Transcriptome Analysis and Identification of Chemosensory Membrane Proteins in the Head of Euplatypus parallelus
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Collection
2.2. RNA Extraction and cDNA Library Construction and Transcriptome Sequencing
2.3. Transcriptome Assembly and Gene Annotation
2.4. Gene Identification and Sequence Analysis
2.5. Phylogenetic Analysis of Chemosensory Membrane Protein
3. Results
3.1. Transcriptome Sequencing and Assembly
3.2. Functional Annotation of the Unigenes in E. parallelus
3.3. Identification and Analysis of Candidate Chemosensory Membrane Protein Genes
3.3.1. The OR Gene Family
3.3.2. The GR Gene Family
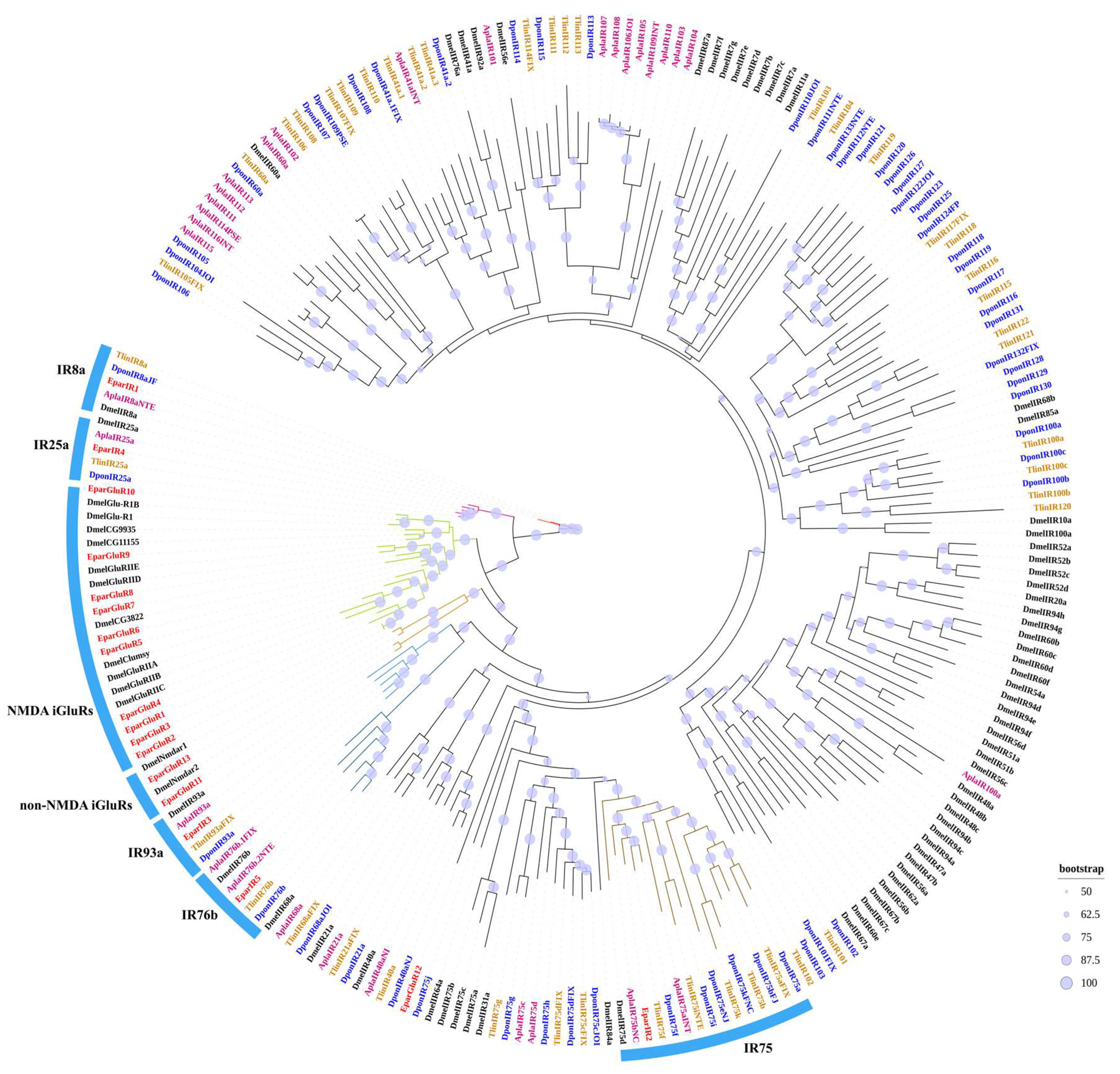
3.3.3. The IR and iGluR Gene Family
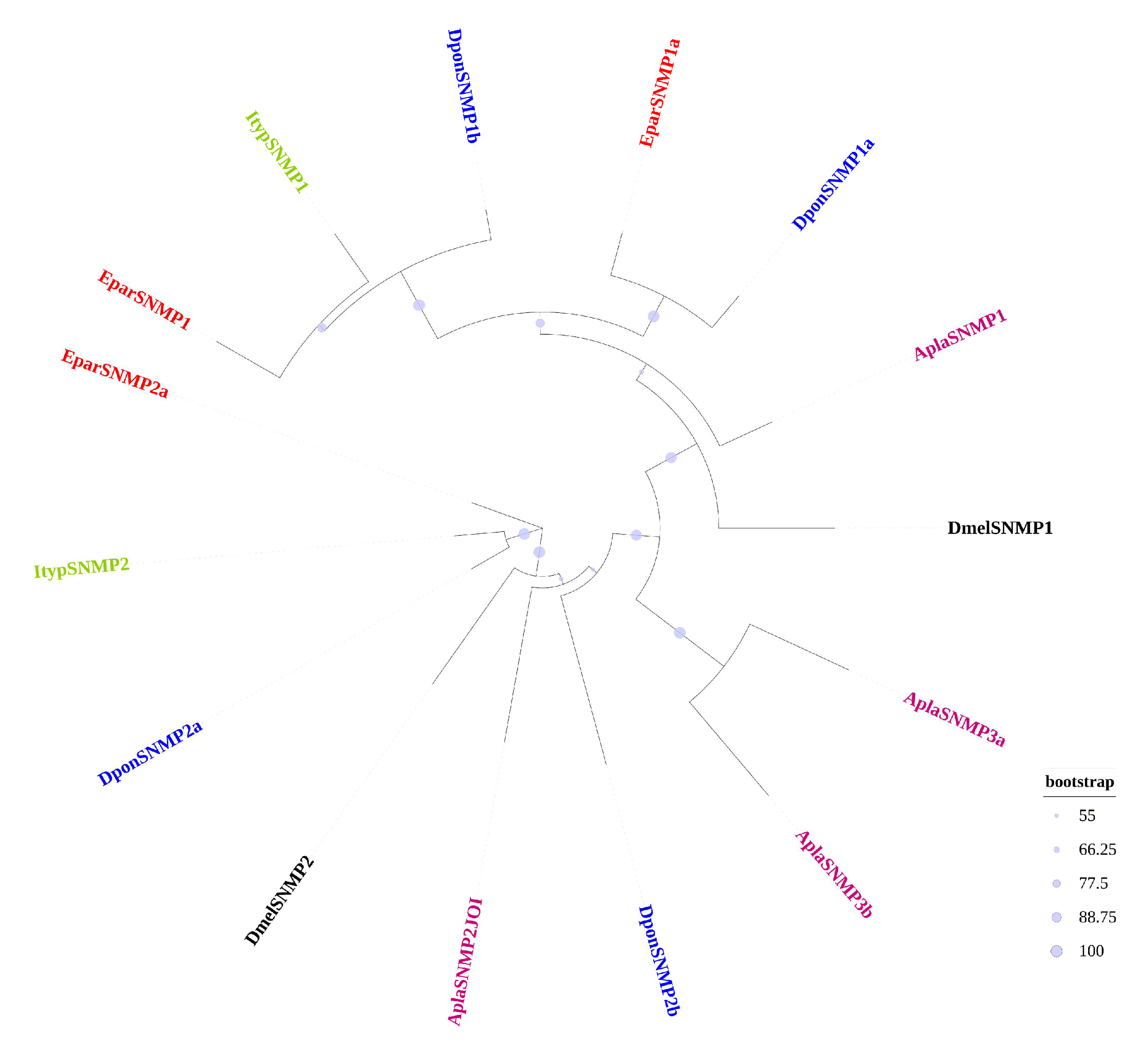
3.3.4. The SNMP Gene Family
3.3.5. Expression Profiles of Chemosensory Membrane Protein Genes Between Sexes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Da Silva, J.; Putz, P.; de Carvalho Silveira, E.; Flechtmann, C.A.H. Biological aspects of Euplatypus parallelus (F.) (Coleoptera, Curculionidae, Platypodinae) attacking Hevea brasiliensis (Willd. ex A. Juss.) in São Paulo northwest) Curculionidae, Brazil. In Proceedings of the 3rd Congresso Brasileiro de Heveicultura, Guarapari, Brazil, 24–26 July 2013; pp. 24–26. [Google Scholar]
- Beaver, R.A. The invasive Neotropical ambrosia beetle Euplatypus parallelus (Fabricius 1801) in the Oriental region and its pest status (Coleoptera: Curculionidae: Platypodinae). Entomol. Mon. Mag. 2013, 149, 143–154. [Google Scholar]
- Gillett, C.P.; Crampton-Platt, A.; Timmermans, M.J.; Jordal, B.H.; Emerson, B.C.; Vogler, A.P. Bulk de novo mitogenome assembly from pooled total DNA elucidates the phylogeny of weevils (Coleoptera: Curculionoidea). Mol. Biol. Evol. 2014, 31, 2223–2237. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, X.; Lai, S.; Yin, T.; Ji, Y.; Wang, S.; Hulcr, J. First record of Euplatypus parallelus (Coleoptera: Curculionidae) in China. Fla. Entomol. 2018, 101, 141–143. [Google Scholar] [CrossRef]
- Onso-Zarazagama, M.A.; Lyal, C.H.C. A catalogue of family and genus group names in Scolytinae and Platypodinae with nomenclatural remarks (Coleoptera: Curculionidae). Zootaxa 2009, 2258, 1–134. [Google Scholar]
- Boa, E.; Kirkendall, L. Sandragon wilt disease, Seychelles. In TCP Final Technical Report; CABI Bioscience, University of Bergen: Bergen, Norway, 2004; pp. 17–79. [Google Scholar]
- Atkinson, T.H.; Martínez-Fernández, E.; Saucedo-Céspedes, E.; Burgos-Solorio, A. Scolytidae y Platypodidae (Coleoptera) asociados a selva baja y comunidades derivadas en el estado de Morelos, México. Folia Entomol. Mex. 1986, 69, 41–82. [Google Scholar]
- Zanuncio, J.C.; Sossai, M.F.; Couto, L.; Pinto, R. Occurrence of Euplatypus parallelus, Euplatypus sp. (col.: Euplatypodidae) and Xyleborus affinis (col.: Scolytidae) in Pinus sp. in Ribas do Rio Pardo, Mato Grosso do Sul, Brazil. Rev. Arvore 2002, 26, 387–389. [Google Scholar] [CrossRef]
- Bumrungsri, S.; Beaver, R.; Phongpaichit, S.; Sittichaya, W. The infestation by an exotic ambrosia beetle, Euplatypus parallelus (F.)(Coleoptera: Curculionidae: Platypodinae) of Angsana trees (Pterocarpus indicus Willd.) in southern Thailand. Songklanakarin J. Sci. Technol. 2008, 30, 5. [Google Scholar]
- Renou, M.; Anton, S. Insect olfactory communication in a complex and changing world. Curr. Opin. Insect Sci. 2020, 42, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Vieira, F.G.; Rozas, J. Comparative genomics of the odorant-binding and chemosensory protein gene families across the Arthropoda: Origin and evolutionary history of the chemosensory system. Genome Biol. Evol. 2011, 3, 476–490. [Google Scholar] [CrossRef]
- Liu, N.Y.; Li, Z.B.; Zhao, N.; Song, Q.S.; Zhu, J.Y.; Yang, B. Identification and characterization of chemosensory gene families in the bark beetle, Tomicus yunnanensis. Comp. Biochem. Physiol. Part D Genom. Proteom. 2018, 25, 73–85. [Google Scholar] [CrossRef]
- Vosshall, L.B.; Stocker, R.F. Molecular architecture of smell and taste in Drosophila. Annu. Rev. Neurosci. 2007, 30, 505–533. [Google Scholar] [CrossRef] [PubMed]
- Stengl, M.; Funk, N.W. The role of the coreceptor Orco in insect olfactory transduction. J. Comp. Physiol. A 2013, 199, 897–909. [Google Scholar] [CrossRef] [PubMed]
- Butterwick, J.A.; Del Mármol, J.; Kim, K.H.; Kahlson, M.A.; Rogow, J.A.; Walz, T.; Ruta, V. Cryo-EM structure of the insect olfactory receptor Orco. Nature 2018, 560, 447–452. [Google Scholar] [CrossRef]
- Nakagawa, T.; Pellegrino, M.; Sato, K.; Vosshall, L.B.; Touhara, K. Amino acid residues contributing to function of the heteromeric insect olfactory receptor complex. PLoS ONE 2012, 7, e32372. [Google Scholar] [CrossRef]
- Del Mármol, J.; Yedlin, M.A.; Ruta, V. The structural basis of odorant recognition in insect olfactory receptors. Nature 2021, 597, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Chen, A.Q.; Ryu, J.; Del Mármol, J. Structural basis of odor sensing by insect heteromeric odorant receptors. Science 2024, 384, 1460–1467. [Google Scholar] [CrossRef]
- Wang, Y.; Qiu, L.; Wang, B.; Guan, Z.; Dong, Z.; Zhang, J.; Cao, S.; Yang, L.; Wang, B.; Gong, Z.; et al. Structural basis for odorant recognition of the insect odorant receptor OR-Orco heterocomplex. Science 2024, 384, 1453–1460. [Google Scholar] [CrossRef]
- Pang, J.X.; Zeng, X.; Zhu, J.Y.; Liu, N.Y. Chemosensory transmembrane protein families in the coffee white stemborer, Xylotrechus quadripes (Coleoptera: Cerambycidae). Environ. Entomol. 2018, 47, 969–981. [Google Scholar] [CrossRef]
- Zhao, H.; Du, Y.; Gao, P.; Wang, S.; Pan, J.; Jiang, Y. Antennal transcriptome and differential expression analysis of five chemosensory gene families from the Asian honeybee Apis cerana cerana. PLoS ONE 2016, 11, e0165374. [Google Scholar] [CrossRef]
- Wicher, D.; Miazzi, F. Functional properties of insect olfactory receptors: Ionotropic receptors and odorant receptors. Cell Tissue Res. 2021, 383, 7–19. [Google Scholar] [CrossRef]
- Nichols, Z.; Vogt, R.G. The SNMP/CD36 gene family in Diptera, Hymenoptera and Coleoptera: Drosophila melanogaster, D. pseudoobscura, Anopheles gambiae, Aedes aegypti, Apis mellifera, and Tribolium castaneum. Insect Biochem. Mol. Biol. 2008, 38, 398–415. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, W.; Nie, R.E.; Li, W.Z.; Segraves, K.A.; Yang, X.K.; Xue, H.J. Comparative transcriptome analysis of chemosensory genes in two sister leaf beetles provides insights into chemosensory speciation. Insect Biochem. Mol. Biol. 2016, 79, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Wang, C.; Fang, C.; Zhang, S.; Cao, Y.; Li, K.; Leal, W.S. Functional characterization of odorant-binding proteins from the scarab beetle Holotrichia oblita based on semiochemical-induced expression alteration and gene silencing. Insect Biochem. Mol. Biol. 2019, 104, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.L.; Yuan, M.L. Progress in insect transcriptomics based on the next-generation sequencing technique. Acta Entomol. Sin. 2013, 56, 1489–1508. [Google Scholar]
- Zhang, Y.N.; Jin, J.Y.; Jin, R.; Xia, Y.H.; Zhou, J.J.; Deng, J.Y.; Dong, S.L. Differential expression patterns in chemosensory and non-chemosensory tissues of putative chemosensory genes identified by transcriptome analysis of insect pest the purple stem borer Sesamia inferens (Walker). PLoS ONE 2013, 8, e69715. [Google Scholar]
- Li, X.M.; Zhu, X.Y.; Wang, Z.Q.; Wang, Y.; He, P.; Chen, G.; Sun, L.; Deng, D.G.; Zhang, Y.N. Candidate chemosensory genes identified in Colaphellus bowringi by antennal transcriptome analysis. BMC Genom. 2015, 16, 1028. [Google Scholar] [CrossRef]
- Leitch, O.; Papanicolaou, A.; Lennard, C.; Kirkbride, K.P.; Anderson, A. Chemosensory genes identified in the antennal transcriptome of the blowfly Calliphora stygia. BMC Genom. 2015, 16, 255. [Google Scholar] [CrossRef]
- Du, L.; Zhao, X.; Liang, X.; Gao, X.; Liu, Y.; Wang, G. Identification of candidate chemosensory genes in Mythimna separata by transcriptomic analysis. BMC Genom. 2018, 19, 518. [Google Scholar] [CrossRef]
- Glaser, N.; Gallot, A.; Legeai, F.; Montagné, N.; Poivet, E.; Harry, M.; Calatayud, P.; Jacquin-Joly, E. Candidate chemosensory genes in the stemborer Sesamia nonagrioides. Int. J. Biol. Sci. 2013, 9, 481. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Deng, Y. Integrated nr database in protein annotation system and its localization. Comput. Eng. 2006, 32, 71. [Google Scholar]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.Z.; Lopez, R.; Magrane, M.; et al. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Koonin, E.V.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Krylov, D.M.; Makarova, K.S.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; Rao, S.B.; et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004, 5, 1–28. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Menda, D.R.; Sunagawa, S.; Kuhn, M.; et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016, 44, D286–D293. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, S.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Eddy, S.R. Profile hidden Markov models. Bioinformatics 1998, 14, 755–763. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Liu, Y.; Wei, J.; Liao, X.; Walker, W.B.; Li, J.; Wang, G. Identification of candidate olfactory genes in Chilo suppressalis by antennal transcriptome analysis. Int. J. Biol. Sci. 2014, 10, 846. [Google Scholar] [CrossRef]
- Mitchell, R.F.; Schneider, T.M.; Schwartz, A.M.; Andersson, M.N.; McKenna, D.D. The diversity and evolution of odorant receptors in beetles (Coleoptera). Insect Mol. Biol. 2020, 29, 77–91. [Google Scholar] [CrossRef]
- Andersson, M.N.; Keeling, C.I.; Mitchell, R.F. Genomic content of chemosensory genes correlates with host range in wood-boring beetles (Dendroctonus ponderosae, Agrilus planipennis, and Anoplophora glabripennis). BMC Genom. 2019, 20, 690. [Google Scholar] [CrossRef] [PubMed]
- Biswas, T.; Vogel, H.; Biedermann, P.H.W.; Lehenberger, M.; Yuvaraj, J.K.; Andersson, M.N. Few chemoreceptor genes in the ambrosia beetle Trypodendron lineatum may reflect its specialized ecology. BMC Genom. 2024, 25, 764. [Google Scholar] [CrossRef]
- Zhao, Y.J.; Li, G.C.; Zhu, J.Y.; Liu, N.Y. Genome-based analysis reveals a novel SNMP group of the Coleoptera and chemosensory receptors in Rhaphuma horsfieldi. Genomics 2020, 112, 2713–2728. [Google Scholar] [CrossRef]
- Buck, L.; Axel, R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell 1991, 65, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.N.; Grosse-Wilde, E.; Keeling, C.I.; Bengtsson, J.M.; Yuen, M.M.; Li, M.; Hillbur, Y.; Bohlmann, J.; Hansson, B.S.; Schlyter, F. Antennal transcriptome analysis of the chemosensory gene families in the tree killing bark beetles, Ips typographus and Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytinae). BMC. Genom. 2013, 14, 198. [Google Scholar] [CrossRef]
- Johny, J.; Große-Wilde, E.; Kalinová, B.; Roy, A. Antennal Transcriptome Screening and Identification of Chemosensory Proteins in the Double-Spine European Spruce Bark Beetle, Ips duplicatus (Coleoptera: Scolytinae). Int. J. Mol. Sci. 2024, 25, 9513. [Google Scholar] [CrossRef]
- Yuvaraj, J.K.; Roberts, R.E.; Sonntag, Y.; Hou, X.Q.; Grosse-Wilde, E.; Machara, A.; Zhang, D.D.; Hansson, B.S.; Johanson, U.; Löfstedt, C.; et al. Putative ligand binding sites of two functionally characterized bark beetle odorant receptors. BMC Biol. 2021, 19, 16. [Google Scholar] [CrossRef]
- Antony, B.; Soffan, A.; Jakše, J.; Abdelazim, M.M.; Aldosari, S.A.; Aldawood, A.S.; Pain, A. Identification of the genes involved in odorant reception and detection in the palm weevil Rhynchophorus ferrugineus, an important quarantine pest, by antennal transcriptome analysis. BMC Genom. 2016, 17, 69. [Google Scholar] [CrossRef] [PubMed]
- Benton, R. Multigene family evolution: Perspectives from insect chemoreceptors. Trends Ecol. Evol. 2015, 30, 590–600. [Google Scholar] [CrossRef]
- Nei, M.; Niimura, Y.; Nozawa, M. The evolution of animal chemosensory receptor gene repertoires: Roles of chance and necessity. Nat. Rev. Genet. 2008, 9, 951–963. [Google Scholar] [CrossRef] [PubMed]
- Smadja, C.; Shi, P.; Butlin, R.K.; Robertson, H.M. Large gene family expansions and adaptive evolution for odorant and gustatory receptors in the pea aphid, Acyrthosiphon pisum. Mol. Biol. Evol. 2009, 26, 2073–2086. [Google Scholar] [CrossRef]
- Kandasamy, D.; Gershenzon, J.; Andersson, M.N.; Hammerbacher, A. Volatile organic compounds influence the interaction of the Eurasian spruce bark beetle (Ips typographus) with its fungal symbionts. ISME J. 2019, 13, 1788–1800. [Google Scholar] [CrossRef]
- Blanchette, R.A.; Farrell, R.L.; Burnes, T.A.; Wendler, P.A.; Zimmerman, W. Biological control of pitch in pulp and paper production by Ophiostoma piliferum. Tappi J. 1992, 75, 102–106. [Google Scholar]
- Kandasamy, D.; Zaman, R.; Nakamura, Y.; Zhao, T.; Hartmann, H.; Andersson, M.N.; Hammerbacher, A.; Gershenzon, J. Conifer-killing bark beetles locate fungal symbionts by detecting volatile fungal metabolites of host tree resin monoterpenes. PLoS Biol. 2023, 21, e3001887. [Google Scholar] [CrossRef] [PubMed]
- Ji, T.; Xu, Z.; Jia, Q.; Wang, G.; Hou, Y. Non-palm plant volatile α-pinene is detected by antenna-biased expressed odorant receptor 6 in the Rhynchophorus ferrugineus (Olivier) (Coleoptera: Curculionidae). Front. Physiol. 2021, 12, 701545. [Google Scholar] [CrossRef] [PubMed]
- Biswas, T.; Sims, C.; Yuvaraj, J.K.; Roberts, R.E.; Löfstedt, C.; Andersson, M.N. Functional characterization supports multiple evolutionary origins of pheromone re-ceptors in bark beetles. Mol. Biol. Evol. 2024, 41, msae196. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.E.; Biswas, T.; Yuvaraj, J.K.; Grosse-Wilde, E.; Powell, D.; Hansson, B.S.; Löfstedt, C.; Andersson, M.N. Odorant receptor orthologues in conifer-feeding beetles display conserved responses to ecologically relevant odours. Mol. Ecol. 2022, 31, 3693–3707. [Google Scholar] [CrossRef]
- Zhang, Q.H.; Schlyter, F. Olfactory recognition and behavioural avoidance of angiosperm nonhost volatiles by conifer-inhabiting bark beetles. Agric. For. Entomol. 2004, 6, 1–20. [Google Scholar] [CrossRef]
- Rainho, H.L.; Silva, W.D.; Bento, J.M.S. Semiochemical-based attractant for the ambrosia pinhole borer Euplatypus parallelus. Agronomy 2021, 11, 266. [Google Scholar] [CrossRef]
- Wang, X.; Wang, S.; Yi, J.; Li, Y.; Liu, J.; Wang, J.; Xi, J. Three host plant volatiles, hexanal, lauric acid, and tetradecane, are detected by an antenna-biased expressed odorant receptor 27 in the dark black chafer Holotrichia parallela. J. Agric. Food. Chem. 2020, 68, 7316–7323. [Google Scholar] [CrossRef]
- Li, R.; Song, X.; Shan, S.; Hussain Dhiloo, K.; Wang, S.; Yin, Z.; Lu, Z.; Khashaveh, A.; Zhang, Y. Female-Biased Odorant Receptor MmedOR48 in the Parasitoid Microplitis mediator Broadly Tunes to Plant Volatiles. J. Agric. Food Chem. 2024, 72, 17617–17625. [Google Scholar] [CrossRef]
- Anderson, A.R.; Wanner, K.W.; Trowell, S.C.; Warr, C.G.; Jaquin-Joly, E.; Zagatti, P.; Robertson, H.; Newcomb, R.D. Molecular basis of female-specific odorant responses in Bombyx mori. Insect Biochem. Mol. Biol. 2009, 39, 189–197. [Google Scholar] [CrossRef]
- Weiss, L.A.; Dahanukar, A.; Kwon, J.Y.; Banerjee, D.; Carlson, J.R. The molecular and cellular basis of bitter taste in Drosophila. Neuron 2011, 69, 258–272. [Google Scholar] [CrossRef]
- Johny, J.; Nihad, M.; Alharbi, H.A.; AlSaleh, M.A.; Antony, B. Silencing sensory neuron membrane protein RferSNMPu1 impairs pheromone detection in the invasive Asian Palm Weevil. Sci. Rep. 2024, 14, 16541. [Google Scholar] [CrossRef] [PubMed]
- Cassau, S.; Krieger, J. The role of SNMPs in insect olfaction. Cell Tissue Res. 2021, 383, 21–33. [Google Scholar] [CrossRef] [PubMed]



| Sample | Read Number | Base Number | GC Content | % ≥ Q30 |
|---|---|---|---|---|
| Epar-F-1 | 21,805,037 | 6,525,067,675 | 35.34% | 92.18% |
| Epar-F-2 | 22,408,048 | 6,693,904,718 | 34.73% | 92.74% |
| Epar-F-3 | 24,058,796 | 7,197,880,307 | 35.34% | 94.02% |
| Epar-M-1 | 25,618,478 | 7,661,751,802 | 34.45% | 92.94% |
| Epar-M-2 | 20,465,126 | 6,125,589,791 | 35.11% | 92.78% |
| Epar-M-3 | 24,621,535 | 7,367,886,987 | 34.75% | 92.92% |
| Length Range | Transcript | Unigene |
|---|---|---|
| 200–300 | 11,098 (19.02%) | 9699 (29.59%) |
| 300–500 | 8685 (14.88%) | 6056 (18.48%) |
| 500–1000 | 11,779 (20.18%) | 6454 (19.69%) |
| 1000–2000 | 12,723 (21.80%) | 5341 (16.30%) |
| 2000+ | 14,077 (24.12%) | 5225 (15.94%) |
| Total Number | 58,362 | 32,775 |
| Total Length | 83,550,564 | 35,529,222 |
| N50 Length | 2494 | 2104 |
| Mean Length | 1431.59 | 1084.03 |
| Database | Number of Annotated Unigenes | 300 ≤ Length < 1000 | Length ≥ 1000 |
|---|---|---|---|
| COG_Annotation | 4608 | 1180 | 3009 |
| GO_Annotation | 14,295 | 4202 | 8068 |
| KEGG_Annotation | 11,014 | 2742 | 7259 |
| KOG_Annotation | 10,113 | 2575 | 6536 |
| Pfam_Annotation | 12,919 | 3566 | 8037 |
| Swissprot_Annotation | 7769 | 1803 | 5365 |
| eggNOG_Annotation | 11,575 | 3040 | 7515 |
| nr_Annotation | 14,557 | 4152 | 8808 |
| All_Annotated | 17,761 | 5658 | 9296 |
| Gene Name | Best Blast Mach | Gene Description | Acc. Number | E-Value | Identity (%) |
|---|---|---|---|---|---|
| EparORco | Pachyrhinus yasumatsui | odorant receptor co-receptor | UUW42911.1 | 0.00 | 86.1 |
| EparOR1 | Eucryptorrhynchus scrobiculatus | odorant receptor 13 | QXE93194.1 | 4.00 × 10−81 | 42.75 |
| EparOR2 | Pachyrhinus yasumatsui | odorant receptor 19 | WJJ63319.1 | 1.00 × 10−147 | 57.07 |
| EparOR3 | Eucryptorrhynchus scrobiculatus | odorant receptor 19 | QXE93200.1 | 2.00 × 10−137 | 52.84 |
| EparOR4 | Pachyrhinus yasumatsui | odorant receptor 10 | WJJ63310.1 | 8.00 × 10−127 | 50 |
| EparOR5 | Anthonomus grandis grandis | gustatory and odorant receptor 22-like | XP_050295302.1 | 0.00 | 80.09 |
| EparOR6 | Eucryptorrhynchus scrobiculatus | odorant receptor 3 | QXE93184.1 | 3.00 × 10−155 | 52.47 |
| EparOR7 | Pachyrhinus yasumatsui | odorant receptor 7 | WJJ63307.1 | 1.00 × 10−76 | 36.48 |
| EparOR8 | Pachyrhinus yasumatsui | odorant receptor 4 | WJJ63304.1 | 1.00 × 10−86 | 38.38 |
| EparOR9 | Euwallacea similis | gustatory and odorant receptor 22-like isoform X1 | KAF7267635.1 | 0.00 | 73.95 |
| EparOR10 | Euwallacea similis | odorant receptor 10-like | XP_066258781.1 | 1.00 × 10−31 | 29.32 |
| EparOR11 | Sitophilus oryzae | gustatory and odorant receptor 22 | XP_030746043.1 | 0.00 | 74.81 |
| EparOR12 | Eucryptorrhynchus scrobiculatus | odorant receptor 1 | QXE93182.1 | 5.00 × 10−115 | 49.34 |
| EparOR13 | Pachyrhinus yasumatsui | odorant receptor 24 | WJJ63324.1 | 2.00 × 10−80 | 36.81 |
| EparOR14 | Cylas formicarius | odorant receptor 49b-like isoform X2 | XP_060524832.1 | 3.00 × 10−24 | 32.01 |
| EparOR15 | Pachyrhinus yasumatsui | odorant receptor 26 | WJJ63326.1 | 5.00 × 10−31 | 28.28 |
| EparOR16 | Cylas formicarius | odorant receptor 49b-like isoform X3 | XP_060520304.1 | 2.00 × 10−55 | 32.5 |
| EparOR17 | Dendroctonus ponderosae | odorant receptor 47b | XP_019753281.2 | 2.00 × 10−79 | 39.9 |
| EparOR18 | Cylas formicarius | odorant receptor 49b-like isoform X3 | XP_060520304.1 | 6.00 × 10−62 | 34.58 |
| EparOR19 | Rhynchophorus ferrugineus | odorant receptor | QCS37751.1 | 4.00 × 10−24 | 31.52 |
| EparOR20 | Sitophilus oryzae | odorant receptor 94a-like | XP_030753544.1 | 5.00 × 10−99 | 43.82 |
| EparOR21 | Sitophilus oryzae | odorant receptor 49b-like | XP_030764608.1 | 1.00 × 10−72 | 37.42 |
| EparOR22 | Sitophilus oryzae | odorant receptor 49b-like | XP_030764608.1 | 8.00 × 10−116 | 61.7 |
| EparOR23 | Sitophilus oryzae | odorant receptor 67c-like | XP_030758890.1 | 3.00 × 10−71 | 56.25 |
| EparOR24 | Cylas formicarius | odorant receptor 4-like isoform X2 | XP_060520303.1 | 8.00 × 10−56 | 34.51 |
| EparOR25 | Pachyrhinus yasumatsui | odorant receptor 1 | WJJ63301.1 | 9.00 × 10−75 | 50.92 |
| EparOR26 | Cylas formicarius | odorant receptor 45b-like isoform X1 | XP_060530731.1 | 6.00 × 10−32 | 29.54 |
| EparOR27 | Sitophilus oryzae | odorant receptor 46a-like | XP_030763857.1 | 3.00 × 10−30 | 37.64 |
| EparOR28 | Sitophilus oryzae | odorant receptor 67c-like | XP_030750008.1 | 6.00 × 10−63 | 46.03 |
| EparOR29 | Eucryptorrhynchus scrobiculatus | odorant receptor 5 | QXE93186.1 | 5.00 × 10−78 | 38.94 |
| EparOR30 | Pachyrhinus yasumatsui | odorant receptor 7 | WJJ63307.1 | 9.00 × 10−59 | 36.84 |
| EparOR31 | Eucryptorrhynchus brandti | odorant receptor 26 | QXE93252.1 | 5.00 × 10−72 | 41.95 |
| EparOR32 | Pachyrhinus yasumatsui | odorant receptor 26 | WJJ63326.1 | 2.00 × 10−28 | 33.78 |
| EparOR33 | Eucryptorrhynchus scrobiculatus | odorant receptor 29 | QXE93210.1 | 5.00 × 10−33 | 38.97 |
| EparOR34 | Pachyrhinus yasumatsui | odorant receptor 26 | WJJ63326.1 | 2.00 × 10−36 | 33.61 |
| EparOR35 | Pachyrhinus yasumatsui | odorant receptor 26 | WJJ63326.1 | 1.00 × 10−36 | 35.78 |
| EparOR36 | Sitophilus oryzae | odorant receptor 67a-like | XP_030763199.1 | 1.00 × 10−12 | 62 |
| EparOR37 | Ips typographus | odorant receptor 40 | WZI48996.1 | 7.00 × 10−19 | 39.29 |
| EparOR38 | Sitophilus oryzae | odorant receptor 46a-like | XP_030763857.1 | 4.00 × 10−09 | 33.96 |
| EparOR39 | Sitophilus oryzae | odorant receptor 67a-like | XP_030763199.1 | 1.00 × 10−12 | 62 |
| EparOR40 | Cylas formicarius | odorant receptor Or2-like | XP_060533836.1 | 3.00 × 10−12 | 48.15 |
| Gene Name | Best Blast Mach | Gene Description | Acc. Number | E-Value | Identity (%) |
|---|---|---|---|---|---|
| EparGR1 | Sitophilus oryzae | gustatory and odorant receptor 22 | XP_030746043.1 | 0 | 74.81 |
| EparGR2 | Pyrrhalta aenescens | gustatory receptor 13 | APC94340.1 | 5 × 10−7 | 44.32 |
| EparGR3 | Anthonomus grandis grandis | gustatory and odorant receptor 22-like | XP_050295302.1 | 0 | 80.09 |
| EparGR4 | Pachyrhinus yasumatsui | gustatory receptor 1 | WJJ63341.1 | 0 | 73.79 |
| EparGR5 | Pachyrhinus yasumatsui | gustatory receptor 9 | WJJ63349.1 | 9 × 10−7 | 32.81 |
| EparGR6 | Sitophilus oryzae | gustatory receptor family protein 3-like | XP_030759073.1 | 9 × 10−63 | 68.16 |
| EparGR7 | Anthonomus grandis grandis | gustatory and odorant receptor 24 | XP_050306870.1 | 1 × 10−135 | 85.98 |
| EparGR8 | Colaphellus bowringi | gustatory receptor 1 | ALR72527.1 | 9 × 10−16 | 54.41 |
| EparGR9 | Sitophilus oryzae | gustatory receptor for sugar taste 64a-like | XP_030761257.1 | 3 × 10−31 | 41.71 |
| EparGR10 | Colaphellus bowringi | gustatory receptor 1 | ALR72527.1 | 3 × 10−11 | 63.83 |
| EparGR11 | Anthonomus grandis grandis | gustatory receptor for sugar taste 43a-like isoform X1 | XP_050302234.1 | 1 × 10−39 | 61.64 |
| EparGR12 | Pachyrhinus yasumatsui | gustatory receptor 6 | WJJ63346.1 | 2 × 10−19 | 40.32 |
| Gene Name | Best Blast Mach | Gene Description | Acc. Number | E-Value | Identity (%) |
|---|---|---|---|---|---|
| EparIR1 | Pachyrhinus yasumatsui | ionotropic receptor 8a | WJJ63357.1 | 0 | 68.01 |
| EparIR2 | Pachyrhinus yasumatsui | ionotropic receptor 75s | WJJ63351.1 | 0 | 64.11 |
| EparIR3 | Pachyrhinus yasumatsui | ionotropic receptor 93a | WJJ63358.1 | 0 | 69.4 |
| EparIR4 | Sitophilus oryzae | ionotropic receptor 25a | XP_030756779.1 | 0 | 84.33 |
| EparIR5 | Pachyrhinus yasumatsui | ionotropic receptor 76b | WJJ63350.1 | 0 | 63.75 |
| EparIR6 | Pachyrhinus yasumatsui | ionotropic receptor 64a | WJJ63355.1 | 3 × 10−33 | 69.77 |
| EparIR7 | Sitophilus oryzae | ionotropic receptor 40a-like | XP_030764108.1 | 7 × 10−35 | 61.61 |
| EparIR8 | Cylas formicarius | ionotropic receptor 75a-like isoform X3 | XP_060526926.1 | 5 × 10−24 | 62.16 |
| EparIR9 | Sitophilus oryzae | ionotropic receptor 40a-like | XP_030764108.1 | 2 × 10−23 | 63.75 |
| EparIR10 | Pachyrhinus yasumatsui | ionotropic receptor 8a | WJJ63357.1 | 9 × 10−34 | 48.53 |
| EparIR11 | Pachyrhinus yasumatsui | ionotropic receptor 93a | WJJ63358.1 | 1 × 10−54 | 68.18 |
| EparIR12 | Pachyrhinus yasumatsui | ionotropic receptor 21a | WJJ63354.1 | 5 × 10−127 | 78.76 |
| EparIR13 | Pachyrhinus yasumatsui | ionotropic receptor 93a | WJJ63358.1 | 5 × 10−62 | 60.71 |
| EparIR14 | Euwallacea similis | ionotropic receptor 25a | XP_066256016.1 | 0 | 86.87 |
| EparGluR1 | Pachyrhinus yasumatsui | glutamate receptor ionotropic 4 | WJJ63362.1 | 0 | 63.72 |
| EparGluR2 | Dendroctonus ponderosae | glutamate receptor ionotropic, kainate 2 | XP_048525692.1 | 0 | 88.28 |
| EparGluR3 | Pachyrhinus yasumatsui | glutamate receptor ionotropic 3 | WJJ63361.1 | 0 | 67.75 |
| EparGluR4 | Sitophilus oryzae | glutamate receptor ionotropic, kainate 2-like | XP_030751235.1 | 0 | 62.98 |
| EparGluR5 | Pachyrhinus yasumatsui | glutamate receptor ionotropic 1 | WJJ63359.1 | 0 | 73.84 |
| EparGluR6 | Sitophilus oryzae | glutamate receptor ionotropic, kainate 2 | XP_030751944.1 | 0 | 91.87 |
| EparGluR7 | Sitophilus oryzae | glutamate receptor ionotropic, kainate 2-like | XP_030749785.1 | 0 | 69.54 |
| EparGluR8 | Sitophilus oryzae | glutamate receptor ionotropic, kainate 2-like isoform X1 | XP_030751945.1 | 0 | 70.83 |
| EparGluR9 | Sitophilus oryzae | glutamate receptor ionotropic, kainate 2-like isoform X3 | XP_030751453.1 | 0 | 84.78 |
| EparGluR10 | Sitophilus oryzae | glutamate receptor 1-like isoform X1 | XP_030765898.1 | 0 | 84.6 |
| EparGluR11 | Euwallacea fornicatus | glutamate receptor ionotropic, NMDA 2B isoform X2 | XP_066141506.1 | 0 | 85.9 |
| EparGluR12 | Sitophilus oryzae | glutamate receptor-interacting protein 2 isoform X2 | XP_030749851.1 | 0 | 85.93 |
| EparGluR13 | Sitophilus oryzae | glutamate [NMDA] receptor subunit 1 isoform X1 | XP_030759804.1 | 0 | 89.76 |
| EparGluR14 | Leptinotarsa decemlineata | glutamate receptor 1-like | XP_023011564.1 | 2 × 10−16 | 38.14 |
| EparGluR15 | Zophobas morio | glutamate receptor 3-like | XP_063905986.1 | 1 × 10−07 | 30.48 |
| Gene Name | Best Blast Mach | Gene Description | Acc. Number | E-Value | Identity(%) |
|---|---|---|---|---|---|
| EparSNMP1 | Cyrtotrachelus buqueti | sensory neuron membrane protein 1b | WAQ79968.1 | 0 | 56.67 |
| EparSNMP1a | Pachyrhinus yasumatsui | sensory neuron membrane protein 1a | WJJ63366.1 | 0 | 61.48 |
| EparSNMP2a | Pachyrhinus yasumatsui | sensory neuron membrane protein 2a | WJJ63368.1 | 0 | 51.42 |
| EparSNMP3 | Dendroctonus ponderosae | sensory neuron membrane protein 2 | XP_019770844.2 | 2 × 10−44 | 56.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Q.; Zhou, X.; Xu, Z.; Zhang, X.; Yuan, H.; Guo, J. Transcriptome Analysis and Identification of Chemosensory Membrane Proteins in the Head of Euplatypus parallelus. Insects 2025, 16, 504. https://doi.org/10.3390/insects16050504
Wu Q, Zhou X, Xu Z, Zhang X, Yuan H, Guo J. Transcriptome Analysis and Identification of Chemosensory Membrane Proteins in the Head of Euplatypus parallelus. Insects. 2025; 16(5):504. https://doi.org/10.3390/insects16050504
Chicago/Turabian StyleWu, Qi, Xiang Zhou, Zheyuan Xu, Xufeng Zhang, Hongchao Yuan, and Jixing Guo. 2025. "Transcriptome Analysis and Identification of Chemosensory Membrane Proteins in the Head of Euplatypus parallelus" Insects 16, no. 5: 504. https://doi.org/10.3390/insects16050504
APA StyleWu, Q., Zhou, X., Xu, Z., Zhang, X., Yuan, H., & Guo, J. (2025). Transcriptome Analysis and Identification of Chemosensory Membrane Proteins in the Head of Euplatypus parallelus. Insects, 16(5), 504. https://doi.org/10.3390/insects16050504







