Cryptic Diversity and Climatic Niche Divergence of Brillia Kieffer (Diptera: Chironomidae): Insights from a Global DNA Barcode Dataset
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
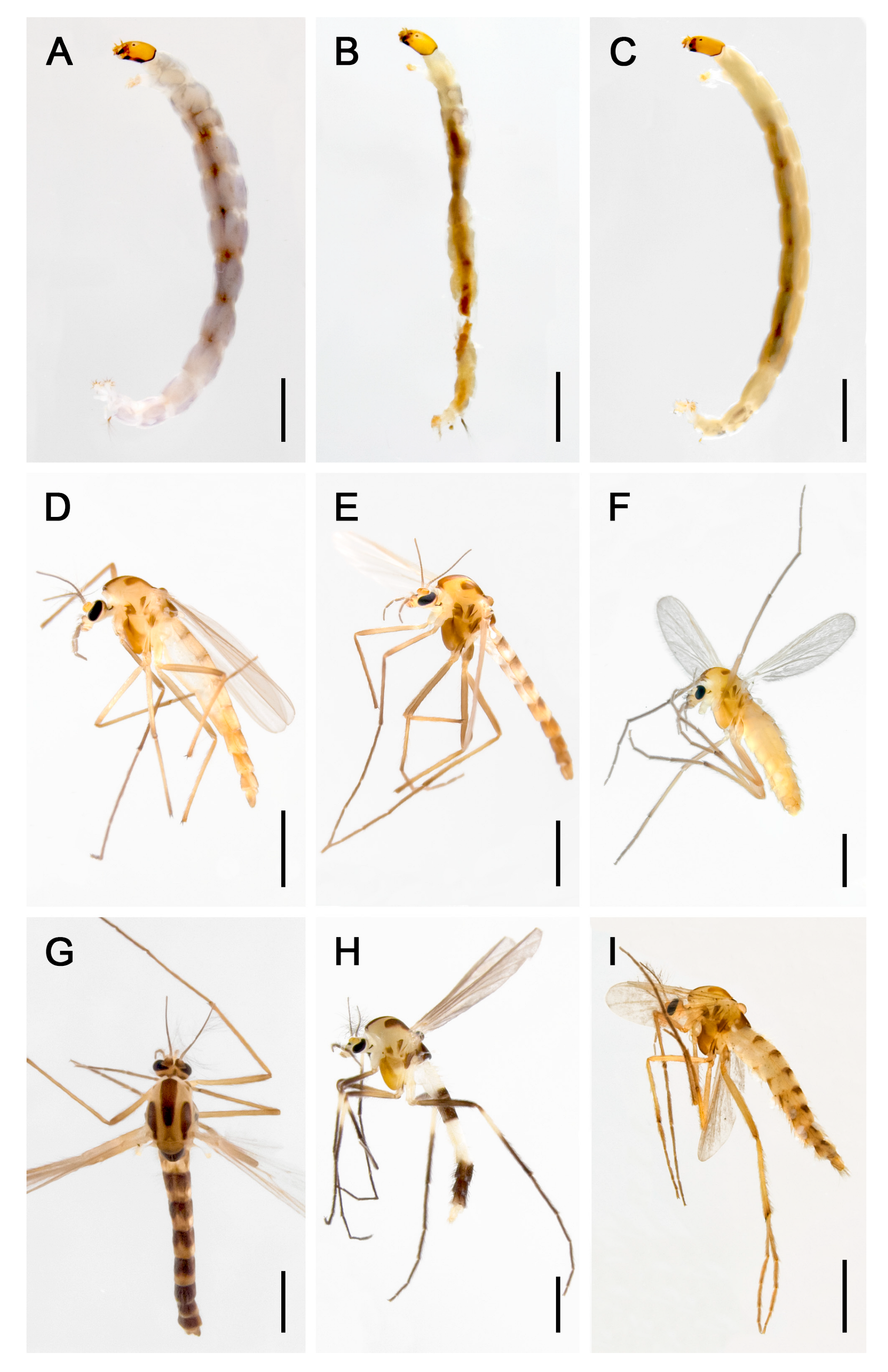
3. Results
3.1. Dataset Characteristics and Global Distribution
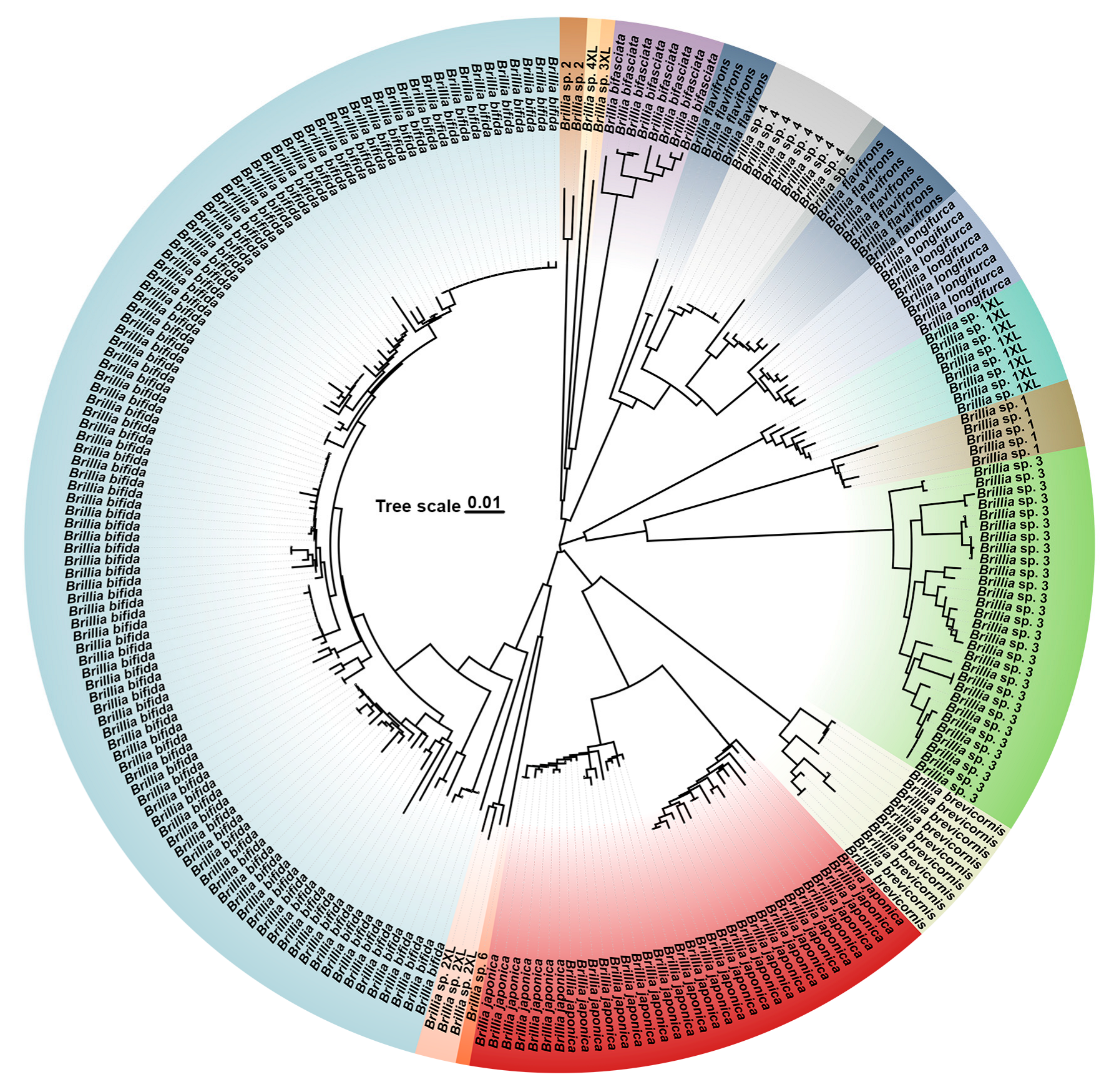
3.2. Species Clustering
3.3. Species Delimitation and Genetic Divergence Revealed by ABGD Analysis
3.4. Haplotype Diversity and Geographic Specificity
3.5. Niche Differentiation of Brillia Geographic Groups
4. Discussion
4.1. Geographic Distribution of Brillia
4.2. Taxonomic Challenges Revealed by ABGD and NJ Analysis
4.3. Geographic Differentiation and Localized Haplotypes in Brillia
4.4. Ecological Niche Divergence Driven by Climatic and Topographic Factors
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Butchart, S.H.; Walpole, M.; Collen, B.; Van Strien, A.; Scharlemann, J.P.; Almond, R.E.; Baillie, J.E.; Bomhard, B.; Brown, C.; Bruno, J.; et al. Global biodiversity: Indicators of recent declines. Science 2010, 328, 1164–1168. [Google Scholar] [CrossRef] [PubMed]
- Newbold, T.; Hudson, L.N.; Hill, S.L.; Contu, S.; Lysenko, I.; Senior, R.A.; Börger, L.; Bennett, D.J.; Choimes, A.; Collen, B.; et al. Global effects of land use on local terrestrial biodiversity. Nature 2015, 520, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Díaz, S.; Fargione, J.; Chapin, F.S., III; Tilman, D. Biodiversity loss threatens human well-being. PLoS Biol. 2006, 4, e277. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.P.; Port, J.A.; Yamahara, K.M.; Martone, R.G.; Lowell, N.; Thomsen, P.F.; Mach, M.E.; Bennett, M.; Prahler, E.; Caldwell, M.R.; et al. Harnessing DNA to improve environmental management. Science 2014, 344, 1455–1456. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Willerslev, E. Environmental DNA–An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015, 183, 4–18. [Google Scholar] [CrossRef]
- Vié, J.-C.; Hilton-Taylor, C.; Stuart, S.N. Wildlife in a Changing World: An Analysis of the 2008 IUCN Red List of Threatened Species, 1st ed.; IUCN: Gland, Switzerland, 2009; pp. 156–160. [Google Scholar]
- Mirone, E. Non-invasive Monitoring of Freshwater Species in Central and Southern Italian Basins: From Standard Survey to Environmental DNA. Ph.D. Thesis, University of Molise, Pesche, Italy, 2024. [Google Scholar]
- Hebert, P.D.N.; Penton, E.H.; Burns, J.M.; Janzen, D.H.; Hallwachs, W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. USA 2004, 101, 14812–14817. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. Ser. B. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Hajibabaei, M.; Baird, D.J.; Fahner, N.A.; Beiko, R.; Golding, G.B. A new way to contemplate Darwin’s tangled bank: How DNA barcodes are reconnecting biodiversity science and biomonitoring. Philos. Trans. R. Soc. B. 2016, 371, 20150330. [Google Scholar] [CrossRef]
- Kanturski, M.; Lee, Y.; Choi, J.; Lee, S. DNA barcoding and a precise morphological comparison revealed a cryptic species in the Nippolachnus piri complex (Hemiptera: Aphididae: Lachninae). Sci. Rep. 2018, 8, 8998. [Google Scholar] [CrossRef]
- Young, M.R.; Proctor, H.C.; deWaard, J.R.; Hebert, P.D.N. DNA barcodes expose unexpected diversity in Canadian mites. Mol. Ecol. 2019, 28, 5347–5359. [Google Scholar] [CrossRef]
- Taberlet, P.; Coissac, E.; Hajibabaei, M.; Rieseberg, L.H. Environmental DNA. Mol. Ecol. 2012, 21, 1789–1793. [Google Scholar] [CrossRef] [PubMed]
- Deiner, K.; Bik, H.M.; Mächler, E.; Seymour, M.; Lacoursière-Roussel, A.; Altermatt, F.; Creer, S.; Bista, I.; Lodge, D.M.; De Vere, N.; et al. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 2017, 26, 5872–5895. [Google Scholar] [CrossRef] [PubMed]
- Macher, J.N.; Vivancos, A.; Piggott, J.J.; Centeno, F.C.; Matthaei, C.D.; Leese, F. Comparison of environmental DNA and bulk-sample metabarcoding using highly degenerate cytochrome c oxidase I primers. Mol. Ecol. Resour. 2018, 18, 1456–1468. [Google Scholar] [CrossRef]
- Leese, F.; Bouchez, A.; Abarenkov, K.; Altermatt, F.; Borja, Á.; Bruce, K.; Ekrem, T.; Čiampor, F., Jr.; Čiamporová-Zat’ovičová, Z.; Costa, F.O.; et al. Why we need sustainable networks bridging countries, disciplines, cultures and generations for aquatic biomonitoring 2.0: A perspective derived from the DNAqua-Net COST action. Adv. Ecol. Res. 2018, 58, 63–99. [Google Scholar] [CrossRef]
- Morinière, J.; Hendrich, L.; Balke, M.; Beermann, A.J.; König, T.; Hess, M.; Koch, S.; Müller, R.; Leese, F.; Hebert, P.D.; et al. A DNA barcode library for Germany’s mayflies, stoneflies and caddisflies (Ephemeroptera, Plecoptera and Trichoptera). Adv. Ecol. Res. 2017, 17, 1293–1307. [Google Scholar] [CrossRef]
- Weigand, H.; Beermann, A.J.; Čiampor, F.; Costa, F.O.; Csabai, Z.; Duarte, S.; Geiger, M.F.; Grabowski, M.; Rimet, F.; Rulik, B.; et al. DNA barcode reference libraries for the monitoring of aquatic biota in Europe: Gap-analysis and recommendations for future work. Sci. Total Environ. 2019, 678, 499–524. [Google Scholar] [CrossRef]
- Lin, X.-L.; Stur, E.; Ekrem, T. Exploring genetic divergence in a species-rich insect genus using 2790 DNA Barcodes. PLoS ONE 2015, 10, e0138993. [Google Scholar] [CrossRef]
- Zhou, X.; Kjer, K.M.; Morse, J.C. Associating larvae and adults of Chinese Hydropsychidae caddisflies (Insecta: Trichoptera) using DNA sequences. J. N. Am. Benthol. Soc. 2007, 26, 719–742. [Google Scholar] [CrossRef]
- Carew, M.E.; Nichols, S.J.; Batovska, J.; St Clair, R.; Murphy, N.P.; Blacket, M.J.; Shackleton, M.E. A DNA barcode database of Australia’s freshwater macroinvertebrate fauna. Mar. Freshwater Res. 2017, 68, 1788–1802. [Google Scholar] [CrossRef]
- Galimberti, A.; Assandri, G.; Maggioni, D.; Ramazzotti, F.; Baroni, D.; Bazzi, G.; Chiandetti, I.; Corso, A.; Ferri, V.; Galuppi, M.; et al. Italian odonates in the Pandora’s box: A comprehensive DNA barcoding inventory shows taxonomic warnings at the Holarctic scale. Mol. Ecol. Resour. 2021, 21, 183–200. [Google Scholar] [CrossRef]
- Wang, H.; Xu, H.F.; Li, C.H.; Zhang, H.X.; Zhang, Y.P.; Sun, B.J.; Lin, X.L. Towards a Comprehensive DNA Barcode Library of Stenochironomus Kieffer, 1919 (Diptera: Chironomidae) from China. Diversity. 2024, 16, 257. [Google Scholar] [CrossRef]
- Yao, Y.; Chen, J.Y.; Gong, X.L.; Li, C.H.; Liu, Z.; Lin, X.L. Species Delimitation and Cryptic Diversity in Rheotanytarsus Thienemann & Bause, 1913 (Diptera: Chironomidae) Based on DNA Barcoding. Insects 2025, 16, 370. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.-L.; Yu, H.-J.; Wang, Q.; Bu, W.-J.; Wang, X.-H. DNA barcodes and morphology confirm a new species of Rheocricotopus (Psilocricotopus) orientalis group (Diptera: Chironomidae). Zootaxa. 2020, 4768, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Li, K.; Liu, X. Comprehensive DNA barcode reference library and optimization of genetic divergence threshold facilitate the exploration of species diversity of green lacewings (Neuroptera: Chrysopidae). Insect Sci. 2024, 31, 613–632. [Google Scholar] [CrossRef]
- Lai, Y.; Liu, Y.; Liu, X. Elevational Diversity Patterns of Green Lacewings (Neuroptera: Chrysopidae) Uncovered With DNA Barcoding in a Biodiversity Hotspot of Southwest China. Front. Ecol. Evol. 2021, 9, 778686. [Google Scholar] [CrossRef]
- Lin, X.-L.; Stur, E.; Ekrem, T. DNA barcodes and morphology reveal unrecognized species in Chironomidae (Diptera). Insect Syst. Evol. 2018, 49, 329–398. [Google Scholar] [CrossRef]
- Lin, X.L.; Mo, L.; Bu, W.J.; Wang, X.H. The first comprehensive DNA barcode reference library of Chinese Tanytarsus (Diptera: Chironomidae) for environmental DNA metabarcoding. Divers. Distrib. 2021, 27, 1932–1941. [Google Scholar] [CrossRef]
- Ashe, P.; O’Connor, J.P. A World Catalogue of Chironomidae (Diptera), Part 1: Buchonomyiinae, Chilenomyiinae, Podonominae, Aphroteniinae, Tanypodinae, Usambaromyiinae, Diamesinae, Prodiamesinae and Telmatogetoninae, 1st ed.; Irish Biogeographical Society: Dublin, Ireland, 2009; p. 445. Available online: https://irishbiogeographicalsociety.com/listings.html (accessed on 11 May 2025).
- Ferrington, L.C. Global diversity of non-biting midges (Chironomidae; Insecta-Diptera) in freshwater. Hydrobiologia 2008, 595, 447–455. [Google Scholar] [CrossRef]
- Pape, T.; Evenhuis, N. Systema Dipterorum. Biodivers. Inf. Sci. Stand. 2023, 7, e111959. [Google Scholar] [CrossRef]
- Medeiros, A.S.; Milošević, Đ.; Francis, D.R.; Maddison, E.; Woodroffe, S.; Long, A.; Walker, I.R.; Hamerlík, L.; Quinlan, R.; Langdon, P.; et al. Arctic chironomids of the northwest North Atlantic reflect environmental and biogeographic gradients. J. Biogeogr. 2021, 48, 511–525. [Google Scholar] [CrossRef]
- Rico, E.; Quesada, A. Distribution and ecology of chironomids (Diptera, Chironomidae) on Byers Peninsula, Maritime Antarctica. Antarct. Sci. 2013, 25, 288–291. [Google Scholar] [CrossRef]
- Brundin, L. Transantarctic relationships and their significance, as evidenced by chironomid midges with a monograph of the subfamilies Podonominae and Aphroteniinae and the austral Heptagynae. Kongl. Sv. Vet.-Akad. Handl. 1966, 11, 1–472. [Google Scholar]
- Cranston, P.S.; Hardy, N.B.; Morse, G.E. A dated molecular phylogeny for the Chironomidae (Diptera). Syst. Entomol. 2012, 37, 172–188. [Google Scholar] [CrossRef]
- Krosch, M.; Cranston, P.S. Not drowning, (hand) waving? Molecular phylogenetics, biogeography and evolutionary tempo of the ‘Gondwanan’ midge Stictocladius Edwards (Diptera: Chironomidae). Mol. Phylogenet. Evol. 2013, 68, 595–603. [Google Scholar] [CrossRef]
- Lin, X.-L.; Stur, E.; Ekrem, T. Molecular phylogeny and temporal diversification of Tanytarsus van der Wulp (Diptera: Chironomidae) support generic synonymies, a new classification and centre of origin. Syst. Entomol. 2018, 43, 659–677. [Google Scholar] [CrossRef]
- Hamilton, C.A.; Hendrixson, B.E.; Brewer, M.S.; Bond, J.E. An evaluation of sampling effects on multiple DNA barcoding methods leads to an integrative approach for delimiting species: A case study of the North American tarantula genus Aphonopelma (Araneae, Mygalomorphae, Theraphosidae). Mol. Phylogenet Evol. 2014, 71, 79–93. [Google Scholar] [CrossRef]
- Chimeno, C.; Rulik, B.; Manfrin, A.; Kalinkat, G.; Hölker, F.; Baranov, V. Facing the infinity: Tackling large samples of challenging Chironomidae (Diptera) with an integrative approach. PeerJ 2023, 11, e15336. [Google Scholar] [CrossRef]
- Armitage, P.D.; Pinder, L.; Cranston, P. The Chironomidae: Biology and Ecology of Non-Biting Midges, 1st ed.; Springer Science & Business Media: Dordrecht, The Netherlands, 2012; p. 572. Available online: https://link.springer.com/book/10.1007/978-94-011-0715-0 (accessed on 11 May 2025).
- Padial, J.M.; Miralles, A.; De la Riva, I.; Vences, M. The integrative future of taxonomy. Front. Zool. 2010, 7, 16. [Google Scholar] [CrossRef]
- Schlick-Steiner, B.C.; Steiner, F.M.; Seifert, B.; Stauffer, C.; Christian, E.; Crozier, R.H. Integrative taxonomy: A multisource approach to exploring biodiversity. Annu. Rev. Entomol. 2010, 55, 421–438. [Google Scholar] [CrossRef]
- Oliver, D.R.; Roussel, M.E. Redescription of Brillia Kieffer (Diptera, Chironomidae) with Descriptions of Nearctic Species. Can. Entomol. 1983, 115, 257–279. [Google Scholar] [CrossRef]
- Sæther, O.A. Some Nearctic Podonominae, Diamesinae, and Orthocladiinae (Diptera: Chironomidae). Bull. Fish. Res. Board. Can. 1969, 170, 1–154. [Google Scholar]
- Wang, X.; Zheng, L.; Ji, B. A taxonomic study on orthodadiinae (Diptera: Chironomidae) of China. II. genus Brillia kieffer. Acta Entomol. Sin. 1994, 37, 359–363. [Google Scholar]
- Zakrzewska, M.; Andersen, T.; Gilka, W. Mimes of the past: Eocene midges of the tribe Pseudochironomini (Chironomidae, Diptera) reveal their peculiarities. PLoS ONE 2023, 18, e0295841. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Tang, Y.; Nie, J.; Liu, W.; Yan, C. Brillia litangensis (Insecta, Diptera, Chironomidae), a new species from Sichuan, China. Ann. Zool. Fenn. 2025, 62, 65–69. [Google Scholar] [CrossRef]
- Cobo, F.; Gonzalez, M.; Vieira-Lanero, R. Notes on some taxonomic problems in the Iberian species of Brillia Kieffer, 1913 (Diptera: Chironomidae), with a description of B. pudorosa sp.n. Ann. Limnol.-Int. J. Lim. 1995, 31, 245–252. [Google Scholar] [CrossRef]
- Larocque-Tobler, I. The Polish sub-fossil chironomids. Palaeontol. Electron. 2014, 17, 1–28. [Google Scholar] [CrossRef][Green Version]
- Sæther, O.A.; Wang, X.H. Euryhapsis fuscipropes sp.n. from China and Tokyobrillia anderseni sp.n. from Tanzania, with a review of genera near Irisobrillia Oliver (Diptera: Chironomidae). Ann. Limnol.-Int. J. Lim. 1992, 28, 209–223. [Google Scholar] [CrossRef]
- Ashe, P.; O’Connor, J.P. A World Catalogue of Chironomidae (Diptera). Part 2. Orthocladiinae, 1st ed.; Irish Biogeographical Society: Dublin, Ireland, 2012; p. 968. [Google Scholar]
- Grove, S.J.; Hanula, J.L. Insect Biodiversity and Dead Wood: Proceedings of a Symposium for the 22nd International Congress of Entomology, Brisbane, Australia, 15–21 August 2004, 1st ed.; US Department of Agriculture, Forest Service, Southern Research Station: Asheville, NC, USA, 2006; p. 109. [Google Scholar]
- Sæther, O.A. Female genitalia in Chironomidae and other Nematocera: Morphology, phylogenies, keys. Bull. Fish. Res. Board. Can. 1977, 197, 1–209. [Google Scholar]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data System (www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics, 1st ed.; Oxford University Press: New York, NY, USA, 2000; pp. 49–83. [Google Scholar]
- Cock, P.J.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422. [Google Scholar] [CrossRef] [PubMed]
- Van Rossum, G.; Drake, F.L. Python 3 Reference Manual, 1st ed.; CreateSpace: Scotts Valley, CA, USA, 2009; pp. 1–242. [Google Scholar]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. Notes 2012, 21, 1864–1877. [Google Scholar] [CrossRef]
- Clement, M.; Snell, Q.; Walker, P.; Posada, D.; Crandall, K. TCS: Estimating gene genealogies. In Proceedings of the 16th International Parallel and Distributed Processing Symposium, Fort Lauderdale, FL, USA, 15–19 April 2002. [Google Scholar]
- Leigh, J.W.; Bryant, D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Karger, D.N.; Conrad, O.; Böhner, J.; Kawohl, T.; Kreft, H.; Soria-Auza, R.W.; Zimmermann, N.E.; Linder, H.P.; Kessler, M. Climatologies at high resolution for the earth’s land surface areas. Sci. Data 2017, 4, 170122. [Google Scholar] [CrossRef]
- Global Soil Water Balance Geospatial Database. CGIAR Consortium for Spatial Information. CGIAR-CSI GeoPortal. Available online: https://csidotinfo.wordpress.com/data/global-high-resolution-soil-water-balance/ (accessed on 18 May 2025).
- Wilson, A.M.; Jetz, W. Remotely sensed high-resolution global cloud dynamics for predicting ecosystem and biodiversity distributions. PLoS Biol. 2016, 14, e1002415. [Google Scholar] [CrossRef]
- Robinson, N.; Regetz, J.; Guralnick, R.P. EarthEnv-DEM90: A nearly-global, void-free, multi-scale smoothed, 90m digital elevation model from fused ASTER and SRTM data. ISPRS J. Photogramm. Remote Sens. 2014, 87, 57–67. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. 2024. Available online: https://www.R-project.org/ (accessed on 20 May 2025).
- Meier, R.; Shiyang, K.; Vaidya, G.; Ng, P.K. DNA barcoding and taxonomy in Diptera: A tale of high intraspecific variability and low identification success. Syst. Biol. 2006, 55, 715–728. [Google Scholar] [CrossRef]
- Fiscus, C.J. Causes and Consequences of Plant Genome Evolution. Ph.D. Thesis, University of California, Riverside, CA, USA, 2022; p. 138. [Google Scholar]
- Ma, C.; Yang, P.; Jiang, F.; Chapuis, M.P.; Shali, Y.; Sword, G.A.; Kang, L.E. Mitochondrial genomes reveal the global phylogeography and dispersal routes of the migratory locust. Mol. Ecol. 2012, 21, 4344–4358. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, J. DNA barcoding a nightmare taxon: Assessing barcode index numbers and barcode gaps for sweat bees. Genome 2018, 61, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Wolton, R.J.; Bentley, H.; Chandler, P.J.; Drake, C.M.; Kramer, J.; Plant, A.R.; Stubbs, A.E. The diversity of Diptera associated with a British hedge. Dipterists Digest. 2014, 21, 1–36. [Google Scholar]
- Spies, M.; Saether, O.A. Notes and recommendations on taxonomy and nomenclature of Chironomidae (Diptera). Zootaxa 2004, 752, 1–90. [Google Scholar] [CrossRef]
- Roháček, J.; Tóthová, A. Morphology versus DNA-what will bring clarity to the relationships of phylogenetically unclear genera of Anthomyzidae (Diptera)? Arthropod. Syst. Phylogeny 2014, 72, 165–176. [Google Scholar] [CrossRef]
- Huemer, P.; Mutanen, M. An incomplete European barcode library has a strong impact on the identification success of Lepidoptera from Greece. Diversity 2022, 14, 118. [Google Scholar] [CrossRef]
- Silva-Santos, R.; Machado Cd, B.; Zanata, A.M.; Camelier, P.; Galetti, P.M., Jr.; Freitas, P.D.d. Molecular characterization of Astyanax species (Characiformes: Characidae) from the upper Paraguaçu River basin, a hydrographic system with high endemism. Neotrop. Ichthyol. 2023, 21, e230032. [Google Scholar] [CrossRef]
- Sandanov, D.V.; Kholina, A.B.; Kozyrenko, M.M.; Artyukova, E.V.; Wang, Z. Genetic diversity of Oxytropis species from the center of the genus origin: Insight from molecular studies. Diversity 2023, 15, 244. [Google Scholar] [CrossRef]
- Goodisman, M.A.; Crozier, R.H. Population and colony genetic structure of the primitive termite Mastotermes darwiniensis. Evolution 2002, 56, 70–83. [Google Scholar] [CrossRef]
- Chiari, Y.; van der Meijden, A.; Mucedda, M.; Lourenco, J.M.; Hochkirch, A.; Veith, M. Phylogeography of Sardinian cave salamanders (genus Hydromantes) is mainly determined by geomorphology. PLoS ONE 2012, 7, e32332. [Google Scholar] [CrossRef]
- Ford, B.M.; Cornellas, A.; Leonard, J.A.; Weir, R.D.; Russello, M.A. Spatiotemporal analyses suggest the role of glacial history and the ice-free corridor in shaping American badger population genetic variation. Ecol. Evol. 2020, 10, 8345–8357. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, G.M. The structure of biodiversity–insights from molecular phylogeography. Front. Zool. 2004, 1, 4. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, G.M. Genetic consequences of climatic oscillations in the Quaternary. Philos. Trans. R. Soc. B. 2004, 359, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Thuiller, W.; Lavorel, S.; Midgley, G.; Lavergne, S.; Rebelo, T. Relating plant traits and species distributions along bioclimatic gradients for 88 Leucadendron taxa. Ecology 2004, 85, 1688–1699. [Google Scholar] [CrossRef]
- Doebeli, M.; Dieckmann, U. Speciation along environmental gradients. Nature 2003, 421, 259–264. [Google Scholar] [CrossRef]
- Schneider, R.; Humpert, M.; Stoner, K.; Steinauer, G. The Nebraska Natural Legacy Project, 2nd ed.; Nebraska Game and Parks Commission: Lincoln, NE, USA, 2011; p. 348. [Google Scholar]
- Barraclough, T.G. TheE Biology of Species, 1st ed.; Oxford University Press: Oxford, UK, 2019; p. 271. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, H.-F.; Lv, M.-Y.; Zhao, Y.; Zhang, Z.-C.; Liu, Z.; Lin, X.-L. Cryptic Diversity and Climatic Niche Divergence of Brillia Kieffer (Diptera: Chironomidae): Insights from a Global DNA Barcode Dataset. Insects 2025, 16, 675. https://doi.org/10.3390/insects16070675
Xu H-F, Lv M-Y, Zhao Y, Zhang Z-C, Liu Z, Lin X-L. Cryptic Diversity and Climatic Niche Divergence of Brillia Kieffer (Diptera: Chironomidae): Insights from a Global DNA Barcode Dataset. Insects. 2025; 16(7):675. https://doi.org/10.3390/insects16070675
Chicago/Turabian StyleXu, Hai-Feng, Meng-Yu Lv, Yu Zhao, Zhi-Chao Zhang, Zheng Liu, and Xiao-Long Lin. 2025. "Cryptic Diversity and Climatic Niche Divergence of Brillia Kieffer (Diptera: Chironomidae): Insights from a Global DNA Barcode Dataset" Insects 16, no. 7: 675. https://doi.org/10.3390/insects16070675
APA StyleXu, H.-F., Lv, M.-Y., Zhao, Y., Zhang, Z.-C., Liu, Z., & Lin, X.-L. (2025). Cryptic Diversity and Climatic Niche Divergence of Brillia Kieffer (Diptera: Chironomidae): Insights from a Global DNA Barcode Dataset. Insects, 16(7), 675. https://doi.org/10.3390/insects16070675





