Selection and Validation of Stable Reference Genes for RT-qPCR in Scotogramma trifolii (Lepidoptera: Noctuidae)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing
2.2. Experimental Design and Sample Collection
2.2.1. Developmental State
2.2.2. Adult Tissues
2.3. Total RNA Extraction and cDNA First Strand Synthesis
2.4. Candidate Reference Genes and Primer Design
2.5. Standard Curve Construction and RT-qPCR
2.6. Stability Analysis
2.7. Stability Validation of Candidate Reference Genes
2.8. Statistical Analysis
3. Results
3.1. Specificity and Amplification Efficiency of RT-qPCR Primers
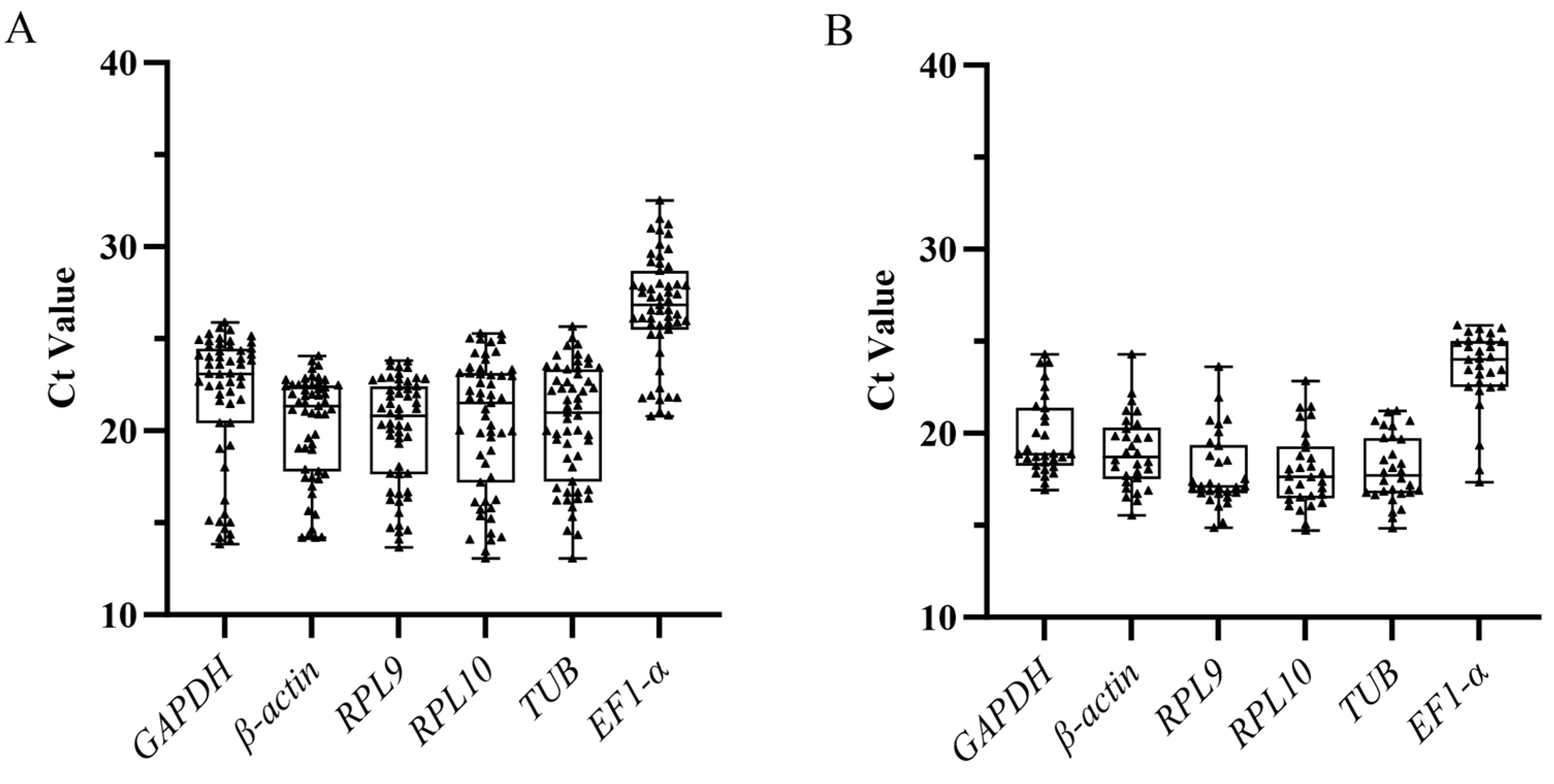
3.2. Expression Level of Six Candidate Reference Genes
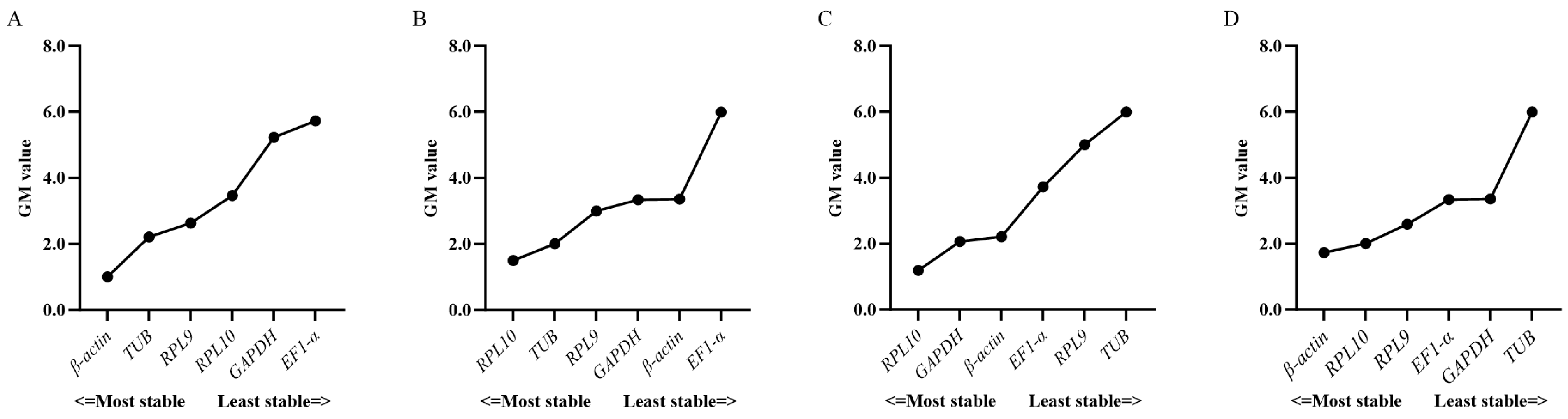
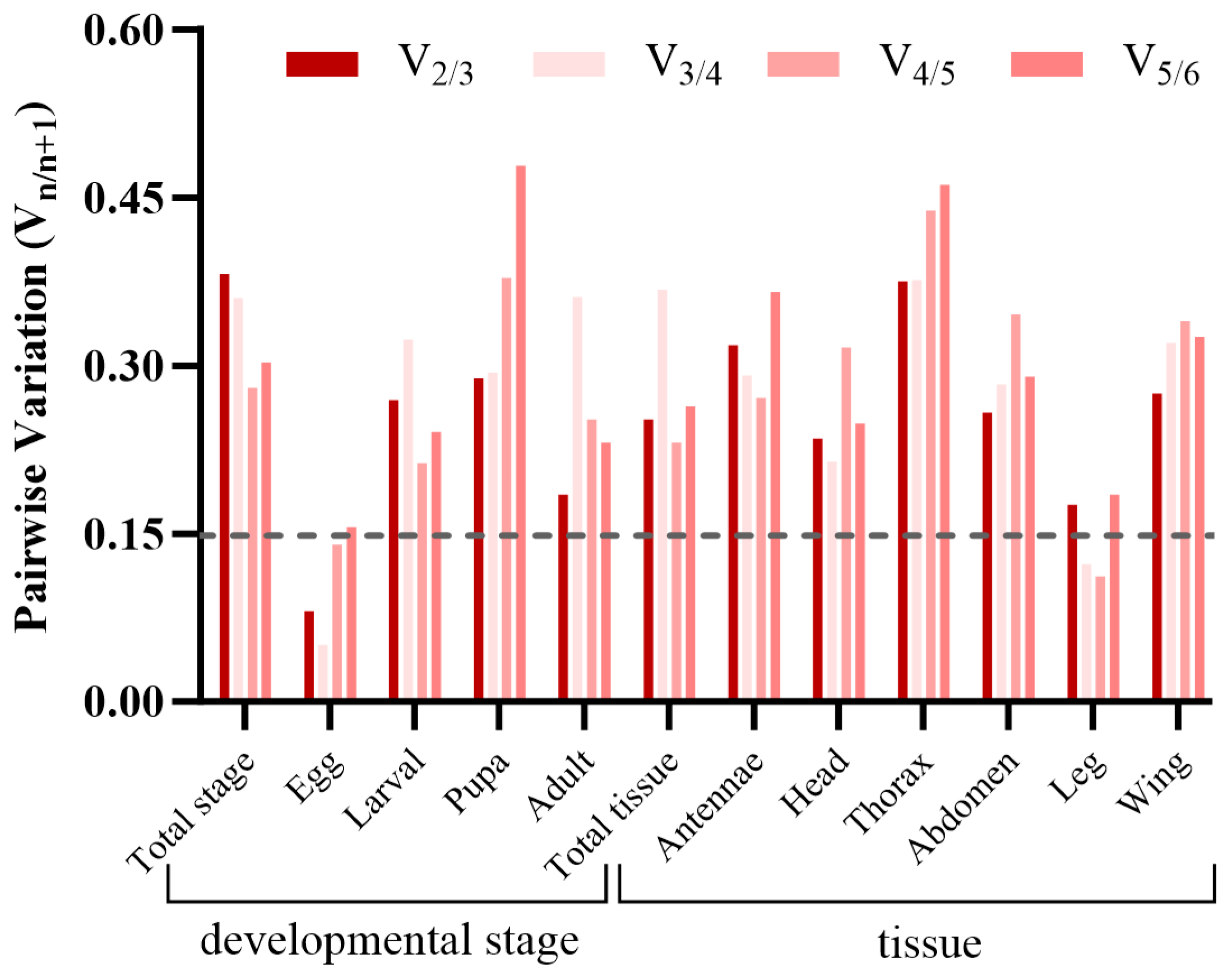
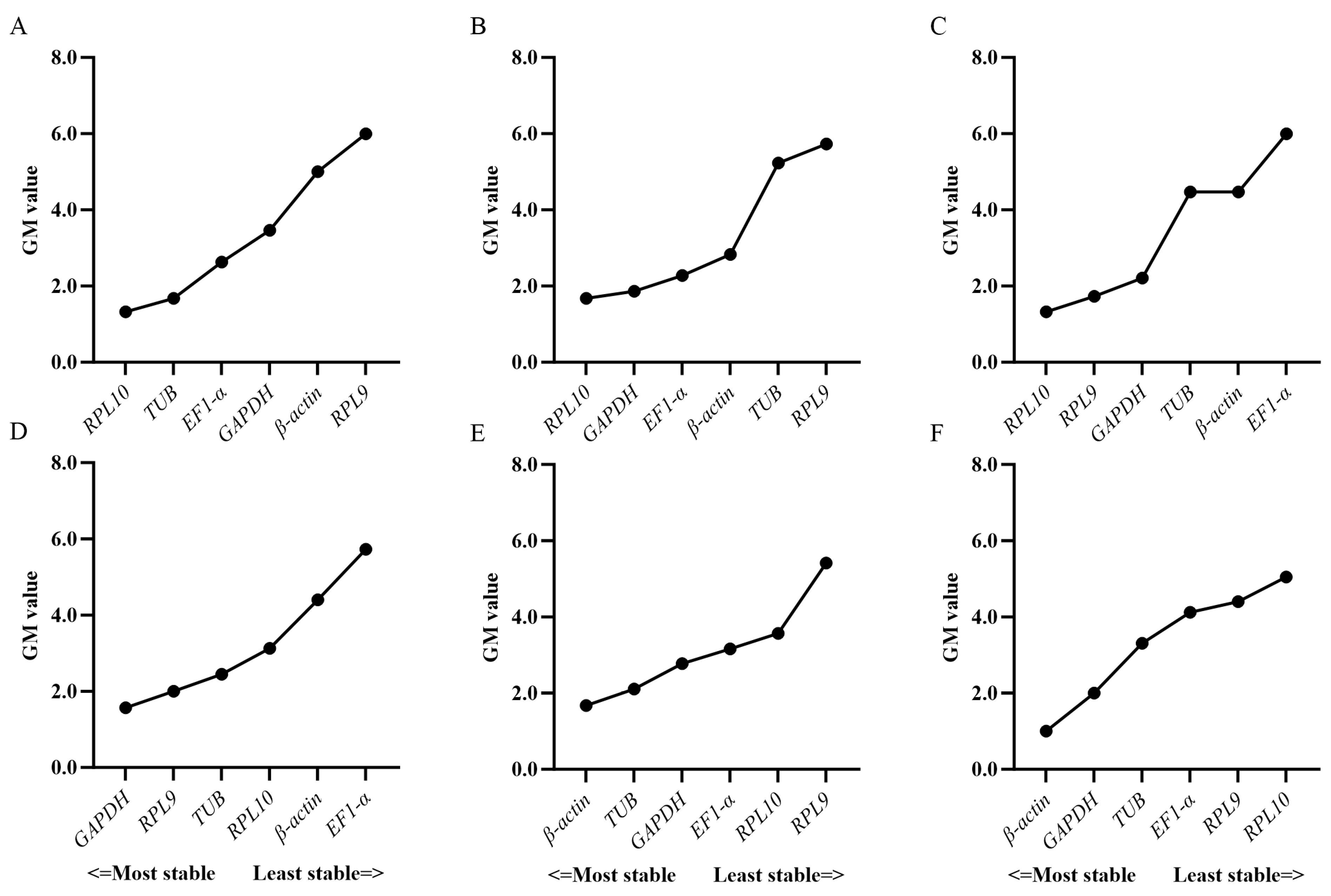
3.3. Expression Stability of Candidate Reference Genes
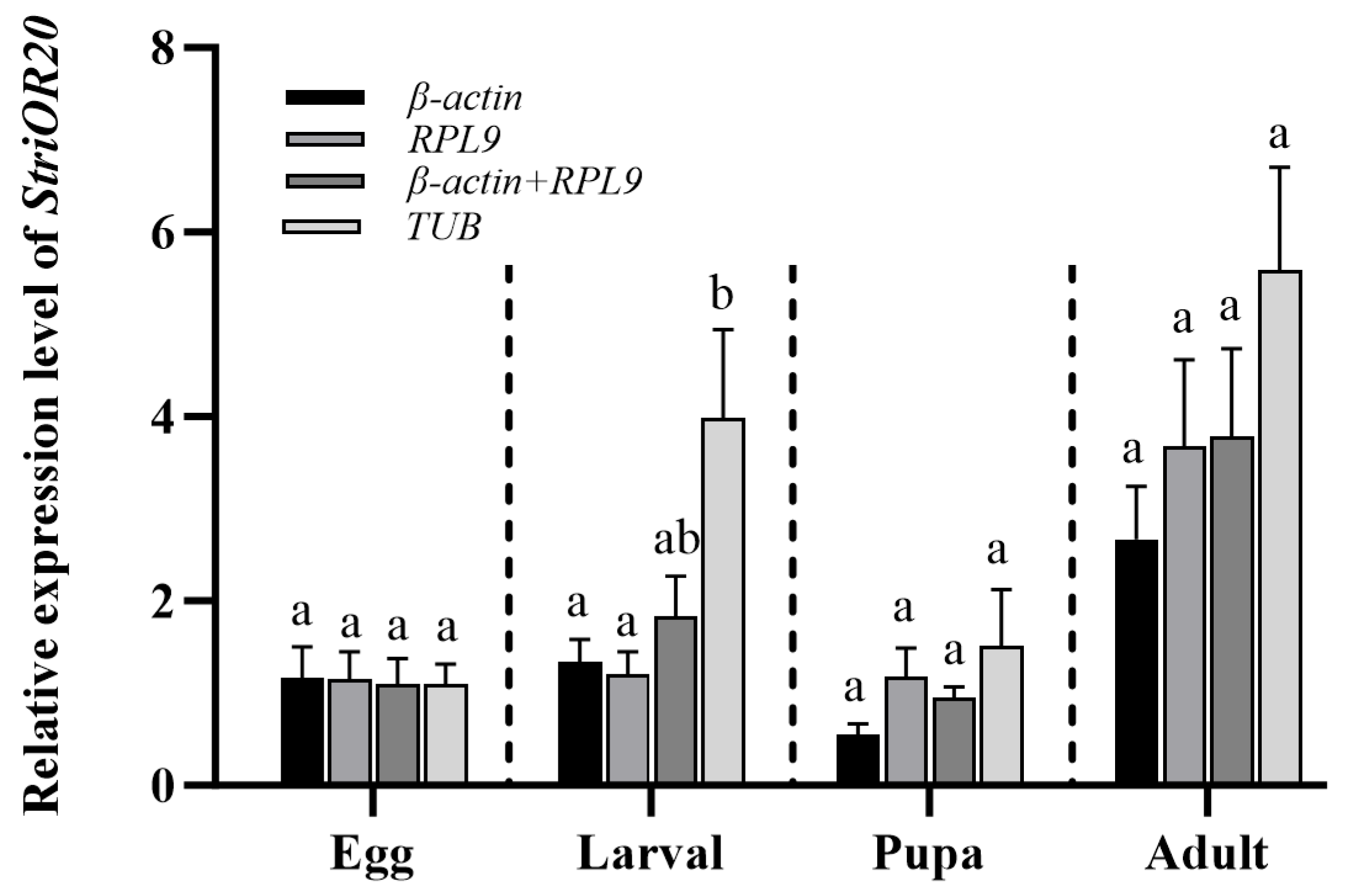
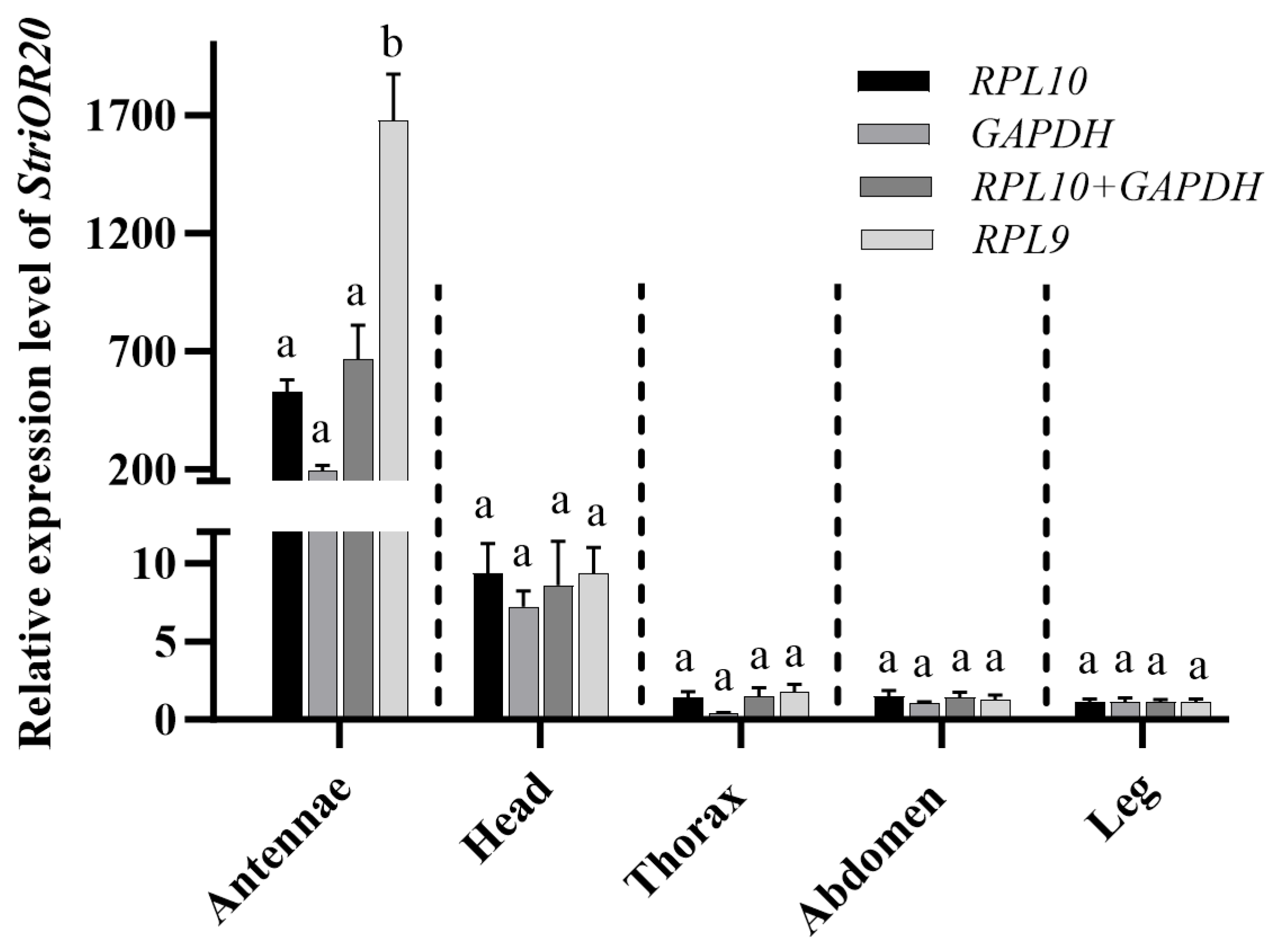
3.4. Verification of Candidate Reference Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Federici, B.A. A new type of insect pathogen in larvae of the clover cutworm, Scotogramma trifolii. J. Invertebr. Pathol. 1982, 40, 41–54. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Cheng, D.F.; Lu, H. Radar observations of the spring migration of spinneret moths in Yanqing, Beijing. In Proceedings of the 2007 Annual Meeting of the Chinese Society for Plant Protection, Guilin, China, 23 November 2007. [Google Scholar]
- Zhao, Z.J.; Chen, E.X.; Zhang, Y. Studies on the biological characteristic of Scotogramma trifolii Rottemberg and its control. Sugar Crops China 1992, 4, 25–28. [Google Scholar]
- Yu, J.N.; Bao, Y.Q. The occurrence of clover cutworm in cotton region of Xinjiang. Xinjiang Agric. Sci. 1996, 1, 34. [Google Scholar]
- Bustin, S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000, 25, 169–193. [Google Scholar] [CrossRef] [PubMed]
- Kubista, M.; Andrade, J.M.; Bengtsson, M.; Forootan, A.; Jonák, J.; Lind, K.; Sindelka, R.; Sjöback, R.; Sjögreen, B.; Strömbom, L.; et al. The real-time polymerase chain reaction. Mol. Asp. Med. 2006, 27, 95–125. [Google Scholar] [CrossRef]
- Derveaux, S.; Vandesompele, J.; Hellemans, J. How to do successful gene expression analysis using real-time PCR. Methods 2010, 50, 227–230. [Google Scholar] [CrossRef]
- Sun, K.Q.; Zhang, Y.J.; D’Alessandro, A.; Nemkov, T.; Song, A.R.; Wu, H.Y.; Liu, H.; Adebiyi, M.; Huang, A.; Wen, Y.E.; et al. Sphingosine-1-phosphate promotes erythrocyte glycolysis and oxygen release for adaptation to high-altitude hypoxia. Nat. Commun. 2016, 7, 12086. [Google Scholar] [CrossRef]
- Guo, J.L.; Ling, H.; Wu, Q.B.; Xu, L.P.; Que, Y.X. The choice of reference genes for assessing gene expression in sugarcane under salinity and drought stresses. Sci. Rep. 2014, 4, 7042. [Google Scholar] [CrossRef]
- Shi, X.-Q.; Guo, W.-C.; Wan, P.-J.; Zhou, L.-T.; Ren, X.-L.; Ahmat, T.; Fu, K.-Y.; Li, G.-Q. Validation of reference genes for expression analysis by quantitative real-time PCR in Leptinotarsa decemlineata (Say). BMC Res. Notes 2013, 6, 93. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Bustin, S.A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Zakrajsek, B.A. Effect of experimental treatment on housekeeping gene expression: Validation by real-time, quantitative RT-PCR. J. Biochem. Biophys. Methods 2000, 46, 69–81. [Google Scholar] [CrossRef]
- Lee, P.D.; Sladek, R.; Greenwood, C.M.; Hudson, T.J. Control genes and variability: Absence of ubiquitous reference transcripts in diverse mammalian expression studies. Genome Res. 2002, 12, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Kozera, B.; Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef]
- Hu, Y.; Fu, H.; Qiao, H.; Sun, S.; Zhang, W.; Jin, S.; Jiang, S.; Gong, Y.; Xiong, Y.; Wu, Y. Validation and Evaluation of Reference Genes for Quantitative Real-Time PCR in Macrobrachium nipponense. Int. J. Mol. Sci. 2018, 19, 2258. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Wu, X.Q.; Lyu, Y.P.; Zhou, H.Z.; Xie, X.Y.; Zhang, X.J.; Yang, H.T. Selection of reliable reference genes for gene expression studies in Botrytis cinerea. J. Microbiol. Methods 2017, 142, 71–75. [Google Scholar] [CrossRef]
- Bagnall, N.H.; Kotze, A.C. Evaluation of reference genes for real-time PCR quantification of gene expression in the Australian sheep blowfly, Lucilia cuprina. Med. Vet. Entomol. 2010, 24, 176–181. [Google Scholar] [CrossRef]
- Bai, Y.; Lv, Y.N.; Zeng, M.; Jia, P.Y.; Lu, H.N.; Zhu, Y.B.; Li, S.; Cui, Y.Y.; Luan, Y.X. Selection of reference genes for normalization of gene expression in Thermobia domestica (Insecta: Zygentoma: Lepismatidae). Genes 2020, 12, 21. [Google Scholar] [CrossRef]
- Scharlaken, B.; de Graaf, D.C.; Goossens, K.; Brunain, M.; Peelman, L.J.; Jacobs, F.J. Reference gene selection for insect expression studies using quantitative real-time PCR: The head of the honeybee, Apis mellifera, after a bacterial challenge. J. Insect Sci. 2008, 8, 33. [Google Scholar] [CrossRef]
- de Boer, M.E.; de Boer, T.E.; Mariën, J.; Timmermans, M.J.T.N.; Nota, B.; van Straalen, N.M.; Ellers, J.; Roelofs, D. Reference genes for QRT-PCR tested under various stress conditions in Folsomia candida and Orchesella cincta (Insecta, Collembola). BMC Mol. Biol. 2009, 10, 54. [Google Scholar] [CrossRef]
- Jiang, H.B.; Liu, Y.H.; Tang, P.A.; Zhou, A.W.; Wang, J.J. Validation of endogenous reference genes for insecticide-induced and developmental expression profiling of Liposcelis bostsrychophila (Psocoptera: Liposcelididae). Mol. Biol. Rep. 2010, 37, 1019–1029. [Google Scholar] [CrossRef]
- Arun, A.; Baumlé, V.; Amelot, G.; Nieberding, C.M. Selection and validation of reference genes for qRT-PCR expression analysis of candidate genes involved in olfactory communication in the butterfly Bicyclus anynana. PLoS ONE 2015, 10, e0120401. [Google Scholar] [CrossRef] [PubMed]
- Koramutla, M.K.; Aminedi, R.; Bhattacharya, R. Comprehensive evaluation of candidate reference genes for qRT-PCR studies of gene expression in mustard aphid, Lipaphis erysimi (Kalt). Sci. Rep. 2016, 6, 25883. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.M.; Chahad-Ehlers, S.; Lima, A.L.; Taniguti, C.H.; Sobrinho, I., Jr.; Torres, F.R.; de Brito, R.A. Reference genes for accessing differential expression among developmental stages and analysis of differential expression of OBP genes in Anastrepha obliqua. Sci. Rep. 2016, 6, 17480. [Google Scholar] [CrossRef]
- Chapman, J.R.; Waldenström, J. With reference to reference genes: A systematic review of endogenous controls in gene expression studies. PLoS ONE 2015, 10, e0141853. [Google Scholar] [CrossRef]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef] [PubMed]
- Han, S.P.; Qin, Q.J.; Wang, D.; Zhou, Y.Y.; He, Y.Z. Selection and evaluation of reference genes for qRT-PCR in Spodoptera frugiperda (Lepidoptera: Noctuidae). Insects 2021, 12, 902. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.L.; Wang, J.Y.; Zhang, B.H. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genom. 2023, 23, 125. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Fu, W.; Xie, W.; Zhang, Z.; Wang, S.; Wu, Q.; Liu, Y.; Zhou, X.; Zhou, X.; Zhang, Y. Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (Lepidoptera: Plutellidae). Int. J. Biol. Sci. 2013, 9, 792–802. [Google Scholar] [CrossRef]
- Zhu, X.; Yuan, M.; Shakeel, M.; Zhang, Y.; Wang, S.; Wang, X.; Zhan, S.; Kang, T.; Li, J. Selection and evaluation of reference genes for expression analysis using qRT-PCR in the beet armyworm Spodoptera exigua (Hübner) (Lepidoptera: Noctuidae). PLoS ONE 2014, 9, e84730. [Google Scholar] [CrossRef]
- Li, H.B.; Dai, C.G.; Zhang, C.R.; He, Y.F.; Ran, H.Y.; Chen, S.H. Screening potential reference genes for quantitative real-time PCR analysis in the oriental armyworm, Mythimna separata. PLoS ONE 2018, 13, e0195096. [Google Scholar] [CrossRef]
- Zhang, S.; An, S.; Li, Z.; Wu, F.; Yang, Q.; Liu, Y.; Cao, J.; Zhang, H.; Zhang, Q.; Liu, X. Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae). Gene 2015, 555, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Yuan, M.; Gao, X.; Kang, T.; Zhan, S.; Wan, H.; Li, J. Identification and validation of reference genes for gene expression analysis using quantitative PCR in Spodoptera litura (Lepidoptera: Noctuidae). PLoS ONE 2013, 8, e68059. [Google Scholar] [CrossRef]
- Xu, J.; Lu, M.X.; Cui, Y.D.; Du, Y.Z. Selection and evaluation of reference genes for expression analysis using qRT-PCR in Chilo suppressalis (Lepidoptera: Pyralidae). J. Econ. Entomol. 2017, 110, 683–691. [Google Scholar] [CrossRef]
- Yang, Q.; Li, Z.; Cao, J.; Zhang, S.; Zhang, H.; Wu, X.; Zhang, Q.; Liu, X. Selection and assessment of reference genes for quantitative PCR normalization in migratory locust Locusta migratoria (Orthoptera: Acrididae). PLoS ONE 2014, 9, e98164. [Google Scholar] [CrossRef]
- Shu, B.; Zhang, J.; Cui, G.; Sun, R.; Sethuraman, V.; Yi, X.; Zhong, G. Evaluation of reference genes for Real-Time Quantitative PCR analysis in Larvae of Spodoptera litura exposed to azadirachtin stress conditions. Front. Physiol. 2018, 9, 372. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Zhou, X.R.; Pang, B.P. Reference gene selection and evaluation for expression analysis using qRT-PCR in Galeruca daurica (Joannis). Bull. Entomol. Res. 2017, 107, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Kariyanna, B.; Prabhuraj, A.; Asokan, R.; Babu, P.; Jalali, S.K.; Venkatesan, T.; Gracy, R.G.; Mohan, M. Identification of suitable reference genes for normalization of RT-qPCR data in eggplant fruit and shoot borer (Leucinodes orbonalis Guenée). Biologia 2020, 75, 289–297. [Google Scholar] [CrossRef]
- Yan, X.; Zhang, Y.; Xu, K.; Wang, Y.; Yang, W. Selection and validation of reference genes for gene expression analysis in Tuta absoluta Meyrick (Lepidoptera: Gelechiidae). Insects 2021, 12, 589. [Google Scholar] [CrossRef]
- Sun, M.; Lu, M.X.; Tang, X.T.; Du, Y.Z. Exploring valid reference genes for quantitative real-time PCR analysis in Sesamia inferens (Lepidoptera: Noctuidae). PLoS ONE 2015, 10, e0115979. [Google Scholar] [CrossRef] [PubMed]
- Leal, W.S. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef]
- Fleischer, J.; Pregitzer, P.; Breer, H.; Krieger, J. Access to the odor world: Olfactory receptors and their role for signal transduction in insects. Cell. Mol. Life Sci. 2018, 75, 485–508. [Google Scholar] [CrossRef]


| Gene | Primer Sequences (5′–3′) | Tm (°C) | Length (bp) |
|---|---|---|---|
| TUB | F:CCGACCTCTTCGTAGTCCTTCTC | 60 | 1202 |
| R:ATGCCCTCGGACAAGACCC | |||
| β-actin | F:TGTGTGACGAGGATGTTGCT | 60 | 892 |
| R:GTGTTGGCGTACAGGTCCTT | |||
| RPL9 | F:CAAATAGTTGCGAACCAAAAAGT | 56 | 559 |
| R:CATCCAGTTCAACAGTAGTCTTCTC | |||
| RPL10 | F:CACGGTTCTGTCGTGGTGTA | 58 | 459 |
| R:AGCCCCATTTCTTGGAGACG | |||
| GAPDH | F:TCCAAAATCGGTATCAACGG | 57 | 970 |
| R:TGAGATCGATGACGCGGT | |||
| EF1-α | F:GAGGCACGCTACGAGGAGATTA | 59 | 767 |
| R:GGGAACTCCTGGAAAGACTCCA |
| Gene | Primer Sequences (5′–3′) | Tm (°C) | Length (bp) | Eff (%) | R2 | Standard Curve Equation |
|---|---|---|---|---|---|---|
| TUB | F:GGCGATGGCGGTGGTGTTAGAC | 60 | 108 | 90.951 | 0.999 | Y = −3.560x + 42.985 |
| R:GGATTCAAGGTGGGCATCAACT | ||||||
| β-actin | F:GTATCCTCACGCTCAAGTA | 60 | 89 | 94.115 | 0.990 | Y = −3.353x + 40.171 |
| R:GTTGTAGAAGGTGTGGTG | ||||||
| RPL9 | F:GAGTGCTGAAGGTAGAGAA | 60 | 161 | 108.777 | 0.990 | Y = −3.128x + 37.734 |
| R:AGTGGTGACACAGTTGAT | ||||||
| RPL10 | F:GCAGTTCTTGACGAGGTA | 60 | 87 | 90.799 | 0.994 | Y = −3.564x + 41.675 |
| R:CTGGTGTCTGACGAGTAC | ||||||
| GAPDH | F:TGCCAAGAAGGTCATCAT | 60 | 79 | 94.201 | 0.990 | Y = −3.469x + 49.458 |
| R:GTCGTATGCCTCAAGGTT | ||||||
| EF1-α | F:GCGATCCAGCCCCCTTC | 60 | 119 | 102.27 | 0.992 | Y = −3.269x + 46.702 |
| R:TTGAGGATGCCGGTCTCAAC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, A.; Zhang, H.; Bai, W.; Ding, R.; Li, W.; Li, G. Selection and Validation of Stable Reference Genes for RT-qPCR in Scotogramma trifolii (Lepidoptera: Noctuidae). Insects 2025, 16, 527. https://doi.org/10.3390/insects16050527
Yang A, Zhang H, Bai W, Ding R, Li W, Li G. Selection and Validation of Stable Reference Genes for RT-qPCR in Scotogramma trifolii (Lepidoptera: Noctuidae). Insects. 2025; 16(5):527. https://doi.org/10.3390/insects16050527
Chicago/Turabian StyleYang, Anpei, Hang Zhang, Weiwei Bai, Ruifeng Ding, Weipeng Li, and Guangkuo Li. 2025. "Selection and Validation of Stable Reference Genes for RT-qPCR in Scotogramma trifolii (Lepidoptera: Noctuidae)" Insects 16, no. 5: 527. https://doi.org/10.3390/insects16050527
APA StyleYang, A., Zhang, H., Bai, W., Ding, R., Li, W., & Li, G. (2025). Selection and Validation of Stable Reference Genes for RT-qPCR in Scotogramma trifolii (Lepidoptera: Noctuidae). Insects, 16(5), 527. https://doi.org/10.3390/insects16050527




