PCR Assay Optimization
To determine the optimal assay conditions, we conducted our real-time PCR experiments on a CFX96™ Touch real-time platform (BioRad Laboratories Inc., Hercules, CA, USA) with different sets of primers and their corresponding probes at various concentrations (250 nM, 300 nM, and 350 nM). Additionally, the assays were tested with different annealing/extension temperatures (58 °C, 60 °C, and 62 °C) in the presence and absence of BSA (10 mg/mL) and MgCl2 (1 mM).
To select the suitable real-time PCR mastermix for our assay, tests were conducted using both PerfeCTa® qPCR ToughMix® (Quanta Bio, Beverly, MA, USA) and SsoAdvanced™ Universal Probes Supermix (BioRad, Laboratories, Hercules, CA, USA). Our new Z. cucurbitae-specific real-time PCR was also evaluated in both singleplex and duplex format, using 18S rRNA as an internal control. All assay optimization tests were conducted in two technical replicates.
2.7. Sensitivity Assessment
To determine the minimum number of target copies that can be detected by the assay, serial dilutions of an Ultramer® DNA Oligo synthetic template (Integrated DNA Technologies, Singapore) were prepared and tested. Sensitivity or limit of detection (LOD) of the Z. cucurbitae COI real-time PCR assay developed was tested with eight serial dilutions (ranging from 108 copies/μL to 10 copies/μL) of the Ultramer® DNA Oligo synthetic templates. All tests for sensitivity were conducted in two technical replicates and the last dilution which resulted in positive amplification in both technical replicates is taken as the LOD for the assay. Furthermore, PCR efficiency, linearity (R2), slope, and y-intercept were also determined as part of this assay development.
2.8. Specificity Assessment
The specificity of the real-time PCR assay was evaluated by using DNA extracted from various target and non-target species (as shown in Table 1) along with synthetic templates of all those closely related fruit flies whose physical specimens were not available for genomic DNA extraction.
2.9. Blind Panel Testing
A total of 34 samples of fruit flies were provided to two scientists for the personalized blind panel testing with no a priori information on their origin or identity. Information about the species and specimens subjected to the blind panel test can be found in Table 3. The samples were diagnosed using the duplex real-time assay format, with each sample tested using technical duplicates. The test included both positive and non-template controls during the assay procedure.

Table 3.
Results for the blind panel test for Z. cucurbitae-specific real-time PCR in duplex format. Samples were tested in technical duplicates for each condition.
3. Results
3.1. Sequence Analyses and Assay Design
A total of 867 COI (5′) gene sequences were downloaded from both NCBI-GenBank and BOLD database (accessed in March 2023), and a total of 13 haplotypes were identified within Z. cucurbitae based on Single Nucleotide Polymorphisms (Appendix A, Figure A1). These identified groups were used for further analyses, including primer and probe design. Both the primers and probes were designed manually, after careful consideration of various parameters such as hairpin, self-dimer, and hetero-dimer. The primers and probes thus designed were finally verified using OligoAnalyzer (https://sg.idtdna.com/calc/analyzer, last accessed on 16th July 2025). Primers were designed to yield an amplicon size of 134 base pairs and the designed probe was positioned closer to the reverse primer. While designing the primers and the probes, utmost care was given to avoid the cross-species amplification by including sufficient nucleotide differences in the regions of the primers/probes, while also aiming to amplify all known thirteen haplotypes within Z. cucurbitae (Figure 1). Our newly designed primers and probe were also subjected to an evaluation process wherein their in silico specificity was assessed against the sequences of phylogenetically related species (Figure 1) to ensure that they have sufficient mismatches to avoid non-specific amplifications. Z. cucurbitae specific primers and probes (Figure 1 and Table 2) thus designed were finally employed in the validation experiments.
Non-target sequences with less than or equal to four mismatches to the probe sequence were selected for further validation to assess the cross-reactivity. Four Zeugodacus species, such as Z. chorista, Z. synnephes, Z. cucumis, and Z. tau, which are phylogenetically closer to Z. cucurbitae, were selected for further specificity testing. Among these, Z. chorista and Z. synnephes specimens were not available to us, and consequently, a synthetic template was generated for the COI region encompassing both the forward and reverse primers. Moreover, the plasmids of Z. cucumis and Z. tau, which carried the COI region as inserts, were utilized in the specificity assessment. The synthetic templates for Z. cucurbitae positive control and non-target species were designed based on the target sequence, primers, and probes. Sequence variations were introduced in the synthetic templates so that they can be identified by sequencing at any later date, if required (Appendix A, Figure A2). The synthetic Z. cucurbitae positive control was used for assay optimization and was also standardized to use as a positive control in future assays.
3.2. Assay Optimization
Both master mixes, namely PerfeCTa® qPCR Toughmix® (QuantaBio, Beverly, MA, USA) and SsoAdvanced™ Universal Probes Supermix (Bio-Rad, Hercules, CA, USA), employed in the initial tests, worked well on the samples tested. Z. cucurbitae samples were identified as positive and non-target samples were identified as negative (Appendix B, Table A1 and Table A2). The newly developed Z. cucurbitae-specific real-time PCR was further optimized for various primer and probe concentrations (200 nM, 250 nM, and 300 nM) and the ideal concentration of the forward and reverse primers was observed to be 250 nM and that of the probe to be 300 nM. We also tested our assay with and without BSA (10 mg/mL); with and without additional MgCl2 (1 mM) as part of optimizing the PCR conditions. Our results indicated that BSA (10 mg/mL) can enhance the assay by preventing non-target amplification, while extra MgCl2 had no effect.
Among the various annealing/extension temperatures (58 °C, 60 °C, and 62 °C) tested, 62 °C was found to be the most suited, exhibiting no cross-reactions at all for any of the non-target DNA tested, including Z. cucumis plasmid, even at 107 copies/µL concentration. The thermal cycling parameters established for our new assay consist of an initial denaturation phase at 95 °C for 2 min, followed by 40 cycles of the denaturation step at 95 °C for 15 s and an annealing/ extension phase at 62 °C for 45 s, and the assay can be run as a singleplex or duplex format by incorporating 18S rRNA primers and probes from Applied Biosystems™ TaqMan™ Ribosomal RNA Control Reagents (Applied Biosystems, Cat # 4308329, Foster City, CA, USA).
3.3. Assay Specificity
The specificity of the newly developed Z. cucurbitae-specific real-time PCR was investigated against a range of genomic DNA (newly and previously extracted), plasmid DNA containing the target region, and synthetic construct for target and non-target Tephritids. Most of the specimens employed in the specificity testing originated from different geographical locations. For example, target genomic DNA (including a few from various life stages) of Z. cucurbitae was prepared from specimens collected from the USA, India, Malaysia, and Indonesia. A total of 32 non-target Tephritids (as listed in Table 1) have been tested, in both singleplex (Appendix B, Table A1) and duplex (including non-target species tested in blind panel test, Table 3) assay format.
All Z. cucurbitae tested produced positive results (with a Cq ≤ 36) and non-target samples produced negative results with no cross-reaction (Table 3). The duplex real-time PCR clearly showed that both target and non-target genomic DNA employed in the specificity assay are PCR-competent (with a Cq value ≤ 30) for 18S rRNA as an internal control (Table 3). Therefore, the newly developed Z. cucurbitae-specific real-time PCR is precise enough to identify Z. cucurbitae specimens in any given life stage.
3.4. Assay Sensitivity
A total of a 169 bp (Appendix A, Figure A2) fragment of the COI gene from Z. cucurbitae was chemically synthesized and resuspended in Tris EDTA (TE) buffer (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. This resuspended synthetic control was used to calculate the copy number and was diluted to 108 copies/µL with TE buffer. From this, ten-fold dilutions were prepared, yielding a final concentration of ten copies, which were used in the Z. cucurbitae-specific real-time PCR testing.
The amplification curves, standard curve (Figure 2a,b), and Cq values (Appendix B, Table A3) for the newly developed Z. cucurbitae-specific real-time PCR sensitivity test indicate amplification signals observed for the synthetic templates down to 10 copies (though it was amplified at later cycles) (Appendix B, Table A3). The 95% confidence limits of the linear dynamic range are plotted in Figure 2b, with a correlation coefficient, R2 of 0.991. The calibration curves also indicated that Z. cucurbitae-specific real-time PCR was able to detect the 10 copies/µL concentration of the synthetic template. Insect samples with Cq ≤ 36 are considered positive for Z. cucurbitae. Samples with Cq values between 36 and 40 should be further investigated for their accurate species identity. Mostly, while performing the real-time PCR, the Cq values for the positive samples are ≤30 (Table 3). Considering the above fact, we have decided to select 1000 copies of COI target/µL concentration of the synthetic template to provide 100% confidence with the limit of detection (LOD).
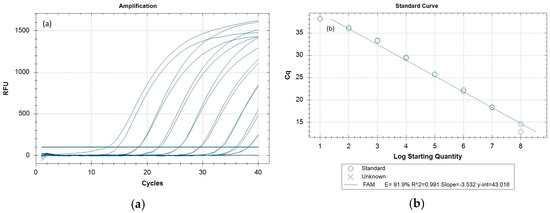
Figure 2.
(a) Amplification curves for the ten-fold dilution series of the Z. cucurbitae synthetic control (108 to 10 copies/µL), employing the PerfeCTa® qPCR Toughmix® (QuantaBio). The baseline threshold was set at 100. (b) Standard curve indicating the efficiency of the Z. cucurbitae-specific real-time PCR. The standard curve was drawn from the Cq values against the log copy number of 108 to 10 copies/µL of the synthetic control fragments.
3.5. Blind Panel Evaluation
A total of 34 insect samples were subjected to blind panel evaluation by two scientists trained in molecular techniques at PHEL-MPI, Auckland. The blind panel tests successfully amplified the target species (Z. cucurbitae), as shown in Table 3, while no amplification was observed in any of the non-target species tested. All tested target and non-target genomic DNAs were PCR-competent as demonstrated with a Cq of less than 30 cycles for the 18S rRNA as an internal control gene.
4. Discussion
Fruit flies that belong to the order Tephritidae are recognized worldwide as major invasive pests, posing a significant threat to agricultural production across diverse climates and regions. Increased transboundary movement of agricultural produce has resulted in the chance introduction of many invasive species that include Zeugodacus and Bactrocera mainly in their immature stages [30,31,32]. Our records at PHEL-MPI show that Tephritid samples are regularly intercepted at the New Zealand border every year as various immature life stages in shipments of fruits and vegetables, which prove to be difficult to identify to the species or even genus level morphologically. Despite numerous identification techniques available for the discrimination of the intercepted pests at immature life stages, morphology-based taxonomic tools are often unreliable and time-consuming, causing significant delays in decision making at the border [24,33]. This demands a quick and accurate species diagnosis method at the port of entry, where the classical taxonomic method has a limited role, as it requires the presence of adult specimens. In this scenario, the molecular methods will be extremely useful as this method is independent of sex, life stages, colour morphs, etc. Considering the economic importance of the fruit fly, several approaches have been tried and tested in the recent past, such as conventional PCR, species-specific PCR, and PCR-RFLP, which have been effective in many ways [19,34,35]. However, such methods are less appropriate for high-throughput or real-time diagnostics due to their labour-intensive nature and the necessity for post-PCR processing. On the other hand, molecular diagnostics based on real-time PCR platforms have transformed the species identification process due to their fast turnaround times, minimal risk of contamination, and being utilized frequently to detect invasive alien insect pests globally [36,37,38].
COI-based DNA barcoding is regarded as one of the most reliable approaches in species discrimination [39] of fruit flies and has proven to be highly effective across various insect orders, including Diptera: Tephritidae [40,41]. Furthermore, it is not merely an identification tool but also enables researchers to verify morphologically similar species and aids in uncovering cryptic species that may be overlooked by traditional methods [18,22,42]. Despite the concerns of its limitations in identifying novel species, issues related to hybridization and regional genetic variation [43,44], the COI gene has become the standardized approach in molecular taxonomy since its inception [45]. Recent research has proven effective in employing the COI gene for the identification of fruit fly species (Diptera: Tephritidae) [40,41,46], and we also endeavoured to apply the same gene for creating species-specific primers and probes for our real-time PCR assay.
The species-specific real-time PCR assays provide a rapid and accurate diagnosis [25], as such assays are equipped with the MIQE guideline for qualitative assays [47]. Several such real-time PCR assays have been developed so far on various fruit fly species including Bactrocera and Zeugodacus species [25,48]. Nevertheless, there are currently no real-time PCR assays available for Z. cucurbitae, an economically important invasive insect pest that poses challenges in identification during its immature life stages.
The presence of common COI haplotypes across different species of fruit flies introduces additional challenges to ultimate species discrimination [49,50,51]. In the present study, primers and probes were meticulously designed for an amplicon size of 134 base pairs in the COI gene region from Z. cucurbitae haplotypes, ensuring the complete absence of any non-specific amplification even with the closely related Tephritid species such as Z. cucumis, by maximizing in silico exclusivity during primer and probe design [52]. Thus designed, the specificity of our primers and probes ensured that only Z. cucurbitae DNA is amplified, avoiding the cross-amplification of DNA from other closely related fruit fly species tested [53].
SYBR green chemistry is known for its simplicity and cost-effectiveness [54,55]. However, the propensity for non-specific amplification remains a significant drawback. Probe-based assays such as those in the present study are known to overcome this issue effectively, and various assays have been successfully developed for many other fruit fly species, such as Z. cucumis, B. jarvisi, B. zonata, and B. dorsalis [25,26,56], even though it pushes the cost a bit higher. The Z. cucurbitae-specific real-time PCR described here provides a swift and precise way of discrimination of these nasty pest species during an interception event, enabling rapid biosecurity decision making. High specificity (no cross-reactions with closely related Tephritids) and sensitivity of the developed assay will be extremely useful in discriminating Z. cucurbitae from the other Tephritids, like how other assays have been designed to differentiate other fruit flies [25,48].
The sensitivity of an assay can be defined as its capacity to identify target DNA at low concentrations, which is of paramount importance for early and reliable identification of invasive pests such as fruit flies. Thus, the high sensitivity reported from our Z. cucurbitae-specific real-time PCR facilitates the identification of minimal pest populations prior to the occurrence of outbreaks, thereby enabling timely intervention in its management and eradication [56].
The Z. cucurbitae-specific real-time PCR developed in this study is appropriate for any organization carrying out routine diagnostics and facilitating trade. This assay will allow for rapid and accurate discrimination of Z. cucurbitae and is fully optimized for immediate deployment at the port of entry in any part of the world, as it can detect the target species from any geographical origin irrespective of its life stages [26,57]. Additionally, the Z. cucurbitae-specific real-time PCR developed in this study at PHEL-MPI, New Zealand, received international accreditation and authorization for application through International Accreditation New Zealand (IANZ—ISO 17025) [58].
5. Conclusions
Effective biosecurity decisions heavily rely on accurate and efficient diagnostic tests that can identify the species in question, particularly with invasive species like fruit flies, irrespective of their life stages, sex, intactness of the specimen, and colour morphs. The sensitivity, specificity, and efficiency of our newly developed assay ensure precise and rapid species identification of Z. cucurbitae from any other Tephritid fruit flies, directly influencing trade, various surveillance programmes, and facilitating effective pest management measures during a response programme. Our assay has shown remarkable suitability for their intended purposes, thereby making them readily adoptable by any other diagnostic laboratories looking to streamline and optimize their Z. cucurbitae diagnostics or screening processes for greater efficiency and accuracy in outcomes. In this domain, we have not only developed a Z. cucurbitae-specific real-time PCR assay but have also attained international accreditation and approval for its utilization through IANZ—ISO 17025.
Author Contributions
Conceptualization, R.K.B., S.G., and S.P.; Formal Analysis, R.K.B. and G.P.; Funding Acquisition, S.G.; Investigation, R.K.B. and G.P.; Data Curation, R.K.B. and D.G.; Methodology, R.K.B., G.P., and D.L.; Project Administration, R.K.B. and S.G.; Resources, S.G. and S.P.; Software, R.K.B.; Supervision, S.G. and S.P.; Validation, R.K.B.; Visualization, R.K.B.; Writing—Original Draft, R.K.B.; Writing–Review and Editing, S.G., G.P., D.L., D.G., and S.P.; All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Data are contained within the article and Appendix A and Appendix B. The original contributions presented in this study are included in the article. All sequences produced from this research have been submitted to NCBI-GenBank (Accession Numbers—PV731299–PV731305). For additional inquiries, please contact the corresponding author.
Acknowledgments
We express our sincere appreciation to Luciano Rigano and Esha Arshad from MPI, for their invaluable and thorough review of the manuscript. Additionally, we are profoundly thankful to Lalith Kumarasinghe, Manager, PHEL-MPI, Auckland for his unwavering support, insightful suggestions, and essential feedback during the work. This study was carried out as a component of the TEPHRIFADE project (Fast detection methods for quarantine Tephritidae—2021-A-373) under Euphresco initiatives—2021 and represents an in-kind contribution from MPI. We would also like to extend our sincere gratitude to both Baldissera Giovani, Euphresco Coordinator, and Negin Ebrahimi, project lead—TEPHRIFADE for their valuable support during the time of publication.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| BOLD | Barcode of Life Data Systems |
| BSA | Bovine Serum Albumin |
| COI | Cytochrome Oxidase I |
| IANZ | International Accreditation New Zealand |
| ISO | International Organization for Standardization |
| MPI | Ministry for Primary Industries |
| NCBI | National Center for Biotechnology Information |
| PCR | Polymerase Chain Reaction |
| PHEL | Plant Health and Environment Laboratory |
| RFLP | Restriction Fragment Length Polymorphism |
| 18S rRNA | 18S ribosomal RNA |
Appendix A

Figure A1.
Sequence alignment of various Z. cucurbitae haplotypes identified (based on COI gene region) within the region of forward and reverse primers (indicated in green). The probe developed in this study is indicated in red. Both the primers and probes matched perfectly to all known haplotypes of Z. cucurbitae.

Figure A2.
Sequence alignment of COI gene region of Z. cucurbitae with that of synthetic template positive control of Z. cucurbitae indicating the introduced variations that can be used to discriminate the positive control contamination, if required later. Forward and reverse primers are marked in green, while the probe is highlighted in red.
Appendix B

Table A1.
Cq values for the newly developed Z. cucurbitae-specific real-time PCR singleplex assay employing the PerfeCTa® qPCR Toughmix® (QuantaBio). Samples were tested in technical duplicates for each condition.
Table A1.
Cq values for the newly developed Z. cucurbitae-specific real-time PCR singleplex assay employing the PerfeCTa® qPCR Toughmix® (QuantaBio). Samples were tested in technical duplicates for each condition.
| Sl. No. | Species ID | Accession Number | Country | Mean Cq (±S.D.) (FAM) | PCR Result |
|---|---|---|---|---|---|
| 1 | Z. cucurbitae | BC_005 | Unknown | 16.15 (±0.287) | ✓ |
| 2 | Z. cucurbitae | BC_009 | Malaysia | 16.21 (±0.211) | ✓ |
| Non-target species | |||||
| 1 | Z. chorista | Not applicable | Synthetic 1 × 108 copies/µL | 0 | × |
| 2 | Z. chorista | Not applicable | Synthetic 1 × 107 copies/µL | 0 | × |
| 3 | B. dorsalis | T18_3481A | Philippines | 0 | × |
| 4 | Z. synnephes | Not applicable | Synthetic 1 × 108 copies/µL | 0 | × |
| 5 | Z. synnephes | Not applicable | Synthetic 1 × 107 copies/µL | 0 | × |
| 6 | B. dorsalis | T18_2947 | India | 0 | × |
| 7 | Z. cucumis | BS_010 | Australia | 0 | × |
| 8 | B. melanotus | BS_012 | Cook Island | 0 | × |
| 9 | Z. cucumis | Not applicable | Plasmid 1 × 109 copies/µL | 0 | × |
| 10 | Z. cucumis | Not applicable | Plasmid 1 × 108 copies/µL | 0 | × |
| 11 | Z. cucumis | Not applicable | Plasmid 1 × 105 copies/µL | 0 | × |
| 12 | B. umbrosa | BS_001 | Malaysia | 0 | × |
| 13 | Z. tau | Not applicable | Plasmid 1 × 107 copies/µL | 0 | × |
| 14 | B. oleae | BS_002 | Greece | 0 | × |
| 15 | B. jarvisi | Not applicable | Plasmid 1 × 105 copies/µL | 0 | × |
| 16 | B. cacuminatus | BS_003 | Australia | 0 | × |
| 17 | B. psidii | BS_004 | Samoa | 0 | × |
| 18 | B. carambolae | BS_006 | Indonesia | 0 | × |
| 19 | B. silvicola | BS_008 | Australia | 0 | × |
| 20 | B. xanthodes | T19_2976 | Fiji | 0 | × |
| 21 | Z. scutellata | T18_2201 | China | 0 | × |
Note: For the Z. cucurbitae assay, Cq (FAM) values ≤ 36 are positive. Samples with Cq (FAM) values between 36 and 40 need to be further investigated (e.g., PCR). ✓: positive; ×: negative.

Table A2.
Cq values for the newly developed Z. cucurbitae-specific real-time PCR duplex assay employing the PerfeCTa® qPCR Toughmix® (QuantaBio). Samples were tested in technical duplicates for each condition.
Table A2.
Cq values for the newly developed Z. cucurbitae-specific real-time PCR duplex assay employing the PerfeCTa® qPCR Toughmix® (QuantaBio). Samples were tested in technical duplicates for each condition.
| Sl. No. | Species ID | Accession Number | Country | Mean Cq (±S.D.) (FAM) | Mean Cq (±S.D.) (VIC) | PCR Result |
|---|---|---|---|---|---|---|
| 1 | Z. cucurbitae | BC_009 | Malaysia | 16.12 (±0.240) | 20.42 (±0.269) | ✓ |
| 2 | Z. cucurbitae | BC_008 | USA | 15.02 (±0.520) | 21.42 (±0.025) | ✓ |
| 3 | Z. cucurbitae | BC_010 | Indonesia | 14.32 (±0.647) | 18.93 (±0.281) | ✓ |
| 4 | Z. cucurbitae | BC_006 | USA | 16.11 (±0.105) | 20.59 (±0.227) | ✓ |
| Non-target species | ||||||
| 1 | B. facialis | T19_00773 | Unknown | 0 | 22.51 (±0.094) | × |
| 2 | B. zonata | T19_00753 | Unknown | 0 | 20.27 (±0.747) | × |
| 3 | B. invadens | BQ_24 | Kenya | 0 | 19.89 (±0.477) | × |
| 4 | B. tryonii | T19_05386 | Unknown | 0 | 23.57 (±0.341) | × |
| 5 | B. tryonii | T19_00487 | Unknown | 0 | 22.39 (±0.482) | × |
| 6 | B. curvipennis | T15_6386 | New Caledonia | 0 | 22.75 (±0.362) | × |
| 7 | C. capitata | CQ_04 | USA | 0 | 28.2 (±0.002) | × |
| 8 | Anastrepha sp. | Ent_38, E6 | Peru | 0 | 28.15 (±0.131) | × |
| 9 | D. melanogaster | T19_3841 | New Zealand | 0 | 28.45 (±0.071) | × |
| 10 | Drosophila sp. | T19_5393 | Mexico | 0 | 21.74 (±0.196) | × |
| 11 | Drosophila sp. | T19_5388 | Mexico | 0 | 21.04 (±0.113) | × |
| 12 | D. suzukii | T19_5260 | Unknown | 0 | 25.06 (±0.095) | × |
| 13 | B. cucumis | BS_010 | Australia | 0 | 22.51 (±0.709) | × |
| 14 | B. passiflora | T19_229 | Fiji | 0 | 24.8 (±0.130) | × |
| 15 | B. jarvisi | T17_4198 | Australia | 0 | 23.21 (±0.178) | × |
| 16 | B. latifrons | T17_711 | Thailand | 0 | 21.12 (±0.019) | × |
| 17 | B. correcta | T15_7487C | Vietnam | 0 | 28.46 (±0.207) | × |
| 18 | C. capitata | DN11 | USA | 0 | 20.71 (±0.317) | × |
| 19 | D. subpulcherella | DD8 | Unknown | 0 | 22.1 (±0.198) | × |
Note: For the Z. cucurbitae assay, Cq (FAM) values ≤ 36 are positive. For internal control (IC), samples with Cq (VIC) values ≤ 30 are PCR-competent. Samples with Cq (FAM) values between 36 and 40 need to be further investigated (e.g., PCR). ✓: positive; ×: negative. S.D.—Standard deviation.

Table A3.
Cq values for the Z. cucurbitae-specific real-time PCR with serially diluted synthetic control from 108 copies to 10 copies/µL. Samples were tested in duplicates for each condition.
Table A3.
Cq values for the Z. cucurbitae-specific real-time PCR with serially diluted synthetic control from 108 copies to 10 copies/µL. Samples were tested in duplicates for each condition.
| Sl. No. | Sample Info | Concentrations | Mean Cq (±S.D.) (FAM) | PCR Result |
|---|---|---|---|---|
| 1 | Synthetic template of Z. cucurbitae | 1 × 108 copies/µL | 13.76 (±1.252) | Positive |
| 2 | Synthetic template of Z. cucurbitae | 1 × 107 copies/µL | 18.37 (±0.028) | Positive |
| 3 | Synthetic template of Z. cucurbitae | 1 × 106 copies/µL | 22.10 (±0.221) | Positive |
| 4 | Synthetic template of Z. cucurbitae | 1 × 105 copies/µL | 25.73 (±0.014) | Positive |
| 5 | Synthetic template of Z. cucurbitae | 1 × 104 copies/µL | 29.45 (±0.147) | Positive |
| 6 | Synthetic template of Z. cucurbitae | 1 × 103 copies/µL | 33.29 (±0.131) | Positive |
| 7 | Synthetic template of Z. cucurbitae | 1 × 102 copies/µL | 36.09 (±0.225) | FTR * |
| 8 | Synthetic template of Z. cucurbitae | 10 copies/µL | 38.17 (±0.000) | FTR * |
| 9 | Water | NTC | 0 | Negative |
Note: All serial dilutions are prepared in TE buffer. FTR *—further testing required for absolute confirmation (for, e.g., PCR). S.D.—standard deviation.
References
- Jacob, V.; Ramiaranjatovo, G.; Persyn, E.; Machara, A.; Kyjaková, P.; Atiama-Nurbel, T.; Pompeiano, A.; Benelli, G.; De Meyer, M.; Vaníčková, L. Female melon fruit flies, Zeugodacus cucurbitae, are attracted to a synthetic chemical blend based on male epicuticular components. J. Pest Sci. 2024, 97, 1395–1415. [Google Scholar] [CrossRef]
- Stark, J.D.; Vargas, R.I. Comparison of sampling methods to estimate the number of oriental fruit fly and melon fly (Diptera: Tephritidae) captured in traps. J. Econ. Entomol. 1990, 83, 2274. [Google Scholar] [CrossRef]
- Ekesi, S.; Samira, A.-E.W. Mass rearing and quality control parameters for tephritid fruit flies of economic importance in Africa. In Insecticides—Pest Engineering; Perveen, F.K., Ed.; InTech: Rijeka, Croatia, 2011. [Google Scholar] [CrossRef]
- Sumathi, E.; Manimaran, R.; Devi, M.N.; Ilamaran, M.; Agila, R. Population dynamics and management of mango fruit fly Bactrocera dorsalis (Hendel) (Diptera: Tephritidae). Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 2705–2710. [Google Scholar] [CrossRef]
- White, I.M.; Elson-Harris, M.M. Fruit Flies of Economic Significance: Their Identification and Bionomics; CAB International: Wallingford, UK, 1992. [Google Scholar]
- Dhillon, M.K.; Singh, R.; Naresh, J.S.; Sharma, H.C. The melon fruit fly, Bactrocera cucurbitae: A review of its biology and management. J. Insect Sci. 2005, 5, 40–46. [Google Scholar] [CrossRef]
- De Meyer, M.; Delatte, H.; Mwatawala, M.; Virgilio, M.; De Villiers, M.; Khamis, F.M.; Ekesi, S. A review of the current knowledge on Zeugodacus cucurbitae (Coquillett) (Diptera, Tephritidae) in Africa, with a list of species included in Zeugodacus. Zookeys 2015, 540, 539–557. [Google Scholar] [CrossRef]
- Halder, J.; Sardana, H.R.; Pandey, M.K.; Nagendran, K.; Bhat, M.N. Synthesis and validation IPM technology and its economic analysis for bottle gourd (Lagenaria siceraria). Indian J. Agric. Sci. 2020, 90, 341–345. [Google Scholar] [CrossRef]
- Rai, A.B.; Halder, J.; Kodandaram, M.H. Emerging insect pest problems in vegetable crops and their management in India: An appraisal. Pest Manag. Hortic. Ecosyst. 2014, 20, 113–122. [Google Scholar]
- Virgilio, M.; Delatte, H.; Backeljau, T.; De Meyer, M. Macrogeographic population structuring in the cosmopolitan agricultural pest Bactrocera cucurbitae (Diptera: Tephritidae). Mol. Ecol. 2010, 19, 2713–2724. [Google Scholar] [CrossRef] [PubMed]
- Doharey, K.L. Bionomics of fruit flies (Dacus spp.) on some fruits. Indian J. Entomol. 1983, 45, 406–413. [Google Scholar]
- Vijay, R.; Keshavareddy, G. Record of melon fruit fly, Zeugodacus cucurbitae (Coquillett) (Diptera: Tephritidae) on tomato: A case of host range expansion. Pest Manag. Hortic. Ecosyst. 2021, 27, 31–36. [Google Scholar] [CrossRef]
- Halder, J.; Rai, A.B.; Deb, D. Distribution and abundance of cucurbit fruit fly Zeugodacus (Bactrocera) cucurbitae in relation to weather parameters. J. Agrometeorol. 2022, 24, 220–222. [Google Scholar] [CrossRef]
- Mehta, S.V.; Haight, R.G.; Homans, F.R.; Polasky, S.; Venette, R.C. Optimal detection and control strategies for invasive species management. Ecol. Econ. 2007, 61, 237–245. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Ratnasingham, S.; deWaard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B 2003, 270 (Suppl. S1), 96–99. [Google Scholar] [CrossRef] [PubMed]
- Buckwalter, J.D.; Angermeier, P.L.; Argentina, J.E.; Wolf, S.L.; Floyd, S.P.; Hallerman, E.M. Drift of larval darters (Family Percidae) in the Upper Roanoke River Basin, USA, characterized using phenotypic and DNA barcoding markers. Fishes 2019, 4, 59. [Google Scholar] [CrossRef]
- Caterino, M.S.; Cho, S.; Sperling, F.A.H. The current state of insect molecular systematics: A thriving tower of Babel. Annu. Rev. Entomol. 2000, 45, 1–54. [Google Scholar] [CrossRef] [PubMed]
- Rebijith, K.B.; Asokan, R.; Krishna, V.; Ranjitha, H.H.; Kumar, N.K.K.; Ramamurthy, V.V. DNA barcoding and elucidation of cryptic diversity in thrips (Thysanoptera). Fla. Entomol. 2014, 97, 1328–1347. [Google Scholar] [CrossRef]
- Asokan, R.; Rebijith, K.B.; Singh, S.K.; Sidhu, A.S.; Siddharthan, S.; Subramanian, S.; Ramamurthy, V. Molecular identification and phylogeny of Bactrocera species (Diptera: Tephritidae). Fla. Entomol. 2011, 94, 1026–1035. [Google Scholar] [CrossRef]
- Burg, T.M.; Catry, P.; Ryan, P.G.; Phillips, R.A. Genetic population structure of Black-browed and Campbell albatrosses, and implications for assigning provenance of birds killed in fisheries. Aquat. Conserv. 2017, 27, 1156–1163. [Google Scholar] [CrossRef]
- Hajibabaei, M.; Janzen, D.H.; Burns, J.M.; Hallwachs, W.; Hebert, P.D.N. DNA barcodes distinguish species of tropical Lepidoptera. Proc. Natl. Acad. Sci. USA 2007, 103, 968–971. [Google Scholar] [CrossRef]
- Rebijith, K.B.; Asokan, R.; Kumar, N.K.; Krishna, V.; Chaitanya, B.N.; Ramamurthy, V.V. DNA barcoding and elucidation of cryptic aphid species (Hemiptera: Aphididae) in India. Bull. Entomol. Res. 2013, 103, 601–610. [Google Scholar] [CrossRef]
- Cebrián-Camisón, S.; la Puente, J.M.; Figuerola, J. A literature review of host feeding patterns of invasive Aedes mosquitoes in Europe. Insects 2020, 11, 848. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, K.F.; Cameron, C.M.; Frampton, E.R.; Suckling, D.M. Aliens at the border and cadavers in the field: A molecular technique for species identification. Proc. N. Z. Plant Prot. Conf. 1997, 50, 316–321. [Google Scholar] [CrossRef]
- Li, D.; Waite, D.W.; Gunawardana, D.N.; McCarthy, B.; Anderson, D.; Flynn, A.; George, S. DNA barcoding and real-time PCR detection of Bactrocera xanthodes (Tephritidae: Diptera) complex. Bull. Entomol. Res. 2018, 108, 1–9. [Google Scholar]
- Koohkanzade, M.; Zakiaghl, M.; Dhami, M.K.; Fekrat, L.; Namaghi, H.S. Rapid identification of Bactrocera zonata (Dip.: Tephritidae) using TaqMan real-time PCR assay. PLoS ONE 2018, 13, e0205136. [Google Scholar] [CrossRef]
- Plant Health Australia. The Australian Handbook for Identification of Fruit Flies; Version 3; Plant Health Australia: Canberra, ACT, Australia, 2018. [Google Scholar]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Barr, N.B.; Islam, M.S.; De Meyer, M.; McPheron, B.A. Molecular identification of Ceratitis capitata (Diptera: Tephritidae) using DNA sequences of the COI barcode region. Ann. Entomol. Soc. Am. 2012, 105, 339–350. [Google Scholar] [CrossRef]
- Blacket, M.; Semeraro, L.; Malipatil, M. Barcoding Queensland fruit flies (Bactrocera tryoni): Impediments and improvements. Mol. Ecol. Resour. 2012, 12, 428–436. [Google Scholar] [CrossRef]
- Hulme, P.E. Trade, transport and trouble: Managing invasive species pathways in an era of globalization. J. Appl. Ecol. 2009, 46, 10–18. [Google Scholar] [CrossRef]
- Dhami, M.K.; Kumarasinghe, L. A HRM real-time PCR assay for rapid and specific identification of the emerging pest spotted-wing Drosophila (Drosophila suzukii). PLoS ONE 2014, 9, e98934. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, K.F.; Ball, S.L. DNA barcodes for biosecurity: Invasive species identification. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005, 360, 1813–1823. [Google Scholar] [CrossRef]
- Chua, T.H.; Chong, Y.V.; Lim, S.H. Species determination of Malaysian Bactrocera pests using PCR-RFLP analyses (Diptera: Tephritidae). Pest Manag. Sci. 2009, 66, 379–384. [Google Scholar] [CrossRef]
- Walsh, K.; Boonham, N.; Barker, I.; Collins, D.W. Development of a sequence-specific real-time PCR to the melon thrips, Thrips palmi (Thysanoptera: Thripidae). J. Appl. Entomol. 2005, 129, 272–279. [Google Scholar] [CrossRef]
- Barr, N.B.; Ledezma, L.A.; Farris, R.E.; Epstein, M.E.; Gilligan, T.M. A multiplex real-time polymerase chain reaction assay to diagnose Epiphyas postvittana (Lepidoptera: Tortricidae). J. Econ. Entomol. 2011, 104, 1706–1719. [Google Scholar] [CrossRef] [PubMed]
- Naaum, A.M.; Foottit, R.G.; Maw, H.E.L.; Hanner, R. Real-time PCR for identification of the soybean aphid, Aphis glycines Matsumura. J. Appl. Entomol. 2014, 138, 485–489. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Stoeckle, M.Y.; Zemlak, T.S.; Francis, C.M. Identification of birds through DNA barcodes. PLoS Biol. 2004, 2, e312. [Google Scholar] [CrossRef]
- Kar, O.; Mukherjee, A.; Ghosh, D.; Mukherjee, K.; Pramanik, D.; Naskar, A.; Banerjee, D. DNA barcoding of economically important fruit flies (Diptera: Tephritidae) from the Lower Gangetic Plains of Eastern India. J. Adv. Biol. Biotechnol. 2024, 27, 587–604. [Google Scholar] [CrossRef]
- Doorenweerd, C.; San Jose, M.; Leblanc, L.; Barr, N.; Geib, S.M.; Chung, A.Y.; Dupuis, J.R.; Ekayanti, A.; Flegalan, E.; Hemachandra, K.S.; et al. DNA barcodes and reliable molecular identifications in a diverse group of invasive pests: Lessons from Bactrocera fruit flies on variation across the COI gene, introgression, and standardization. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ko, H.-L.; Wang, Y.-T.; Chiu, T.; Lee, M.; Leu, M.; Chang, K.-Z.; Chen, W.; Shao, K. Evaluating the accuracy of morphological identification of larval fishes by applying DNA barcoding. PLoS ONE 2013, 8, e53451. [Google Scholar] [CrossRef]
- Becker, S.; Hanner, R.; Steinke, D. Five years of FISH-BOL: Brief status report. Mitochondrial DNA 2011, 22, 3–9. [Google Scholar] [CrossRef]
- Ahmed, S.; Ibrahim, M.; Nantasenamat, C.; Nisar, M.; Malik, A.A.; Waheed, R.; Ahmed, M.Z.; Ojha, S.C.; Alam, M.K. Pragmatic applications and universality of DNA barcoding for substantial organisms at species level: A review to explore a way forward. BioMed Res. Int. 2022, 2022, 1846485. [Google Scholar] [CrossRef] [PubMed]
- Hubert, N.; Hanner, R.; Holm, E.; Mandrak, N.E.; Taylor, E.B.; Burridge, M.; Watkinson, D.A.; Dumont, P.; Curry, R.A.; Bentzen, P.; et al. Identifying Canadian freshwater fishes through DNA barcodes. PLoS ONE 2008, 3, e2490. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekhar, V.; Arya, V.; David, K.J.; Narayana, S. Diagnosis, taxonomic keys, DNA barcoding and molecular phylogeny of economically important fruit fly species (Diptera: Tephritidae). Int. J. Trop. Insect Sci. 2024, 44, 3021–3035. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Dhami, M.K.; Gunawardana, D.N.; Voice, D.; Kumarasinghe, L. A real-time PCR toolbox for accurate identification of invasive fruit fly species. J. Appl. Entomol. 2016, 140, 536–552. [Google Scholar] [CrossRef]
- Doorenweerd, C.; Jose, M.S.; Barr, N.; Rubinoff, D.; Geib, S. Genomic data reveal new species and the limits of mtDNA barcode diagnostics to contain a global pest species complex (Diptera: Tephritidae: Dacinae). Syst. Entomol. 2023, 48, e12616. [Google Scholar] [CrossRef]
- Schutze, M.K.; Aketarawong, N.; Amornsak, W.; Armstrong, K.F.; Augustinos, A.A.; Barr, N.; Bo, W.; Bourtzis, K.; Boykin, L.M.; Cáceres, C. Synonymization of key pest species within the Bactrocera dorsalis species complex (Diptera: Tephritidae): Taxonomic changes based on a review of 20 years of integrative morphological, molecular, cytogenetic, behavioural and chemoecological data. Syst. Entomol. 2015, 40, 456–471. [Google Scholar] [CrossRef]
- Nugnes, F.; Russo, E.; Viggiani, G.; Bernardo, U. First record of an invasive fruit fly belonging to Bactrocera dorsalis complex (Diptera: Tephritidae) in Europe. Insects 2018, 9, 182. [Google Scholar] [CrossRef]
- Cardenas-Cadena, S.A.; Castañeda-Lopez, M.E.; Mollinedo-Montaño, F.E.; Vazquez-Reyes, S.; Lara-Arias, J.; Marino-Martinez, I.A.; Rodriguez-Sanchez, I.P.; Garza-Veloz, I.; Martinez-Fierro, M.L. Tick-borne pathogens screening using a multiplex real-time polymerase chain reaction-based method. Acta Parasitol. 2023, 68, 705–710. [Google Scholar] [CrossRef]
- Tang, M.; Tan, M.; Meng, G.; Yang, S.; Su, X.; Liu, S.; Song, W.; Li, Y.; Wu, Q.; Zhang, A.; et al. Multiplex sequencing of pooled mitochondrial genomes—A crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 2014, 42, e166. [Google Scholar] [CrossRef]
- Yu, D.; Zhang, G.M.; Chen, Z.L.; Zhang, R.J.; Yin, W.Y. Rapid identification of Bactrocera latifrons (Dipt., Tephritidae) by real-time PCR using SYBR Green chemistry. J. Appl. Entomol. 2004, 128, 670–676. [Google Scholar] [CrossRef]
- Yu, D.; Chen, Z.; Zhang, R.; Yin, W. Real-time qualitative PCR for the inspection and identification of Bactrocera phillippinensis and Bactrocera occipitalis (Diptera: Tephritidae) using SYBR green assay. Raffles Bull. Zool. 2005, 53, 73–78. [Google Scholar]
- Rizzo, D.; Zubieta, C.G.; Sacchetti, P.; Marrucci, A.; Miele, F.; Ascolese, R.; Nugnes, F.; Bernardo, U. Diagnostic tool for the identification of Bactrocera dorsalis (Hendel) (Diptera: Tephritidae) using real-time PCR. Insects 2024, 15, 44. [Google Scholar] [CrossRef]
- Asokan, R.; Rebijith, K.B.; Singh, S.K.; Ramamurthy, V. Life stage independent identification of fruit flies (Diptera: Tephritidae) using 28s rDNA sequences. Bioscan 2013, 8, 253–256. [Google Scholar]
- ISO/IEC 17025:2017; General Requirements for the Competence of Testing and Calibration Laboratories. International Organization for Standardization (ISO): Geneva, Switzerland, 2017.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).