Transcriptomic Analysis of Venom Secretion in Achelura yunnanensis: Lipid Metabolism, Redox Reactions, and Structural Adaptations
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
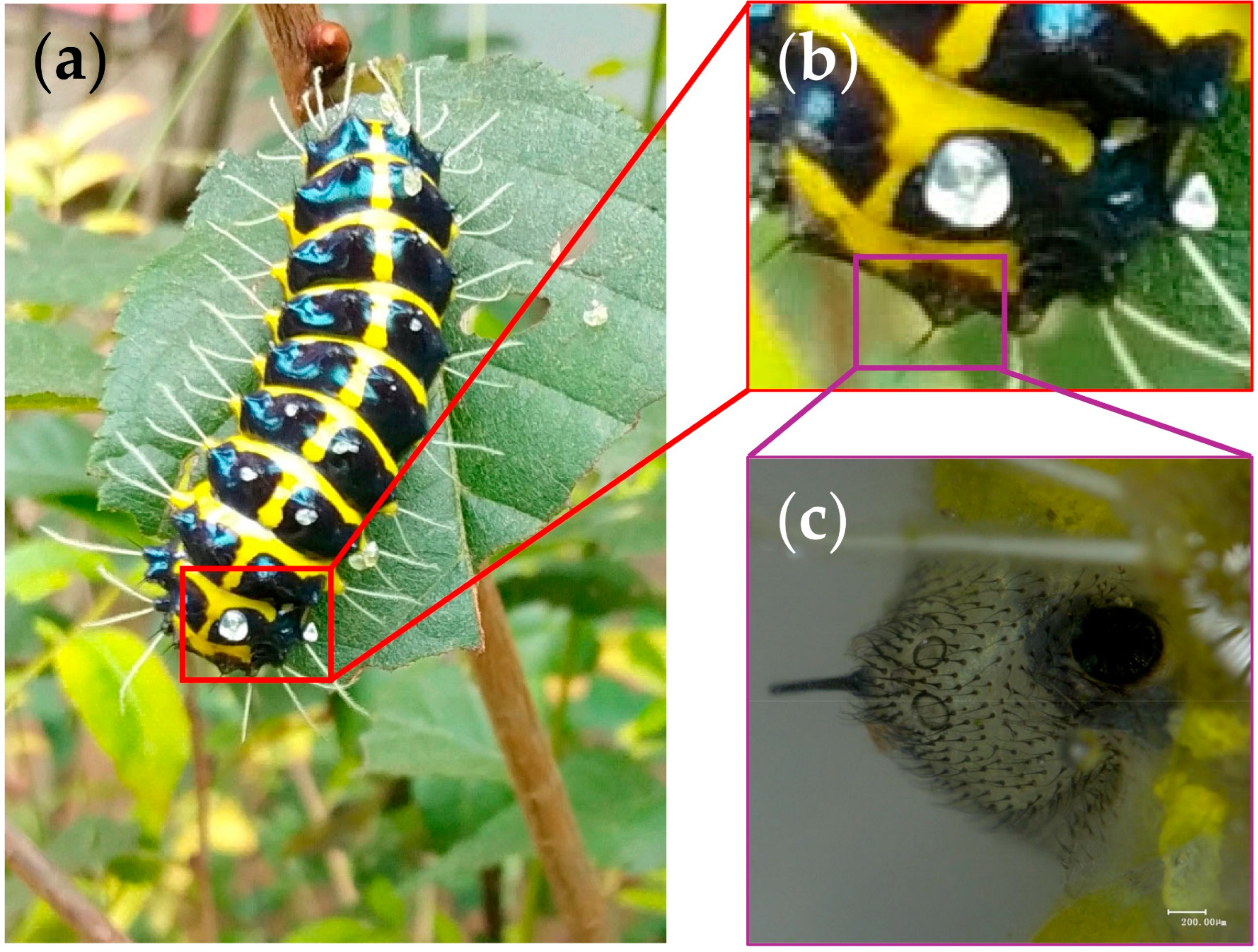
2.1. Insect Collection and Sample Preparation
2.2. RNA Extraction and Sequencing
2.3. Transcriptome Assembly and Annotation
2.4. Differential Expression and Functional Analysis
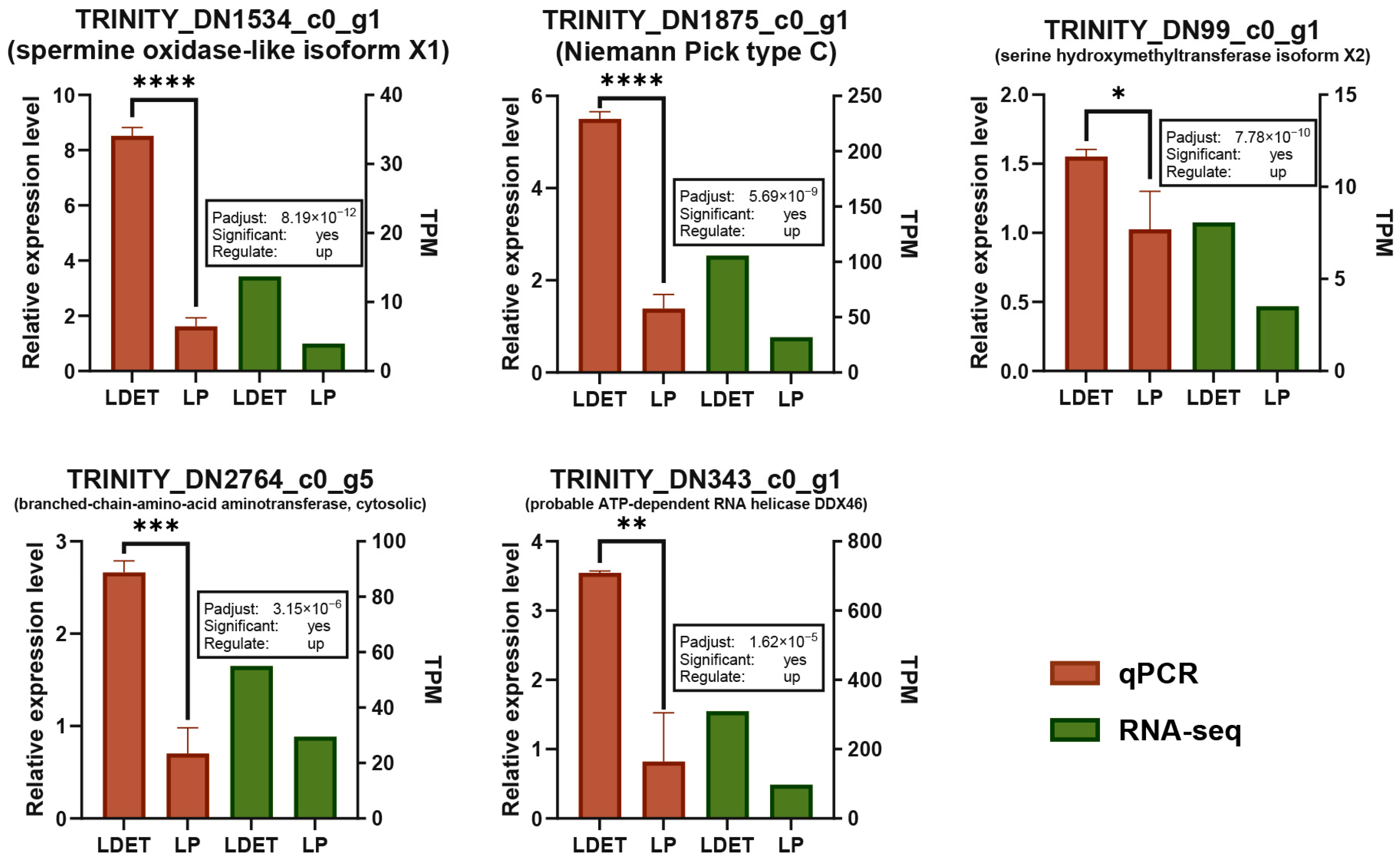
2.5. Validation of RNA-Seq Data by qRT-PCR
2.6. Gene Correlation and Network Analysis
3. Results
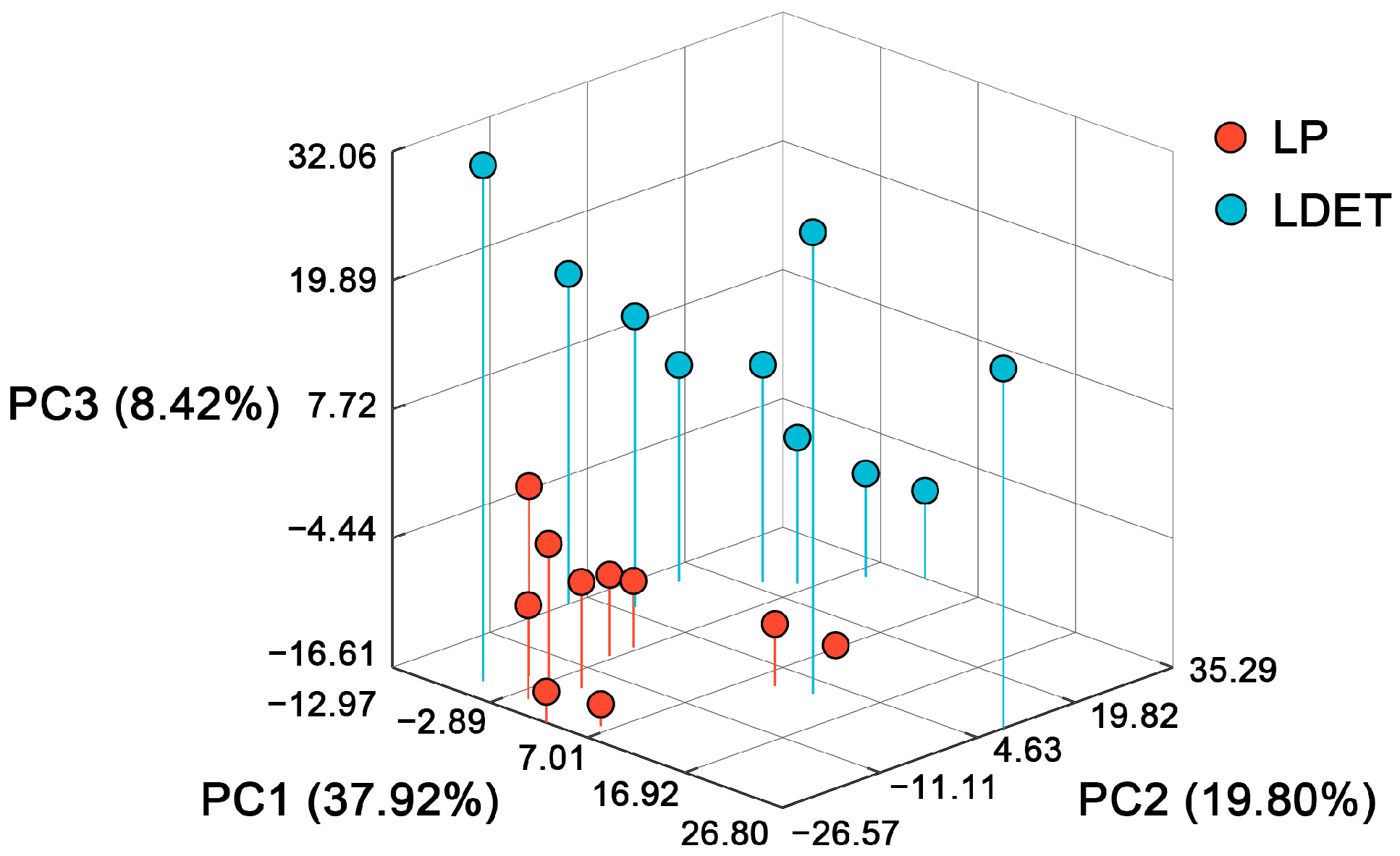
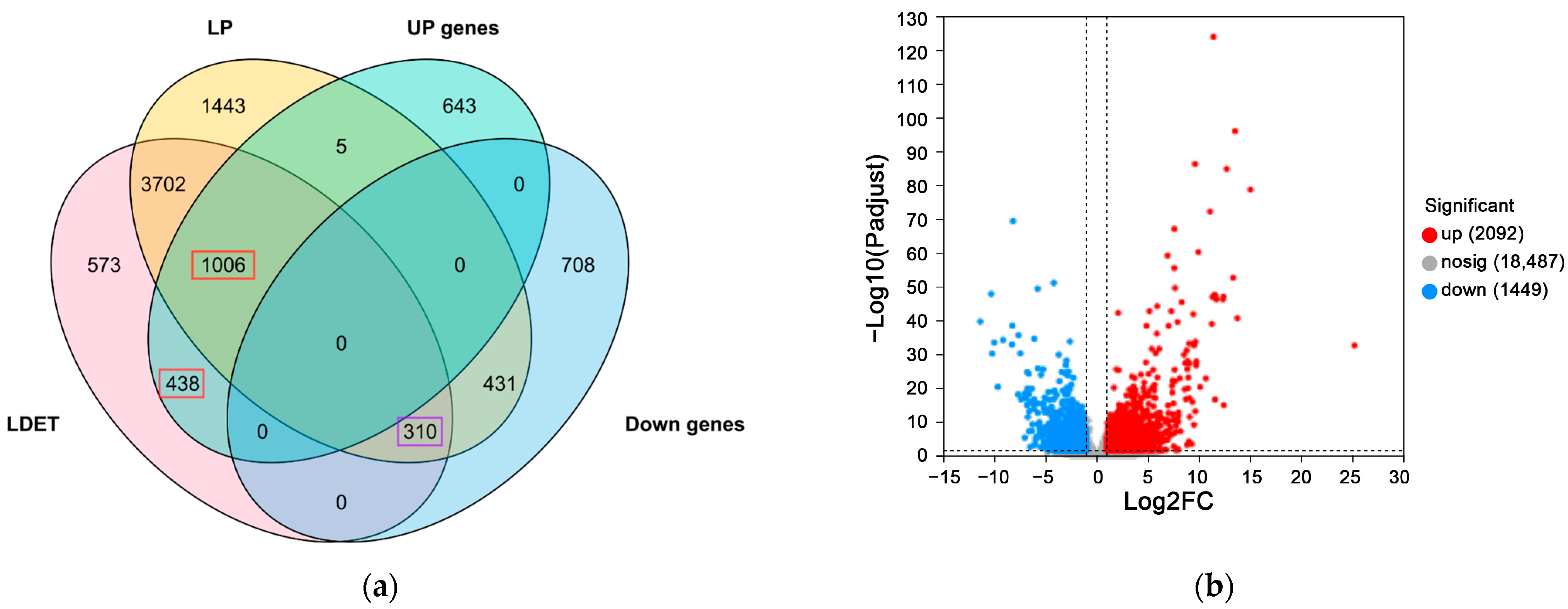
3.1. Distinct Transcriptomic Profiles of Venom-Secreting Tissue
3.2. Unexpected Absence of Cyanogenic Glycoside Biosynthesis Signature
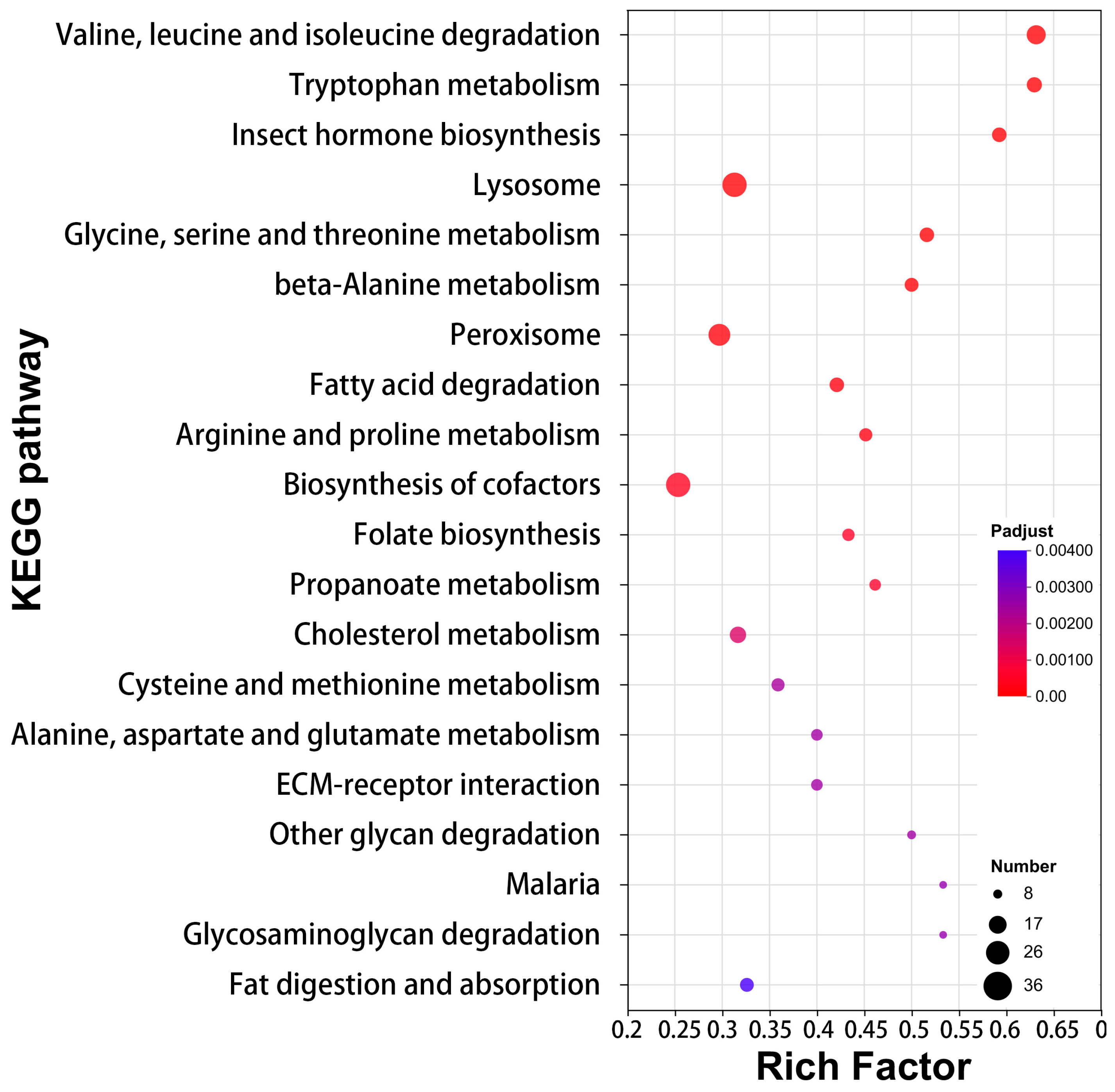
3.3. Lipid Metabolism and Redox Reactions as Pillars of Venom Function
3.4. Structural Adaptations Supporting Venom Secretion
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tian, M.-X.; Rong, C.H.; Bai, B.; Yang, Z.X.; Liu, P. Biological characteristics and occurrence patterns of Achelura yunnanensis (Lepidoptera: Zygaenidae) in Yunnan Province. Plant Prot. 2018, 44, 191–194. [Google Scholar] [CrossRef]
- Huang, S.Z.; Gong, W.C.; Ma, Q.Y.; Niu, Y.; Mei, W.L.; Chen, G.; Zhao, Y.X. Isolation and identification of cyanogenic glycosides from larval secretions of Achelura yunnanensis (Lepidoptera: Zygaenidae) and the bioactivity against Tapinoma melanocephalum (Hymenoptera: Fromicidae). Acta Entomol. Sin. 2013, 56, 207–211. [Google Scholar]
- Rong, C.H.; Wang, S.J.; Yang, F.; Wu, Y. Spatial distribution patterns and sampling techniques of the Achelura yunnanensis Horie &Xue. J. West China For. Sci. 2015, 44, 72–76. [Google Scholar] [CrossRef]
- Fürstenberg-Hägg, J.; Zagrobelny, M.; Jørgensen, K.; Vogel, H.; Møller, B.L.; Bak, S. Chemical defense balanced by sequestration and de novo biosynthesis in a lepidopteran specialist. PLoS ONE 2014, 9, e108745. [Google Scholar] [CrossRef]
- Franzl, S.; Nahrstedt, A.; Naumann, C. Evidence for site of biosynthesis and transport of the cyanoglucosides linamarin and lotaustralin in larvae of Zygaena trifolii (Insecta: Lepidoptera). J. Insect Physiol. 1986, 32, 705–709. [Google Scholar] [CrossRef]
- Nahrstedt, A.; Davis, R.H. Uptake of linamarin and lotaustralin from their foodplant by larvae of Zygaena trifolii. Phytochemistry 1986, 25, 2299–2302. [Google Scholar] [CrossRef]
- Davis, R.H.; Nahrstedt, A. Biosynthesis of cyanogenic glucosides in butterflies and moths: Effective incorporation of 2-methylpropanenitrile and 2-methylbutanenitrile into linamarin and lotaustralin by Zygaena and Heliconius species (Lepidoptera). Insect Biochem. 1987, 17, 689–693. [Google Scholar] [CrossRef]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Smith-Unna, R.; Boursnell, C.; Patro, R.; Hibberd, J.; Kelly, S. TransRate: Reference free quality assessment of de novo transcriptome assemblies. Genome Res. 2016, 34, 1134–1144. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO update: Novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Yu, G.; Shi, C.; Liu, L.; Guo, Q.; Han, C.; Zhang, D.; Zhang, L.; Liu, B.; Gao, H.; et al. Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta 2022, 1, e12. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Tang, Y.; Li, M.; Wang, J.X.; Pan, Y.; Wu, F.X. CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems 2015, 127, 67–72. [Google Scholar] [CrossRef]
- Bittenbinder, M.A.; van Thiel, J.; Cardoso, F.C.; Casewell, N.R.; Gutiérrez, J.-M.; Kool, J.; Vonk, F.J. Tissue damaging toxins in snake venoms: Mechanisms of action, pathophysiology and treatment strategies. Commun. Biol. 2024, 7, 358. [Google Scholar] [CrossRef]
- Zagrobelny, M.; Olsen, C.E.; Pentzold, S.; Fürstenberg-Hägg, J.; Jørgensen, K.; Bak, S.; Møller, B.L.; Motawia, M.S. Sequestration, tissue distribution and developmental transmission of cyanogenic glucosides in a specialist insect herbivore. Insect Biochem. Mol. Biol. 2014, 44, 44–53. [Google Scholar] [CrossRef]
- Arai, H.; Kono, N. α-Tocopherol transfer protein (α-TTP). Free Radic. Biol. Med. 2021, 176, 162–175. [Google Scholar] [CrossRef]
- Takos, A.M.; Knudsen, C.; Lai, D.; Kannangara, R.; Mikkelsen, L.; Motawia, M.S.; Olsen, C.E.; Sato, S.; Tabata, S.; Jørgensen, K.; et al. Genomic clustering of cyanogenic glucoside biosynthetic genes aids their identification in Lotus japonicus and suggests the repeated evolution of this chemical defence pathway. Plant J. 2011, 68, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Millar, J.G. Chemical synthesis of insect cuticular hydrocarbons. In Insect Hydrocarbons: Biology, Biochemistry, and Chemical Ecology; Blomquist, G.J., Bagnères, A.-G., Eds.; Cambridge University Press: Cambridge, UK, 2010; pp. 163–186. [Google Scholar]
- Stearns, S.C. The Evolution of Life Histories; Oxford University Press: Oxford, UK, 1992. [Google Scholar]
- Yu, H.; Yi, L.; Lu, Z. Silencing of chitin-binding protein with PYPV-rich domain impairs cuticle and wing development in the asian citrus psyllid, Diaphorina citri. Insects 2022, 13, 353. [Google Scholar] [CrossRef] [PubMed]




| Gene_ID | NR Description | Mean TPM (LDET) | Mean TPM (LP) | Log2FC (LDET vs. LP) | p-Value | Padj * | Regulation |
|---|---|---|---|---|---|---|---|
| DN1005_c0_g1 | CYP405A2, partial | 213.097 | 470.398 | 1.212 | 0.584 | 0.732 | up |
| DN3986_c0_g1 | CYP332A3 | 53.848 | 76.196 | 2.606 | 0.018 | 0.060 | up |
| DN3987_c0_g2 | CYP332A3 | 69.290 | 109.101 | 1.218 | 0.528 | 0.688 | up |
| DN7282_c0_g3 | UGT33A1 | 1.814 | 3.568 | −0.971 | 0.954 | 0.976 | down |
| DN1642_c0_g1 | UGT33A1 | 1.551 | 2.301 | 1.713 | 0.237 | 0.405 | up |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, P.; Zhu, H.-Q.; Wang, S.-M.; Wang, Y.-Q.; Ruan, Z.-Y.; Qiao, L.; Wu, X.-X.; Yan, Q.-H.; Lu, Y.-P.; Bai, B.; et al. Transcriptomic Analysis of Venom Secretion in Achelura yunnanensis: Lipid Metabolism, Redox Reactions, and Structural Adaptations. Insects 2025, 16, 588. https://doi.org/10.3390/insects16060588
Liu P, Zhu H-Q, Wang S-M, Wang Y-Q, Ruan Z-Y, Qiao L, Wu X-X, Yan Q-H, Lu Y-P, Bai B, et al. Transcriptomic Analysis of Venom Secretion in Achelura yunnanensis: Lipid Metabolism, Redox Reactions, and Structural Adaptations. Insects. 2025; 16(6):588. https://doi.org/10.3390/insects16060588
Chicago/Turabian StyleLiu, Ping, Hui-Qin Zhu, Si-Ming Wang, Yu-Qian Wang, Zhen-Yuan Ruan, Lu Qiao, Xing-Xing Wu, Qing-Hua Yan, Ya-Ping Lu, Bing Bai, and et al. 2025. "Transcriptomic Analysis of Venom Secretion in Achelura yunnanensis: Lipid Metabolism, Redox Reactions, and Structural Adaptations" Insects 16, no. 6: 588. https://doi.org/10.3390/insects16060588
APA StyleLiu, P., Zhu, H.-Q., Wang, S.-M., Wang, Y.-Q., Ruan, Z.-Y., Qiao, L., Wu, X.-X., Yan, Q.-H., Lu, Y.-P., Bai, B., & Ding, W.-F. (2025). Transcriptomic Analysis of Venom Secretion in Achelura yunnanensis: Lipid Metabolism, Redox Reactions, and Structural Adaptations. Insects, 16(6), 588. https://doi.org/10.3390/insects16060588






