1. Introduction
Insect metamorphosis constitutes an evolutionarily successful ecological adaptation strategy that achieves temporal niche partitioning, mitigates predator pressure and seasonal constraints, and enables developmental plasticity in response to environmental variables (e.g., photoperiod, temperature, and food availability), thereby enhancing organismal fitness across heterogeneous habitats. As the most vulnerable phase in an insect’s life cycle, metamorphosis is regulated by a complex interplay of genetic, hormonal, and environmental factors. The precise hormonal regulation of metamorphosis, particularly through the juvenile hormone (JH)-ecdysone signaling cascade, presents a potential target for developing next-generation, eco-conscious insect control strategies.
The silkworm (
B. mori) serves as both a holometabolous lepidopteran model organism and an economically significant species. During metamorphosis, larval tissue degradation occurs primarily through programmed cell death (PCD) mechanisms, particularly apoptosis and autophagy, which mediate the elimination of senescent tissues [
1,
2,
3]. This developmental process is coordinately regulated by 20E through its induction of a downstream transcriptional cascade [
4,
5,
6,
7]. The ecdysone signaling pathway primarily upregulates autophagy-related genes, including the Atg family [
8],
E74, and
Met, etc., while suppressing mTOR activity to induce autophagy. Simultaneously, 20E upregulates pro-apoptotic genes (including
Apaf-1,
Nedd2 like1,
ICE1,
Arp, and
IAP), thereby triggering PCD in the fat body during larval–pupal metamorphosis [
9]. These lines of evidences demonstrate that 20E-mediated apoptosis and autophagy constitute essential mechanisms for larval tissue degradation during metamorphosis. However, the insufficient elucidation of 20E’s molecular mechanisms remains a major barrier to developing targeted, 20E-based insect control strategies. Research on epigenetic-based gene regulation has revealed diverse post-translational modifications (PTMs), including methylation, acetylation, phosphorylation, and ubiquitination, that critically modulate gene expression. These regulatory mechanisms are essential for proper animal development and tissue homeostasis [
10,
11], with demonstrated roles in 20E-mediated gene expression during insect metamorphosis. Notably, the acetylation/deacetylation of Atg proteins regulate autophagy in silkworm cells [
3], while ubiquitination and SUMOylation coordinate 20E-dependent developmental transitions during larval metamorphosis [
10]. While the role of ubiquitin modification in apoptosis regulation is well established [
12], its specific function in 20E-mediated apoptotic pathways within metabolic tissues remains poorly characterized.
The
p53 gene represents a crucial tumor suppressor conserved across metazoans [
13,
14]. Structural and functional analyses reveal that invertebrate
p53 homologs, including insect variants, maintain significant similarity with their vertebrate counterparts [
15], with conserved mechanistic pathways [
16,
17]. In
B. mori,
Bmp53 has been identified as a vertebrate p53 ortholog and characterized as a putative apoptosis regulator [
18]. Our previous research has demonstrated that
Bmp53 can induce apoptosis in silkworm cells and promote the metamorphosis of insect pupae through this effect. Furthermore, we analyzed and predicted the apoptotic regulatory mode of Bmp53, which includes the ubiquitinating structural protein Mdm2-like [
19]. In vertebrates, Mdm2 (mouse double minute 2) serves as the primary negative regulator of p53, mediating ubiquitination and degradation to maintain protein stability [
20,
21]. While most invertebrates, including
Drosophila, lack an
Mdm2 ortholog [
22], the
Drosophila p53 stabilizer, Corp (a companion of reaper), shows partial sequence homology with MDM2 [
23,
24]. However, Corp lacks E3 ubiquitin ligase activity, while our integrated yeast two-hybrid and high-throughput sequencing platform identified MDM2-like as a functional ubiquitin ligase regulator of Bmp53 [
19]. The structural analysis revealed a striking conservation of human MDM2 [
25,
26], suggesting that the ubiquitination regulation of Bmp53 may share greater mechanistic similarity with vertebrate systems than with other insect models like
Drosophila.
Our study shows that Mdm2-like protein mediates Bmp53 ubiquitination, significantly attenuating 20E-dependent apoptotic signaling compared to Bmp53 overexpression alone. These findings establish Bmp53 ubiquitination as a regulatory component of 20E-mediated apoptosis. In other words, we identify the Mdm2-like/Bmp53 ubiquitination pathway as a novel apoptotic regulation mechanism in B. mori, with potential conservation across Lepidoptera, where 20E serves as a critical regulatory factor. These results reveal the role of a novel post-translational modification pathway in the 20E-mediated apoptosis signaling pathway of silkworms, providing a theoretical reference for the development of hormone-based eco-friendly pest control strategies.
2. Materials and Methods
2.1. Reagents and Antibodies
The reagents used in this study were obtained from the following suppliers: 4% Paraformaldehyde Fix Solution (Sangon, Shanghai, China; E672002); BSA (Beyotime, Shanghai, China; ST023); Triton X-100 (Sangon, Shanghai, China; A417820); DAPI (Solarbio, Beijing, China; C0065); One Step Western Kit HRP (Mouse) (CWBio, Taizhou, China; CW2030M); One Step Western Kit HRP (Rabbit) (CWBio, Taizhou, China; CW2029M); Nuclear and Cytoplasmic Extraction Kit (CWBio, Taizhou, China; CW0199S); Protease Inhibitor Cocktail (CWBio, Taizhou, China; CW2200S); DAB Kit (CWBio, Taizhou, China; CW0125M); Protein A/G Resin (Transgen, Beijing, China; DP501); HiFiScript gDNA Removal RT MasterMix (CWBio, Taizhou, China; CW2020M); ChamQ SYBR qPCR Mater Mix (Vazyme, Nanjing, China; Q311); Annexin V-EGFP/PI Cell Apoptosis Detection Kit (Transgen, Beijing, China; FA111); TUNEL BrightRed Apoptosis Detection Kit (Vazyme, Nanjing, China; A113); MG132 (Med Chem Express, New Jersey, USA; HY-13259); 20E was obtained from Zhenjiang Zhongnong Biotechnology Co., Ltd. (Zhenjiang, China). All experiments were performed according to the manufacturers’ protocols. The antibodies used in this study for co-immunoprecipitation and Western blot are as follows: rabbit anti-V5 (Huabio, Hangzhou, China; ET1605-41); rabbit anti-Flag (Sigma-Aldrich, Darmstadt, Germany; F7425-2MG); rabbit anti-ubiquitin (Boster, Wuhan, China; PB9122); iFluorTM 488 goat anti-rabbit IgG H&L (Huabio, Hangzhou, China; HA1121).
2.2. Plasmid Construction
pIZ-V5-His, pIZ-EGFP-V5-His, pIZ-EGFP-Bmp53-V5-His [
19], and pIZ-mCherry-V5-His were preserved in our laboratory. The 3xFlag tag of pIZ-3xFlag was synthesized by Sangon Biotech (Shanghai, China) and constructed by inserting 3xFlag sequences into
SacII and
AgeI cleavage sites on the basis of pIZ-V5-His. The pIZ-Bmp53-3xFlag fusion plasmid was constructed by inserting cloned
Bmp53 into the
BamHI and
XhoI sites of the pIZ-3xFlag vector; the amplified Bmp53 ORF was similarly cloned using Bmp53-F and Bmp53-R(CC) primers [
19]. pIZ-Mdm2-like-V5-His fusion plasmid was constructed by inserting cloned
Mdm2-like into the
EcoRI and
XhoI sites of pIZ-V5-His vector, and the cloning primers were Mdm2-CF: CCGGAATTCATGAACACAACATTTTG and Mdm2-CR: ATACTCGAGTAGCACGACGGCGCGC. The pIZ-mCherry-Mdm2-like-V5-His plasmid was constructed by cloning Mdm2-like into the EcoRI and XhoI sites of the pIZ-mCherry-V5-His vector, using the primers Mdm2-MF (CCGGAATTCaATGAACACAACATTTTGT) and Mdm2-MR (ATActcgagAATAGCACGACGGCGCGC).
2.3. Cell Culture and Treatment
The B. mori ovary epithelial cell line (BmN) was preserved in our laboratory and cultured in TC-100 medium, supplemented with 10% fatal bovine serum (FBS) and incubated at 28 °C.
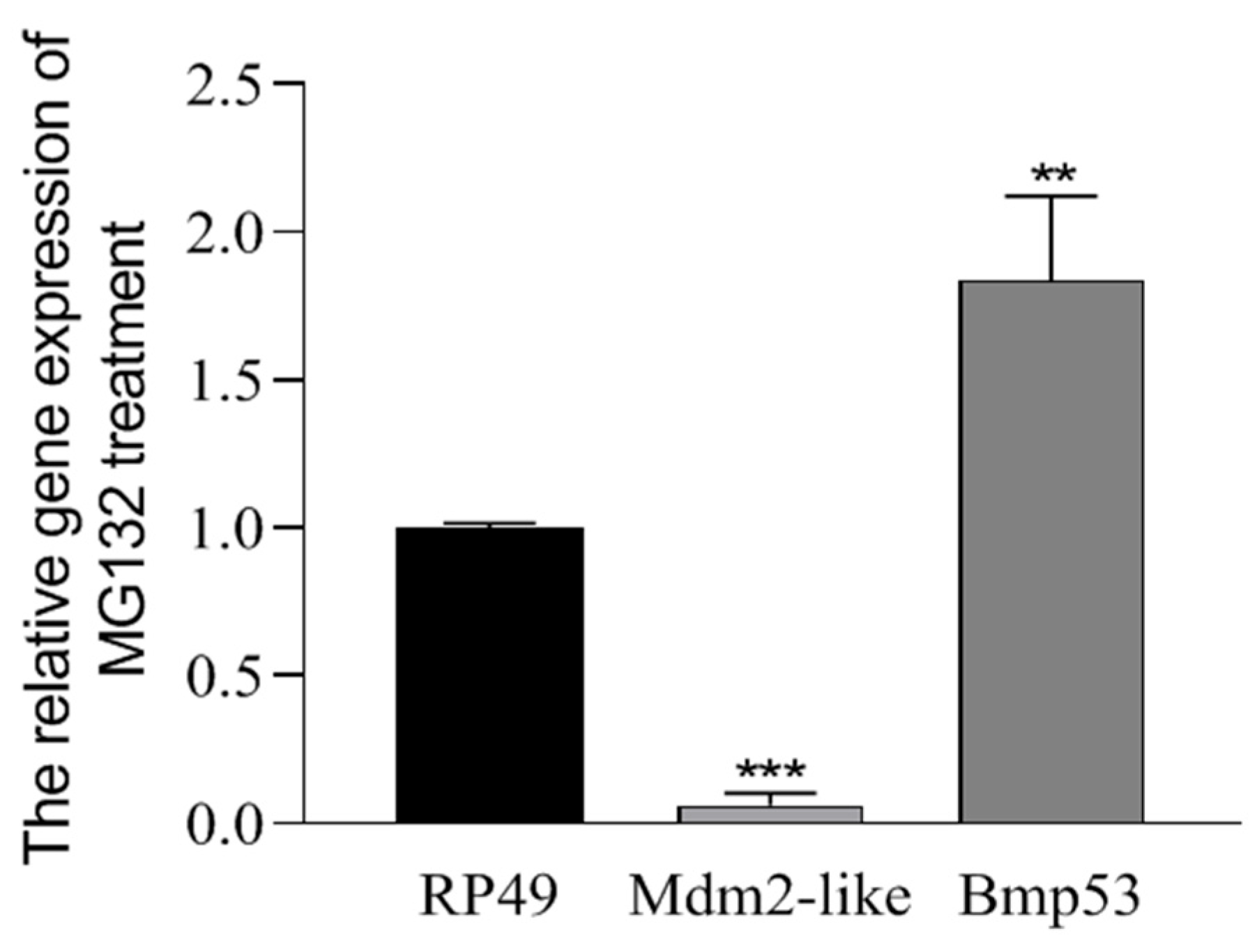
Cells were transfected with the above plasmids using Entranster-H4000 transfection reagent (Engreen, Beijing, China), according to the manufacturer’s instructions. For drug treatment, MG132 (5 µM) or 20-Hydroxyecdysone (20E) (50 ng/µL) were added, whilst the negative control group received an equal volume of DMSO or 20% ethyl alcohol.
2.4. Ubiquitination Detection by Fluorescence Mocroscopy and Western Blotting
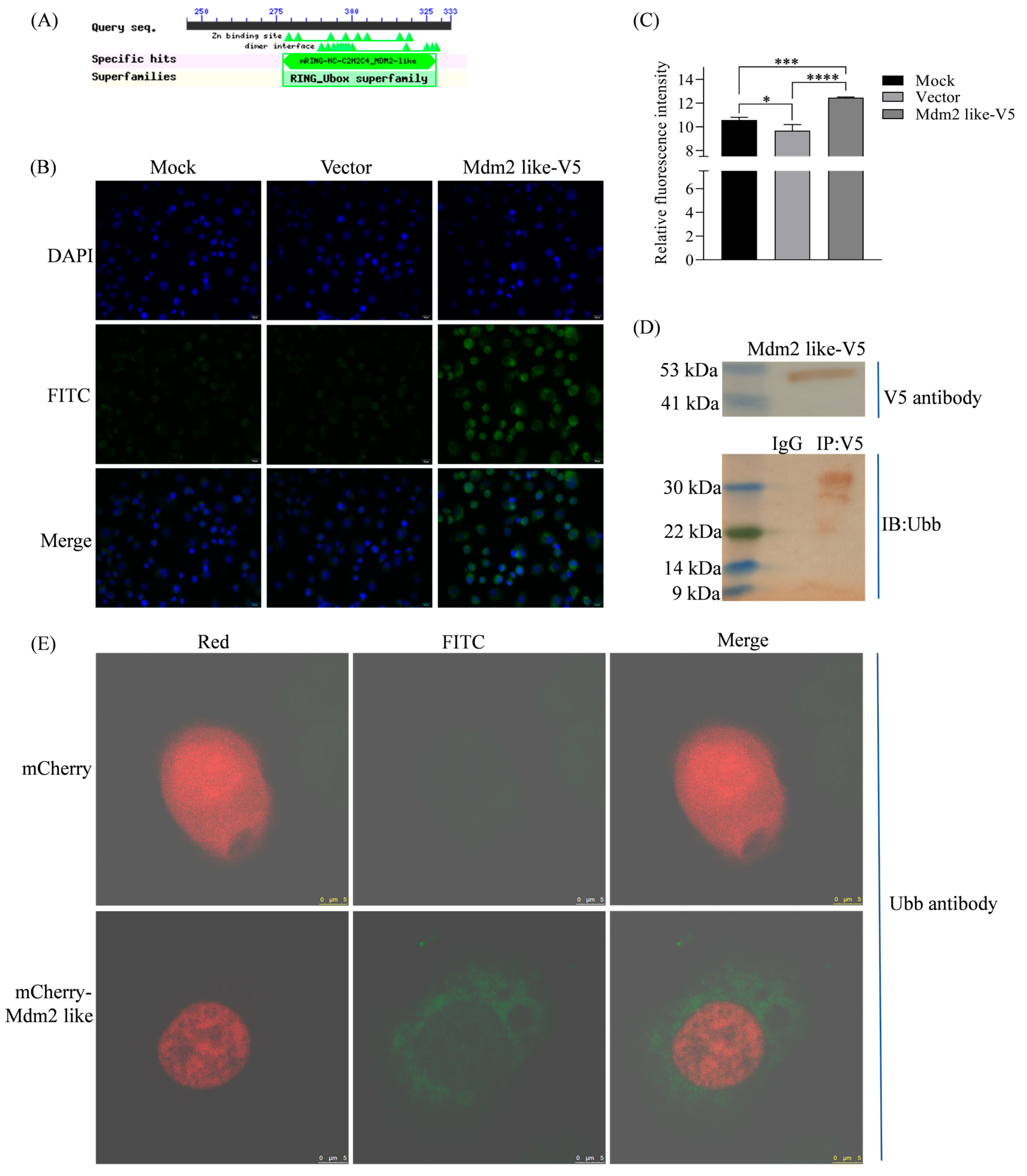
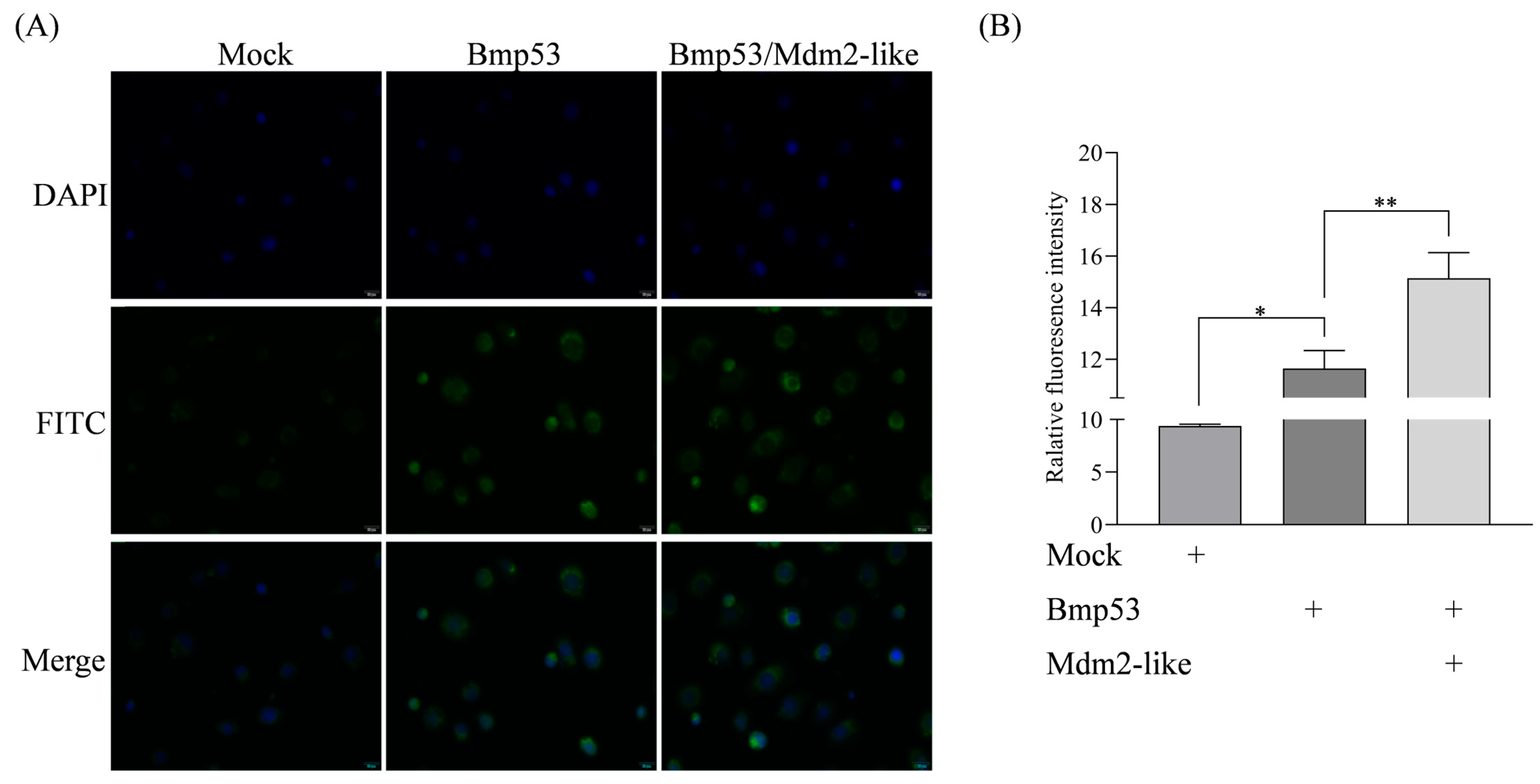
To demonstrate the E3 ubiquitin ligase activity of Mdm2-like, confluent BmN cells were subcultured at a 1:2.5 ratio. After 24 h, cells in six-well plates were transfected with 2 μg of either pIZ-Mdm2-like-V5-His or pIZ-V5-His plasmid (empty vector control) using Entranster™-H4000 transfection reagent. Untransfected BmN cells served as mock controls. Cells were harvested 48 h post-transfection for subsequent analysis. To assess ubiquitination levels mediated by Mdm2-like and Bmp53 interaction, BmN cells in six-well plates were co-transfected with 2 μg each of pIZ-Mdm2-like-V5-His and pIZ-Bmp53-3xFlag plasmids. Control transfections containing 2 μg each of empty vectors (pIZ-V5-His + pIZ-3xFlag) were performed in parallel. All transfections used the same subculture method and Entranster™-H4000 reagent as previously described. Cells were harvested 48 h post-transfection for ubiquitination assays. Then, cells were fixed with 4% paraformaldehyde solution and permeabilized with 0.25% TritonX-100 in PBS for 10 min, and then blocked with 5% BSA for 15 min at room temperature. Cells were probed with the rabbit anti-ubiquitin antibody at 1/200 dilution for overnight at 4 °C. iFluorTM 488 goat anti-rabbit IgG H&L was used as the secondary antibody at 1/500 dilution. DAPI was used as nuclear counterstain. Finally, the fluorescence signal was detected by the fluorescent microscope (Olympus Corporation, Tokyo, Japan, IX83) and confocal microscope (Leica, Wetzlar, Germany, SP8). Fluorescent cell images were processed using Adobe Photoshop (v24.2) (Adobe, San Jose, CA, USA).
BmN cells in 10 cm plates were transfected with 5 μg pIZ-Mdm2-like-V5-His plasmid for 48 h. Nucleoproteins were extracted using a Nuclear and Cytoplasmic Extraction Kit (CWBio, CW0199S) and incubated with rabbit anti-V5 antibody (Huabio, ET1605-41, 1:200 dilution) for 2–4 h at 4 °C. Protein A/G agarose beads (Beyotime, P2055, China) were added and incubated overnight at 4 °C. Beads were pelleted (1000× g, 5 min), washed twice with PBS, and resuspended in 5× protein loading buffer (Beyotime, P0015L, China). For controls, anti-V5 was replaced with species-matched IgG. Samples were denatured (95 °C, 5 min), resolved by 10% SDS-PAGE, and transferred to PVDF membranes. Ubiquitinated proteins were detected using rabbit anti-ubiquitin antibody followed by HRP/Rabbit secondary antibodies.
2.5. Co-Immunoprecipitation (Co-IP)
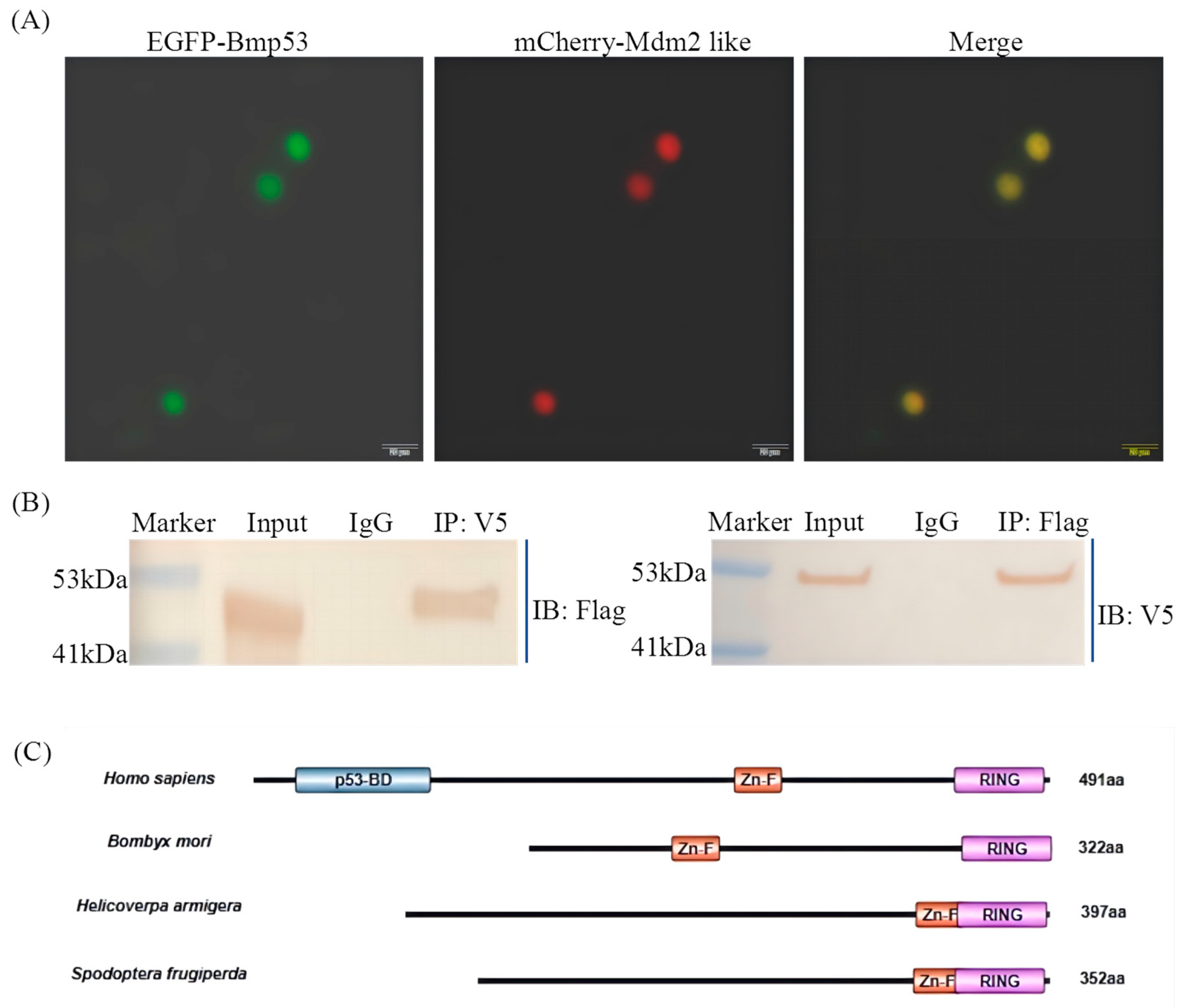
Nuclear protein lysates were extracted from BmN cells using a nucleoprotein extraction kit containing protease inhibitors. The lysate was incubated overnight at 4 °C with 2 µL antibody (rabbit anti-V5 or rabbit anti-Flag). The protein–antibody complex was then mixed with protein A/G agarose beads and incubated for 2 h at 4 °C. After incubation, the beads were washed with PBS, resuspended, and boiled in protein loading buffer. The supernatant was collected by centrifugation for SDS-PAGE analysis. Following electrophoresis, proteins were transferred to a PVDF membrane and detected by immunoblotting.
2.6. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted from cells TRIzol Reagent followed by DNase I treatment (RNase-free) to remove genomic DNA contamination. The first strand cDNA was synthesized from 1 μg total RNA using a HiFiScript gDNA Removal cDNA Synthesis Kit(CWBio, Taizhou, China; CW2020M) according to the manufacturer’s protocol. Quantitative PCR was performed with ChamQ SYBR qPCR Mater Mix using 100 ng cDNA template per reaction. RP49 served as the internal reference gene. The gene-specific primer sequences are provided in
Table 1.
2.7. Flow Cytometry
Apoptosis was assessed using an Annexin V-EGFP/PI Cell Apoptosis Detection Kit (Transgen, Beijing, China; FA111) following the manufacturer’s protocol. Briefly, harvested cells were washed twice with cold PBS and resuspended in 1 × binding buffer. Cells were simultaneously stained with Annexin V-EGFP and propidium iodide (PI) (1:1 v/v) and incubated on ice for 15 min in the dark. Samples were analyzed immediately using a BD FACSVerse flow cytometer (BD Biosciences, New York, NY, USA) with fluorescence detected through FL1 (EGFP) and FL3 (PI) channels. Data were processed using FlowJo software (v10.8.1, BD Life Sciences, Ashland, Oregon, USA;).
2.8. TUNEL Assay
Following PBS washes, the cells were fixed with 4% paraformaldehyde for 30 min and permeabilized with 0.25% Triton X-100 for 30 min. After blocking with 5% BSA for 1 h, the cells were incubated with TUNEL solution for 1 h at 37 °C in a humidified chamber. Nuclei were counterstained with DAPI (1 μg/mL) for 5 min. Fluorescent images were captured using an inverted microscope (IX83, Olympus, Japan) equipped with appropriate filter sets for DAPI and TUNEL detection.
2.9. Statistics
All experiments were performed in triplicate with at least three independent biological replicates. Data are presented as mean ± standard deviation (SD). Statistical analyses were conducted using GraphPad Prism 8, with multiple group comparisons evaluated by one-way ANOVA followed by Tukey’s post hoc test, and pairwise comparisons analyzed by two-tailed Student’s t-test. A threshold of p < 0.05 was considered statistically significant.
4. Discussion
Ubiquitination is a key post-translational modification that modulates the stability and function of proteins critical for cell growth, proliferation, and survival [
29]. Notably, ubiquitination plays a pivotal role in the regulation of cell death, which is essential for animal development as well as multi-tissue physiology and pathology [
10,
12]. Our prior work revealed that the
Bmp53 gene, which is a key regulator of silkworm cell apoptosis and larval–pupal metamorphosis [
19], is likely under the direct control of ubiquitination enzymes.
Bmp53 is a homolog of vertebrate p53 [
18]. The p53 protein plays a crucial role in the apoptotic signaling pathway induced by DNA damage, and its levels are meticulously regulated within the cell [
30,
31], though its regulatory mechanisms in invertebrates remain less defined. In vertebrates, p53 stability is tightly governed by ubiquitination, primarily mediated by the E3 ligase Mdm2, which targets p53 for proteasomal degradation to fine-tune apoptotic responses [
20,
23]. Given this conserved role of ubiquitination in p53 regulation, we hypothesize that Bmp53 may similarly be modulated by ubiquitin-dependent pathways during silkworm development. Notably, some invertebrates, such as
Drosophila, lack a homolog of Mdm2 [
22]. Instead, the
Drosophila protein Corp shares sequence homology with Mdm2 and stabilizes Dp53 [
23,
24]. However, Corp lacks E3 ubiquitin ligase activity, implying divergent regulatory mechanisms between insects and vertebrates. This study demonstrates that an Mdm2-like protein in silkworms possesses intrinsic ubiquitination activity (
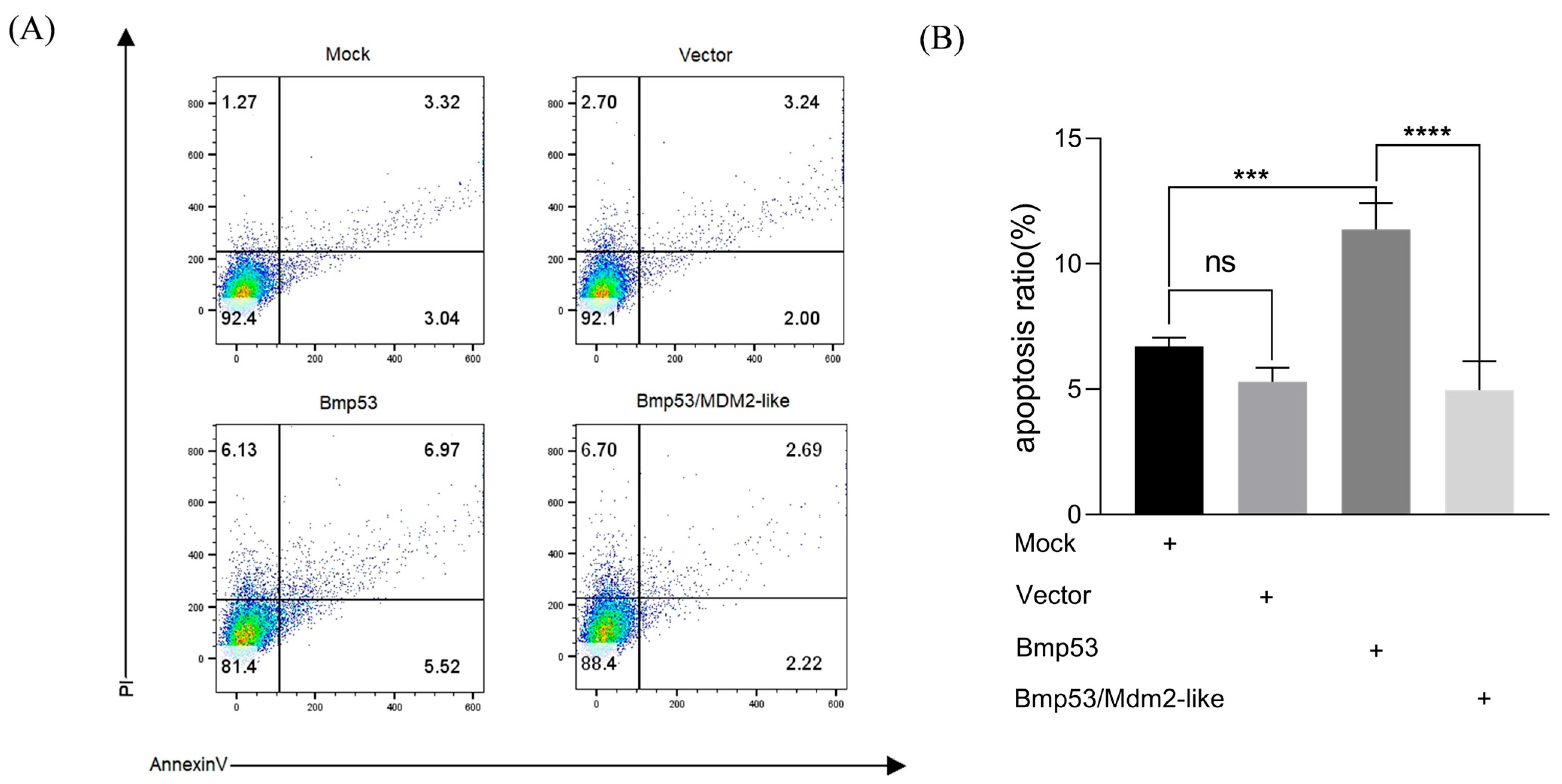
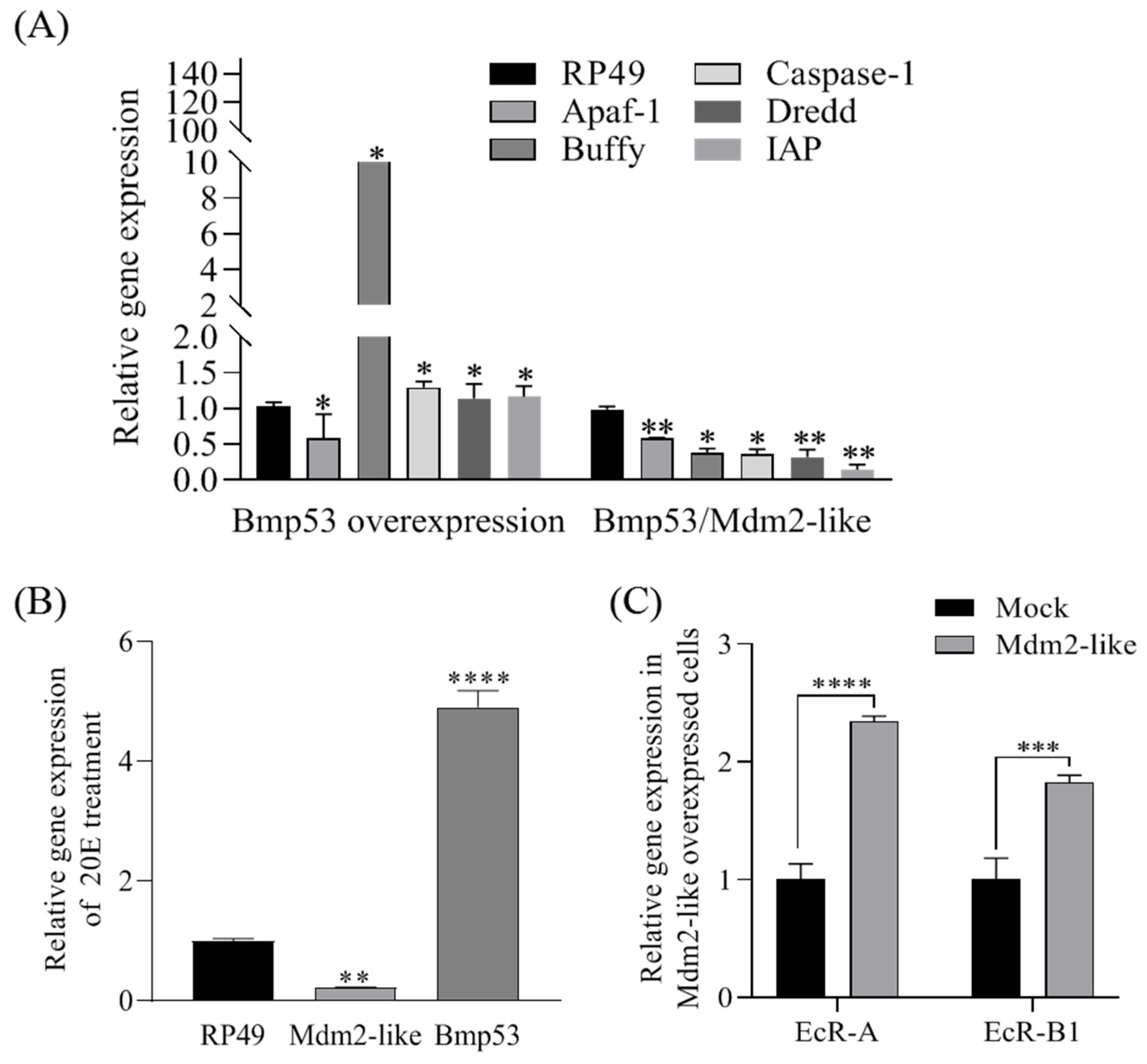
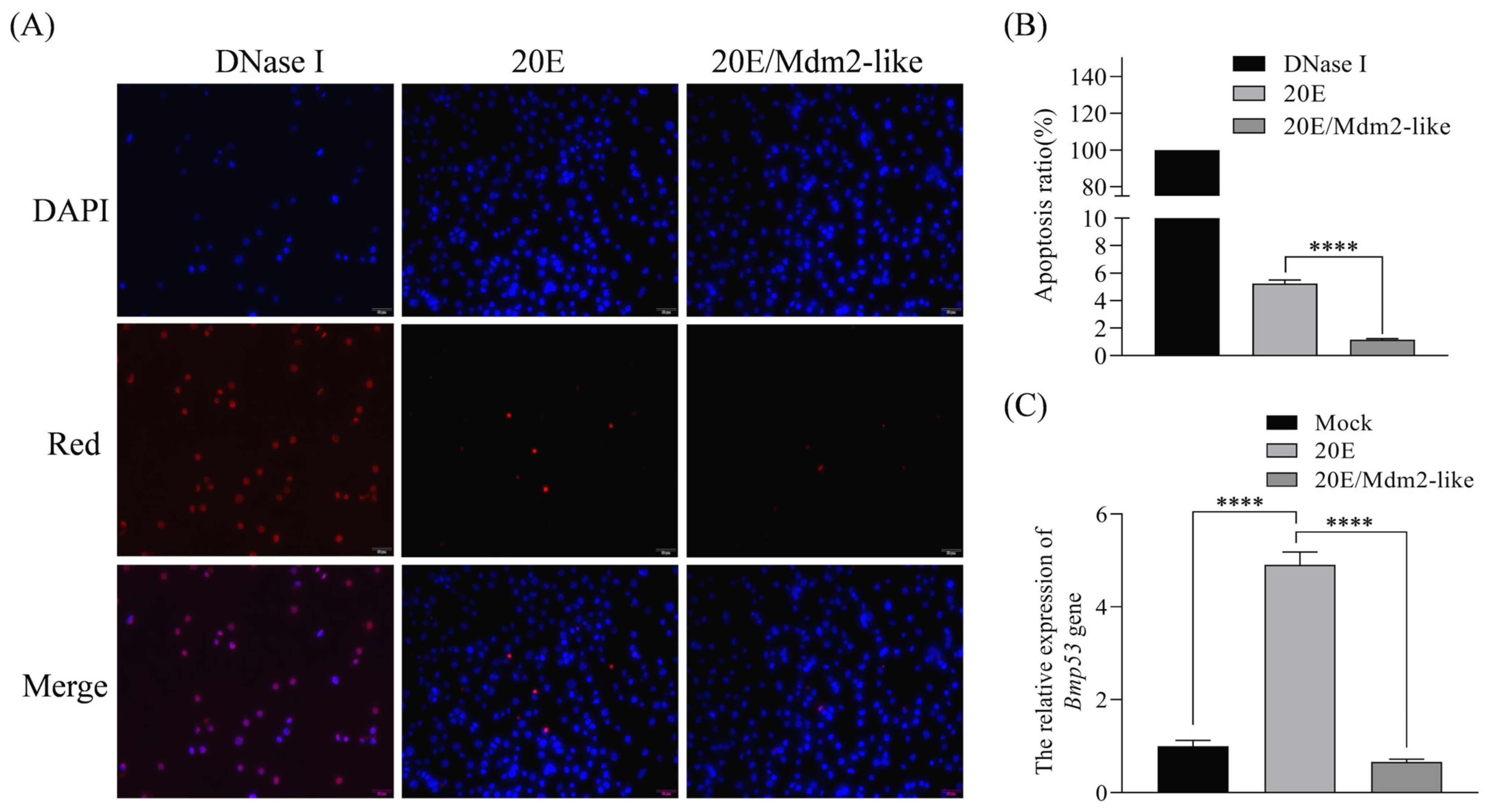
Figure 1) and directly interacts with Bmp53 to suppress its pro-apoptotic function (
Figure 2,
Figure 3,
Figure 4 and
Figure 5). Notably, Mdm2-like lacks the p53-binding domain found in its mammalian homolog, which aligns with previous findings [
25,
26]. Although we could not detect its physical interaction with Bmp53 in whole-cell lysates, subsequent nucleoprotein extraction and testing confirmed their binding. We speculate that this interaction is tightly regulated in a spatiotemporal manner, though the underlying mechanism warrants further investigation. Regardless, these results establish the Mdm2-like–Bmp53 axis as a critical ubiquitination-mediated regulatory mechanism controlling apoptosis in silkworm cells, revealing functional conservation with vertebrate p53 regulation while highlighting insect-specific adaptations. Furthermore, the observed ubiquitination levels upon Mdm2 overexpression, combined with Bmp53’s predicted apoptotic regulatory role, suggest that Mdm2-like proteins likely target additional factors beyond Bmp53 through ubiquitination. In future studies, we will identify and characterize other Mdm2-like regulatory proteins to comprehensively elucidate the ubiquitination-mediated control of apoptosis in silkworms.
During insect metamorphosis, senescent larval tissues are eliminated through programmed cell death (PCD), primarily via apoptosis and autophagy, a process mediated by ecdysone (20-hydroxyecdysone, 20E) and its triggered transcriptional cascades [
6,
9]. 20E exerts its biological function by targeting specific nuclear receptors, primarily the ecdysone receptor (EcR). Upon forming a complex with its heterodimeric partner EcR-USP, 20E triggers the rapid and robust induction of key response genes [
32]. During the larval–pupal transition in
Bombyx mori, 20E upregulates multiple apoptosis-related genes, including
Apaf-1,
Nedd2-like1,
ICE1,
Arp, and
IAP, thereby promoting fat body apoptosis [
9]. Our study reveals that Mdm2-like proteins regulate 20E-mediated apoptosis signaling pathway through Bmp53 ubiquitination. As 20E is a critical hormone governing insect metamorphosis and development, these findings establish a novel link between ubiquitination-dependent protein control and the hormonal regulation of programmed cell death in silkworms. Our current understanding of the ubiquitination axis in metagenesis and 20E signaling is largely based on prior findings [
19]. To further validate this, we performed genetic knockout experiments targeting MDM2-like and Bmp53. However, the high lethality of homozygous mutants has so far prevented the establishment of stable lines. We acknowledge two major challenges: (1) the inherent complexity of 20E pathway regulation [
6] and (2) the difficulty in isolating direct phenotypic effects attributable to the MDM2-like–Bmp53 axis. While we regret the inability to provide organism-level evidence for this mechanism at present, it remains a key focus of our ongoing experimental optimization efforts.
Studies on epigenetic-based gene regulation have unveiled various post-translational modification mechanisms (PTMs), including methylation, acetylation, phosphorylation, and ubiquitination, as well as 20-hydroxyecdysone (20E)-induced gene expression regulation. For example, the acetylation and deacetylation of Atg proteins critically regulate autophagy in silkworm cells [
3], whereas ubiquitination and SUMOylation modulate 20E-dependent larval development [
10]. While the involvement of ubiquitination in apoptosis regulation is well-established, its role in modulating 20E-dependent apoptosis during metamorphic tissue remodeling remains poorly characterized. In conclusion, our findings provide foundational insights into the regulation of silkworm metamorphosis. We demonstrate that Bmp53 promotes developmental progression in
B.
mori, while Mdm2-mediated ubiquitination suppresses
Bmp53 expression and its pro-apoptotic activity under 20E regulation. Importantly, we identify a putative 20E–Mdm2-like–Bmp53 regulatory axis that maintains a dynamic balance to regulate cell apoptosis. This newly discovered mechanism represents a crucial area for future investigation in insect developmental biology.
5. Conclusions
In summary (
Figure 8), our findings demonstrate that Mdm2-like directly participates in silkworm cells apoptosis through the ubiquitination-mediated regulation of Bmp53. This study reveals, for the first time, the Mdm2-like/Bmp53 ubiquitination pathway as a novel apoptotic regulatory mechanism in
B.
mori, with potential implications for other lepidopteran species. Furthermore, this study underscores the critical involvement of the ecdysone signaling molecule 20E in this regulatory pathway. These findings establish a foundation for future research on ubiquitination-mediated control of 20E-dependent metamorphosis and developmental processes. Previous studies have established that the
Bmp53 gene plays a pivotal role in larval–pupal metamorphosis in
B.
mori. Our findings significantly advance the understanding of the genetic regulatory mechanisms governing tissue remodeling during lepidopteran metamorphosis. Moreover, these insights establish a theoretical reference for developing eco-friendly pest control strategies through the targeted disruption of protein modification pathways, thereby contributing to sustainable lepidopteran pest management.
This study provides compelling evidence for the following: Mdm2-like exhibits intrinsic ubiquitin ligase activity and physically interacts with Bmp53 to suppress its pro-apoptotic function; 20E serves as a critical regulator of the Mdm2-like/Bmp53 ubiquitination pathway.