Gut Bacteria Mediate Aggregation Pheromone Release in the Borer Beetle Trigonorhinus sp.
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Collection and Rearing
2.2. Gut Dissection and Sample Preparation
2.3. Antibiotic Treatment Protocol
2.4. Culture-Dependent Isolation and Identification of Gut Bacteria
2.5. 16S rRNA Gene Amplicon Sequencing for Bacterial Community Profiling
2.6. Shotgun Metagenomic Sequencing for Unbiased Community Characterization
2.7. Amplicon and Metagenome Data Processing
2.8. Pheromone Collection and GC-MS Analysis
2.9. Behavioral Bioassays (Y-Tube Olfactometer)
2.10. Statistical Analysis
3. Results
3.1. Gut Microbiota Composition of Trigonorhinus sp.
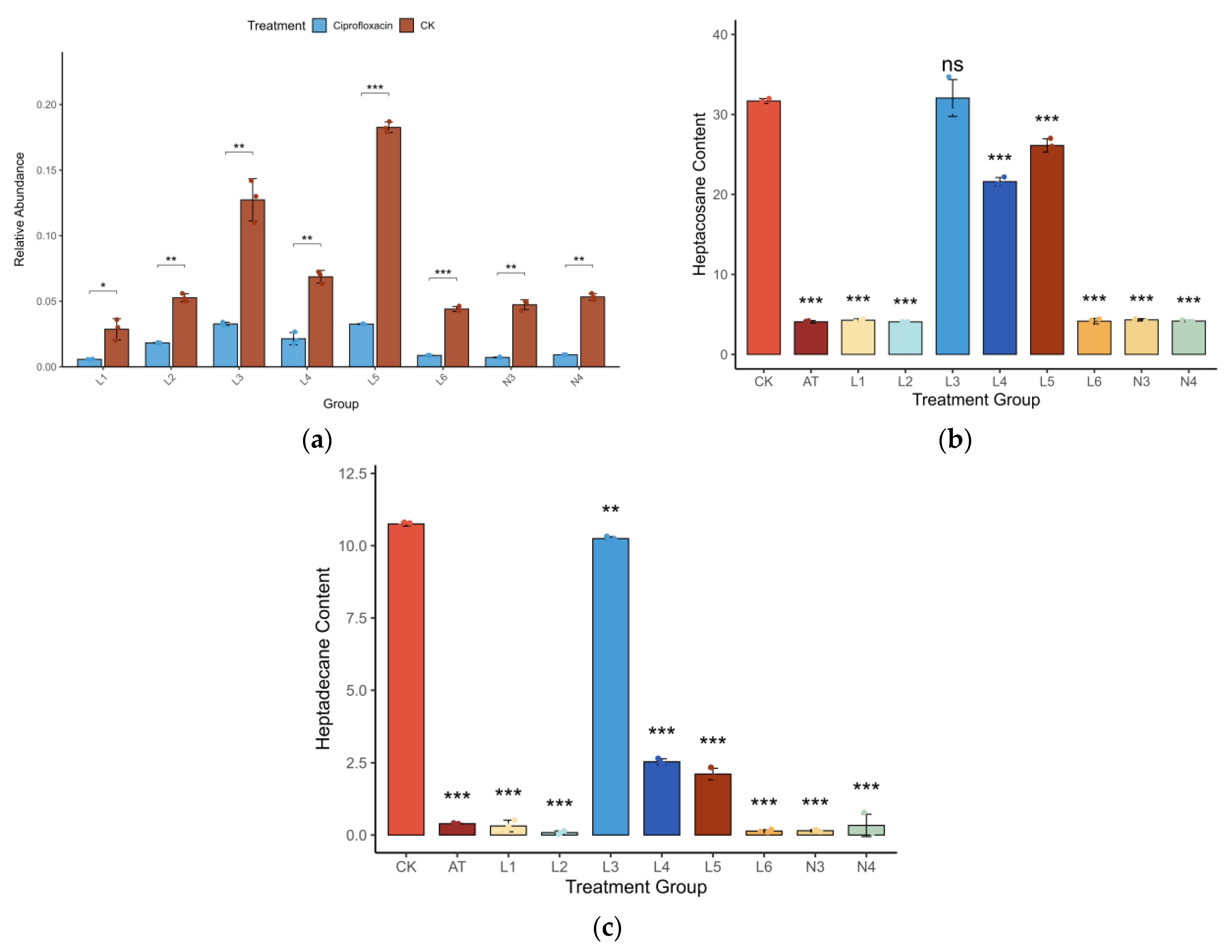
3.2. Gut Bacteria Are Essential for Pheromone Release
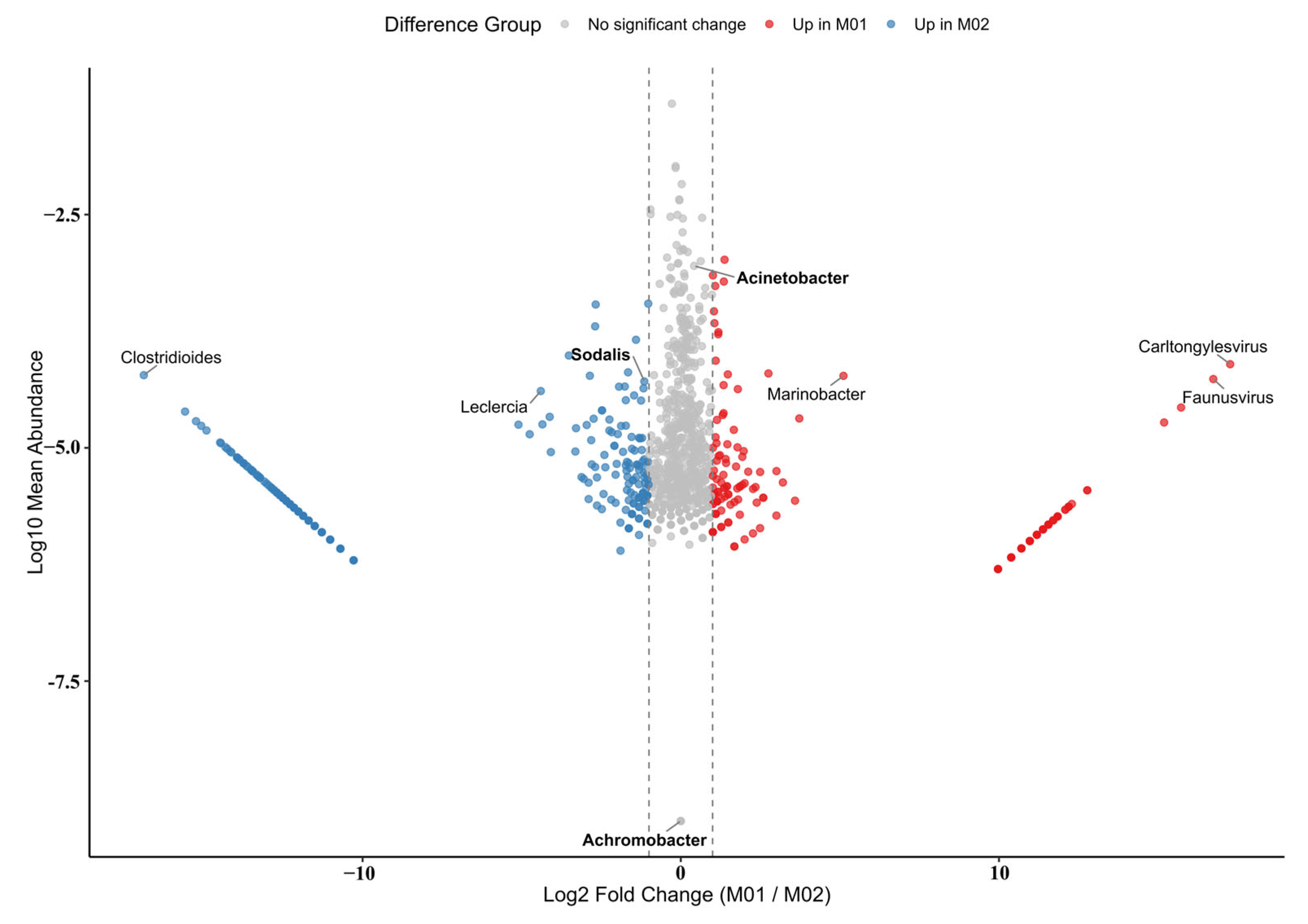
3.3. Characterization of the Gut Microbiome in Trigonorhinus sp.
3.4. Behavioral Consequences of Microbiome Depletion
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Haider, K.; Abbas, D.; Galian, J.; Ghafar, M.A.; Kabir, K.; Ijaz, M.; Hussain, M.; Khan, K.A.; Ghramh, H.A.; Raza, A. The multifaceted roles of gut microbiota in insect physiology, metabolism, and environmental adaptation: Implications for pest management strategies. World J. Microbiol. Biotechnol. 2025, 41, 75. [Google Scholar] [CrossRef]
- Jones, R.M.; Neish, A.S. Gut microbiota in intestinal and liver disease. Annu. Rev. Pathol. Mech. Dis. 2021, 16, 251–275. [Google Scholar] [CrossRef]
- Mondal, S.; Somani, J.; Roy, S.; Babu, A.; Pandey, A.K. Insect microbial symbionts: Ecology, interactions, and biological significance. Microorganisms 2023, 11, 2665. [Google Scholar] [CrossRef]
- Siddiqui, J.A.; Khan, M.M.; Bamisile, B.S.; Hafeez, M.; Qasim, M.; Rasheed, M.T.; Rasheed, M.A.; Ahmad, S.; Shahid, M.I.; Xu, Y. Role of insect gut microbiota in pesticide degradation: A review. Front. Microbiol. 2022, 13, 870462. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, Y.; Jiang, H.; Zhao, S.; Lei, J.; Wang, R.; Chen, Y. Pest characteristics of Trigonorhinus sp. (Coleoptera: Anthribidae) and the developmental process of its galls in Caragana liouana Zhao, Y. Chang and Yakovlev. Can. Entomol. 2023, 155, e14. [Google Scholar] [CrossRef]
- Lei, J.W.; Zhao, S.G.; Li, B.L.; Zhao, P.W.; Zhang, L.Y.; Liu, X.Y.; Hu, Y.R.; Zhang, Y.R. Morphological criteria for sexing adult Trigonorhinus sp. Chin. J. Appl. Entomol. 2023, 60. [Google Scholar]
- Song, X.; Liu, G.; Mo, Y. Trigonorhinus sp.: A new pest in Caragana korshinskii. J. Northwest For. Univ. 2010, 25, 130–131. [Google Scholar]
- Castano-Sanz, V.; Gomez-Mestre, I.; Rodriguez-Exposito, E.; Garcia-Gonzalez, F. Pesticide exposure triggers sex-specific inter-and transgenerational effects conditioned by past sexual selection. Proc. R. Soc. B 2024, 291, 20241037. [Google Scholar] [CrossRef]
- Calixto, E.; IasmimPereira, D. Evidence of climate change effects on insect diversity. Eff. Clim. Change Insects Physiol. Evol. Ecol. Responses 2024, 179. [Google Scholar]
- Abbott, K.C.; Heggerud, C.M.; Lai, Y.-C.; Morozov, A.; Petrovskii, S.; Cuddington, K.; Hastings, A. When and why ecological systems respond to the rate rather than the magnitude of environmental changes. Biol. Conserv. 2024, 292, 110494. [Google Scholar] [CrossRef]
- Engl, T.; Kaltenpoth, M. Influence of microbial symbionts on insect pheromones. Nat. Prod. Rep. 2018, 35, 386–397. [Google Scholar] [CrossRef]
- Delclos, P.J.; Bouldin, T.L.; Tomberlin, J.K. Olfactory choice for decomposition stage in the burying beetle Nicrophorus vespilloides: Preference or aversion? Insects 2020, 12, 11. [Google Scholar] [CrossRef]
- Wada-Katsumata, A.; Zurek, L.; Nalyanya, G.; Roelofs, W.L.; Zhang, A.; Schal, C. Gut bacteria mediate aggregation in the German cockroach. Proc. Natl. Acad. Sci. USA 2015, 112, 15678–15683. [Google Scholar] [CrossRef]
- Wyatt, T.D. Pheromones. Curr. Biol. 2017, 27, R739–R743. [Google Scholar] [CrossRef]
- Ali, A.; Zeb, I.; Zahid, H. Climate Change and Plants: Biodiversity, Growth and Interactions; CRC Press: Boca Raton, FL, USA, 2021; pp. 97–111. [Google Scholar]
- Lavy, O.; Gophna, U.; Gefen, E.; Ayali, A. Locust bacterial symbionts: An update. Insects 2020, 11, 655. [Google Scholar] [CrossRef]
- Dillon, R.; Vennard, C.; Charnley, A. A note: Gut bacteria produce components of a locust cohesion pheromone. J. Appl. Microbiol. 2002, 92, 759–763. [Google Scholar] [CrossRef]
- Raffa, K.F. Terpenes tell different tales at different scales: Glimpses into the chemical ecology of conifer-bark beetle-microbial interactions. J. Chem. Ecol. 2014, 40, 1–20. [Google Scholar] [CrossRef]
- Zhao, T.; Ganji, S.; Schiebe, C.; Bohman, B.; Weinstein, P.; Krokene, P.; Borg-Karlson, A.-K.; Unelius, C.R. Convergent evolution of semiochemicals across Kingdoms: Bark beetles and their fungal symbionts. ISME J. 2019, 13, 1535–1545. [Google Scholar] [CrossRef]
- Douglas, A.E. Multiorganismal insects: Diversity and function of resident microorganisms. Annu. Rev. Entomol. 2015, 60, 17–34. [Google Scholar] [CrossRef]
- Frago, E.; Dicke, M.; Godfray, H.C.J. Insect symbionts as hidden players in insect–plant interactions. Trends Ecol. Evol. 2012, 27, 705–711. [Google Scholar] [CrossRef]
- Busby, P.E.; Soman, C.; Wagner, M.R.; Friesen, M.L.; Kremer, J.; Bennett, A.; Morsy, M.; Eisen, J.A.; Leach, J.E.; Dangl, J.L. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 2017, 15, e2001793. [Google Scholar] [CrossRef]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; Mcveigh, R.; O’Neill, K.; Robbertse, B. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef]
- Biomedicals, M. FastDNA™ SPIN Kit for Soil. Instr. Man. 2021, 1–11. Available online: https://www.mpbio.com/media/document/file/manual/dest/f/a/s/t/d/FastDNA_SPIN_Kit_for_Soil_UM_2021_WEB.pdf (accessed on 13 May 2025).
- Imchen, M.; Kumavath, R.; Vaz, A.B.; Góes-Neto, A.; Barh, D.; Ghosh, P.; Kozyrovska, N.; Podolich, O.; Azevedo, V. 16S rRNA gene amplicon based metagenomic signatures of rhizobiome community in rice field during various growth stages. Front. Microbiol. 2019, 10, 2103. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- R. Core Team, R. R: A Language and Environment for Statistical Computing. 2020. Available online: https://www.R-project.org/ (accessed on 13 May 2025).
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Schloss, P.D.; Delalibera Jr, I.; Handelsman, J.; Raffa, K.F. Bacteria associated with the guts of two wood-boring beetles: Anoplophora glabripennis and Saperda vestita (Cerambycidae). Environ. Entomol. 2006, 35, 625–629. [Google Scholar] [CrossRef]
- Montagna, M.; Chouaia, B.; Mazza, G.; Prosdocimi, E.M.; Crotti, E.; Mereghetti, V.; Vacchini, V.; Giorgi, A.; De Biase, A.; Longo, S. Effects of the diet on the microbiota of the red palm weevil (Coleoptera: Dryophthoridae). PLoS ONE 2015, 10, e0117439. [Google Scholar] [CrossRef]
- Lenka, J.; González-Tortuero, E.; Kuba, S.; Ferry, N. Bacterial community profiling and identification of bacteria with lignin-degrading potential in different gut segments of African palm weevil larvae (Rhynchophorus phoenicis). Front. Microbiol. 2025, 15, 1401965. [Google Scholar] [CrossRef] [PubMed]
- Jing, T.-Z.; Qi, F.-H.; Wang, Z.-Y. Most dominant roles of insect gut bacteria: Digestion, detoxification, or essential nutrient provision? Microbiome 2020, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Edet, U.; Antai, S.; Brooks, A.; Asitok, A.; Enya, O.; Japhet, F. An overview of cultural, molecular and metagenomic techniques in description of microbial diversity. J. Adv. Microbiol. 2017, 7, 1–19. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Bahram, M.; Sanchez-Castro, I.; Dai, D.-Q.; Ariyawansa, K.G.; Jayalal, U.; Suwannarach, N.; Tedersoo, L. Current insight into culture-dependent and culture-independent methods in discovering Ascomycetous Taxa. J. Fungi 2021, 7, 703. [Google Scholar] [CrossRef]
- Forbes, J.D.; Knox, N.C.; Ronholm, J.; Pagotto, F.; Reimer, A. Metagenomics: The next culture-independent game changer. Front. Microbiol. 2017, 8, 1069. [Google Scholar] [CrossRef]
- Garza, D.R.; Dutilh, B.E. From cultured to uncultured genome sequences: Metagenomics and modeling microbial ecosystems. Cell. Mol. Life Sci. 2015, 72, 4287–4308. [Google Scholar] [CrossRef]
- Liu, C.; Song, X.; Liu, J.; Zong, L.; Xu, T.; Han, X.; Li, F.; Li, B.; Zhu, H.; Shi, D. Consistency between metagenomic next-generation sequencing versus traditional microbiological tests for infective disease: Systemic review and meta-analysis. Crit. Care 2025, 29, 55. [Google Scholar] [CrossRef]
- Doughty, E.L.; Sergeant, M.J.; Adetifa, I.; Antonio, M.; Pallen, M.J. Culture-independent detection and characterisation of Mycobacterium tuberculosis and M. africanum in sputum samples using shotgun metagenomics on a benchtop sequencer. PeerJ 2014, 2, e585. [Google Scholar] [CrossRef]
- Singha, L.P.; Kumari, R.; Singha, K.M.; Pandey, P.; Shukla, P. Synergistic co-metabolism enhancing the crude oil degradation by Acinetobacter oleivorans DR1 and its metabolic potential. Microbiol. Spectr. 2025, 13, e0302324. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Q.; Liu, L. Crude oil degrading fingerprint and the overexpression of oxidase and invasive genes for n-hexadecane and crude oil degradation in the Acinetobacter pittii H9-3 strain. Int. J. Environ. Res. Public Health 2019, 16, 188. [Google Scholar] [CrossRef]
- Throne-Holst, M.; Wentzel, A.; Ellingsen, T.E.; Kotlar, H.-K.; Zotchev, S.B. Identification of novel genes involved in long-chain n-alkane degradation by Acinetobacter sp. strain DSM 17874. Appl. Environ. Microbiol. 2007, 73, 3327–3332. [Google Scholar] [CrossRef]
- Salcedo-Vite, K.; Sigala, J.-C.; Segura, D.; Gosset, G.; Martinez, A. Acinetobacter baylyi ADP1 growth performance and lipid accumulation on different carbon sources. Appl. Microbiol. Biotechnol. 2019, 103, 6217–6229. [Google Scholar] [CrossRef]
- Zhao, Y.; Wei, H.-M.; Yuan, J.-L.; Xu, L.; Sun, J.-Q. A comprehensive genomic analysis provides insights on the high environmental adaptability of Acinetobacter strains. Front. Microbiol. 2023, 14, 1177951. [Google Scholar] [CrossRef]
- Jung, J.; Park, W. Acinetobacter species as model microorganisms in environmental microbiology: Current state and perspectives. Appl. Microbiol. Biotechnol. 2015, 99, 2533–2548. [Google Scholar] [CrossRef]
- Lu, M.; Hulcr, J.; Sun, J. The role of symbiotic microbes in insect invasions. Annu. Rev. Ecol. Evol. Syst. 2016, 47, 487–505. [Google Scholar] [CrossRef]
- Lv, C.; Huang, Y.-Z.; Luan, J.-B. Insect–microbe symbiosis-based strategies offer a new avenue for the management of insect pests and their transmitted pathogens. Crop Health 2024, 2, 18. [Google Scholar] [CrossRef]
- Gonella, E.; Alma, A. The role of symbiont-targeted strategies in the management of Pentatomidae and Tephritidae pests under an integrated vision. Agronomy 2023, 13, 868. [Google Scholar] [CrossRef]
- Gonella, E.; Orrù, B.; Marasco, R.; Daffonchio, D.; Alma, A. Disruption of host-symbiont associations for the symbiotic control and management of pentatomid agricultural pests—A review. Frontiers in Microbiology 2020, 11, 547031. [Google Scholar] [CrossRef]
- Wilke, A.B.B.; Marrelli, M.T. Paratransgenesis: A promising new strategy for mosquito vector control. Parasites Vectors 2015, 8, 342. [Google Scholar] [CrossRef]
- Ratcliffe, N.A.; Furtado Pacheco, J.P.; Dyson, P.; Castro, H.C.; Gonzalez, M.S.; Azambuja, P.; Mello, C.B. Overview of paratransgenesis as a strategy to control pathogen transmission by insect vectors. Parasites Vectors 2022, 15, 112. [Google Scholar] [CrossRef] [PubMed]
- Miller, T. Paratransgenesis as a potential tool for pest control: Review of applied arthropod symbiosis. J. Appl. Entomol. 2011, 135, 474–478. [Google Scholar] [CrossRef]

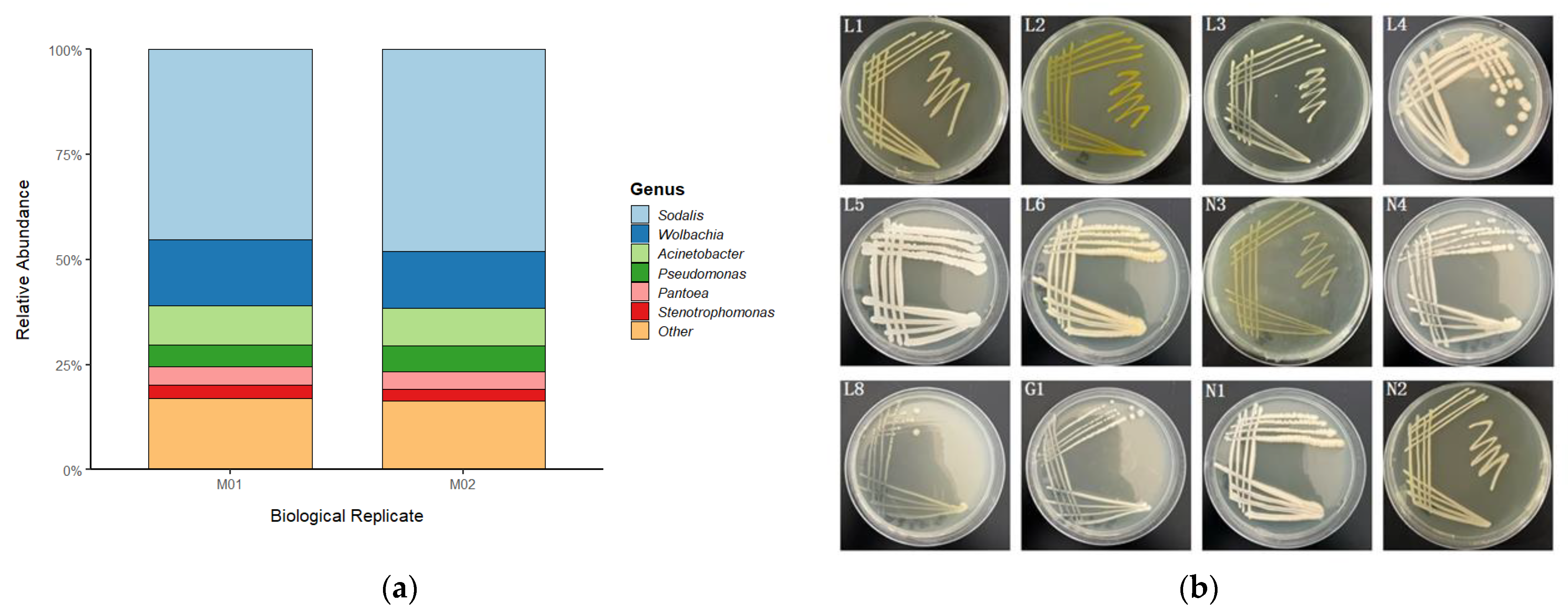
| Strain No. | Most Closely Related Strain | Similarity (%) | Accession Number |
|---|---|---|---|
| L1 | Stenotrophomonas lactitubi | 99.79 | NR179509 |
| L2 | Pantoea plantarum | 99.79 | NR104943 |
| L3 | Acinetobacter guillouiae | 99.50 | NR117626 |
| L4 | Mammaliicoccus sciuri | 100 | NR025520 |
| L5 | Pseudomonas allii | 100 | NR179337 |
| L6 | Comamonas sediminis | 99.14 | NR149789 |
| L8 | Mammaliicoccus sciuri | 100 | NR025520 |
| N1 | Mammaliicoccus sciuri | 100 | NR025520 |
| N2 | Acinetobacter guillouiae | 99.5 | NR117626 |
| N3 | Pantoea endophytica | 99.71 | NR178843 |
| N4 | Brucella pseudogrignonensis | 100 | NR042589 |
| G1 | Acinetobacter guillouiae | 99.5 | NR117626 |
| Treatment Group | Responding Insect | Chose Odor Source | Chose Control Source | No Response | Response Rate (%) | Chi-Square (χ2) | p-Value |
|---|---|---|---|---|---|---|---|
| CG | Female | 63 | 14 | 13 | 81.82 | 31.183 | <0.001 |
| Male | 66 | 15 | 9 | 81.48 | 32.111 | <0.001 | |
| AT | Female | 30 | 32 | 28 | 48.39 | 0.065 | 0.799 |
| Male | 32 | 30 | 28 | 51.61 | 0.065 | 0.799 | |
| L1 | Female | 26 | 28 | 36 | 48.15 | 0.074 | 0.785 |
| Male | 27 | 30 | 33 | 47.37 | 0.158 | 0.691 | |
| L2 | Female | 24 | 22 | 44 | 52.17 | 0.087 | 0.768 |
| Male | 23 | 22 | 45 | 51.11 | 0.022 | 0.881 | |
| L3 | Female | 64 | 16 | 10 | 80 | 28.8 | <0.001 |
| Male | 66 | 15 | 9 | 81.48 | 32.11 | <0.001 | |
| L4 | Female | 48 | 20 | 22 | 70.59 | 11.529 | 0.001 |
| Male | 49 | 16 | 25 | 75.38 | 16.754 | <0.001 | |
| L5 | Female | 58 | 18 | 14 | 76.32 | 21.053 | <0.001 |
| Male | 61 | 20 | 9 | 75.31 | 20.753 | <0.001 | |
| L6 | Female | 29 | 26 | 35 | 52.73 | 0.164 | 0.686 |
| Male | 30 | 28 | 32 | 51.72 | 0.069 | 0.793 | |
| N3 | Female | 31 | 30 | 29 | 50.82 | 0.016 | 0.898 |
| Male | 32 | 30 | 28 | 51.61 | 0.065 | 0.799 | |
| N4 | Female | 27 | 25 | 38 | 51.92 | 0.077 | 0.782 |
| Male | 28 | 25 | 37 | 52.83 | 0.17 | 0.68 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, J.; Yao, X.; Zhang, Y.; Wu, X.; Liu, X.; Zhang, H.; Jiang, H.; Hou, J.; Yan, J.; Sun, J. Gut Bacteria Mediate Aggregation Pheromone Release in the Borer Beetle Trigonorhinus sp. Insects 2025, 16, 999. https://doi.org/10.3390/insects16100999
Dong J, Yao X, Zhang Y, Wu X, Liu X, Zhang H, Jiang H, Hou J, Yan J, Sun J. Gut Bacteria Mediate Aggregation Pheromone Release in the Borer Beetle Trigonorhinus sp. Insects. 2025; 16(10):999. https://doi.org/10.3390/insects16100999
Chicago/Turabian StyleDong, Jinyang, Xiang Yao, Yanru Zhang, Xiuhua Wu, Xinhai Liu, Hongbin Zhang, Haiyan Jiang, Jianli Hou, Jie Yan, and Jianing Sun. 2025. "Gut Bacteria Mediate Aggregation Pheromone Release in the Borer Beetle Trigonorhinus sp." Insects 16, no. 10: 999. https://doi.org/10.3390/insects16100999
APA StyleDong, J., Yao, X., Zhang, Y., Wu, X., Liu, X., Zhang, H., Jiang, H., Hou, J., Yan, J., & Sun, J. (2025). Gut Bacteria Mediate Aggregation Pheromone Release in the Borer Beetle Trigonorhinus sp. Insects, 16(10), 999. https://doi.org/10.3390/insects16100999






