Characterization of Four Complete Mitogenomes of Monolepta Species and Their Related Phylogenetic Implications
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Taxon Sampling and DNA Extraction
2.2. Sequencing and Assembly
2.3. Phylogenetic Analyses
3. Results
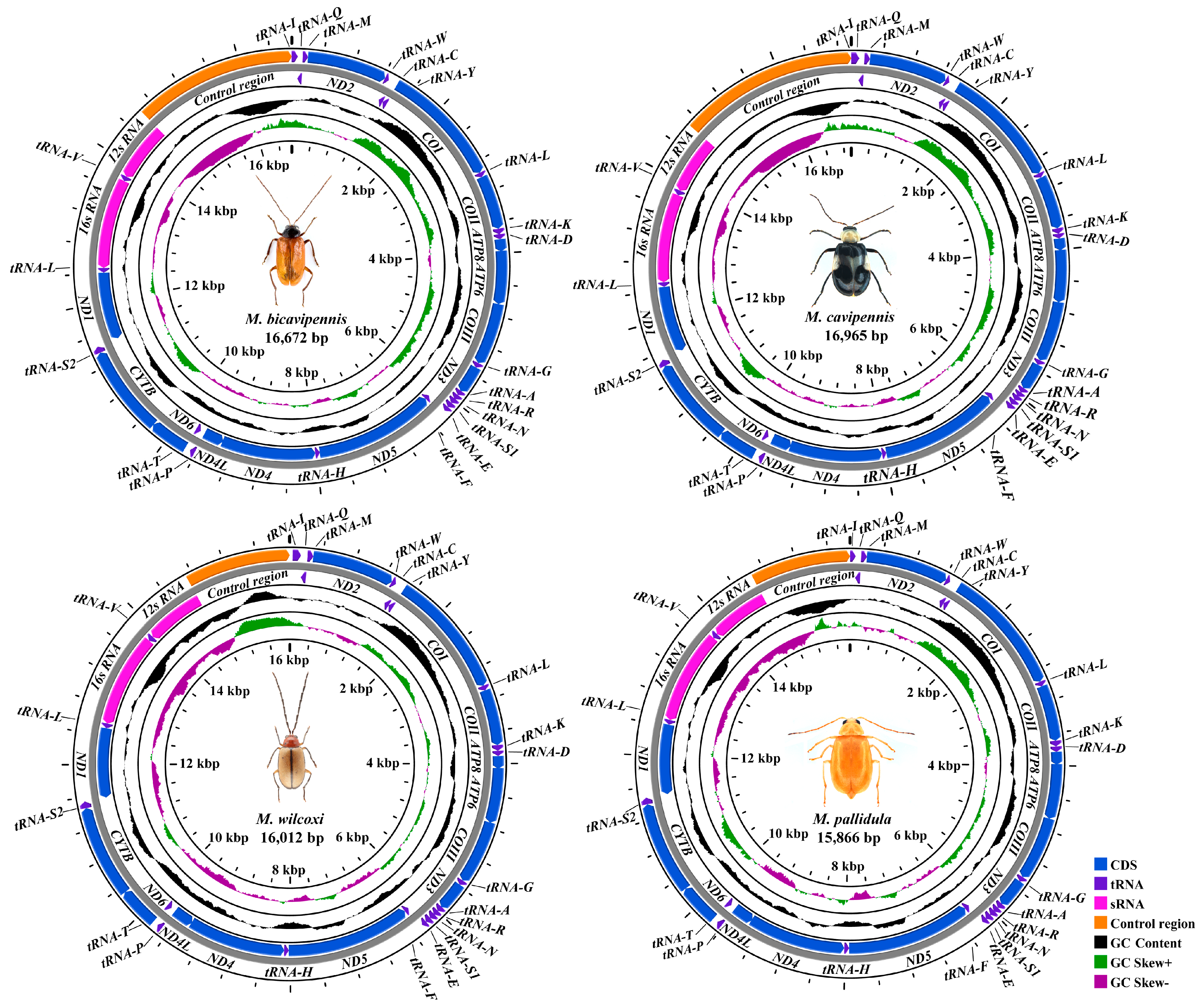
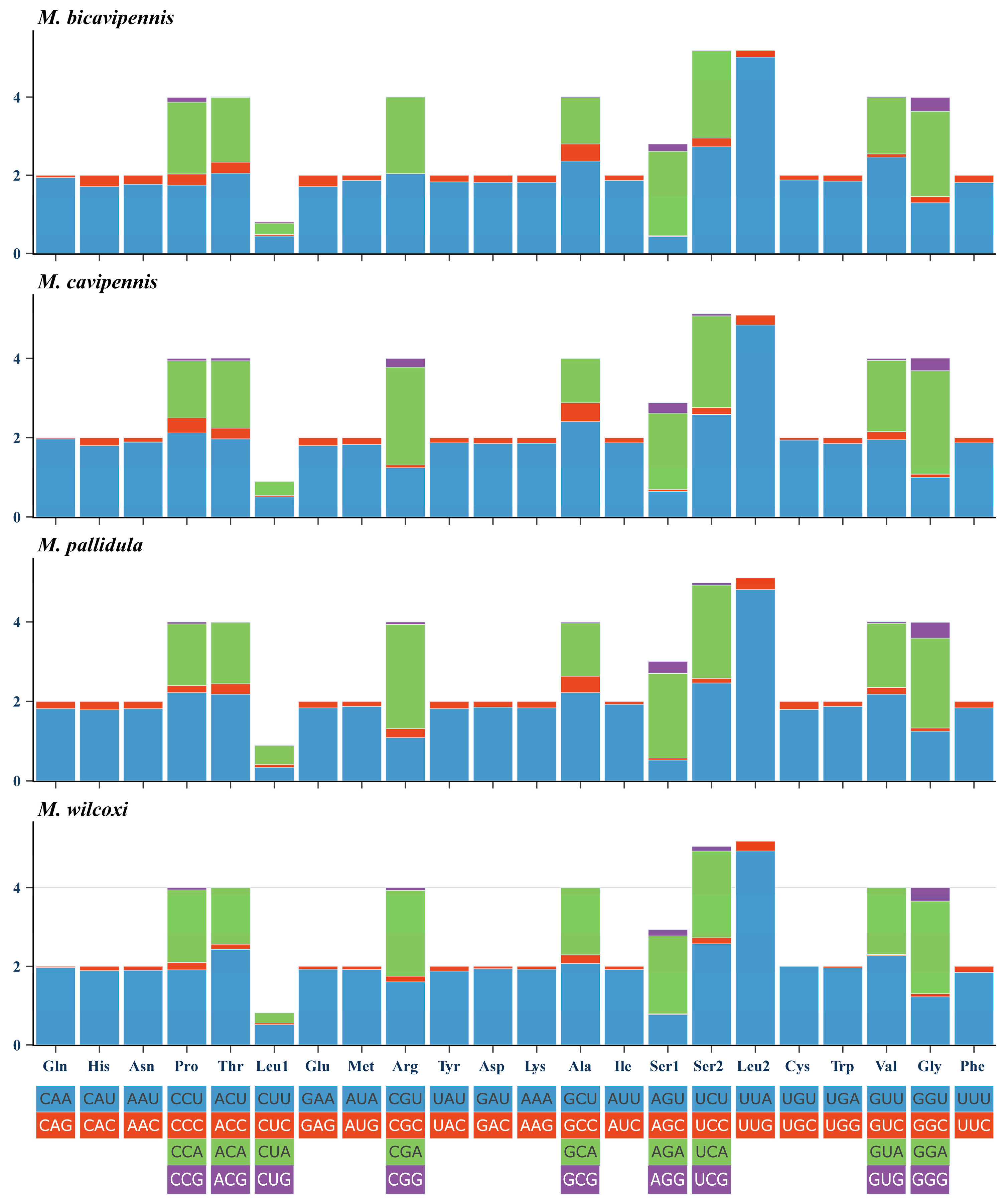
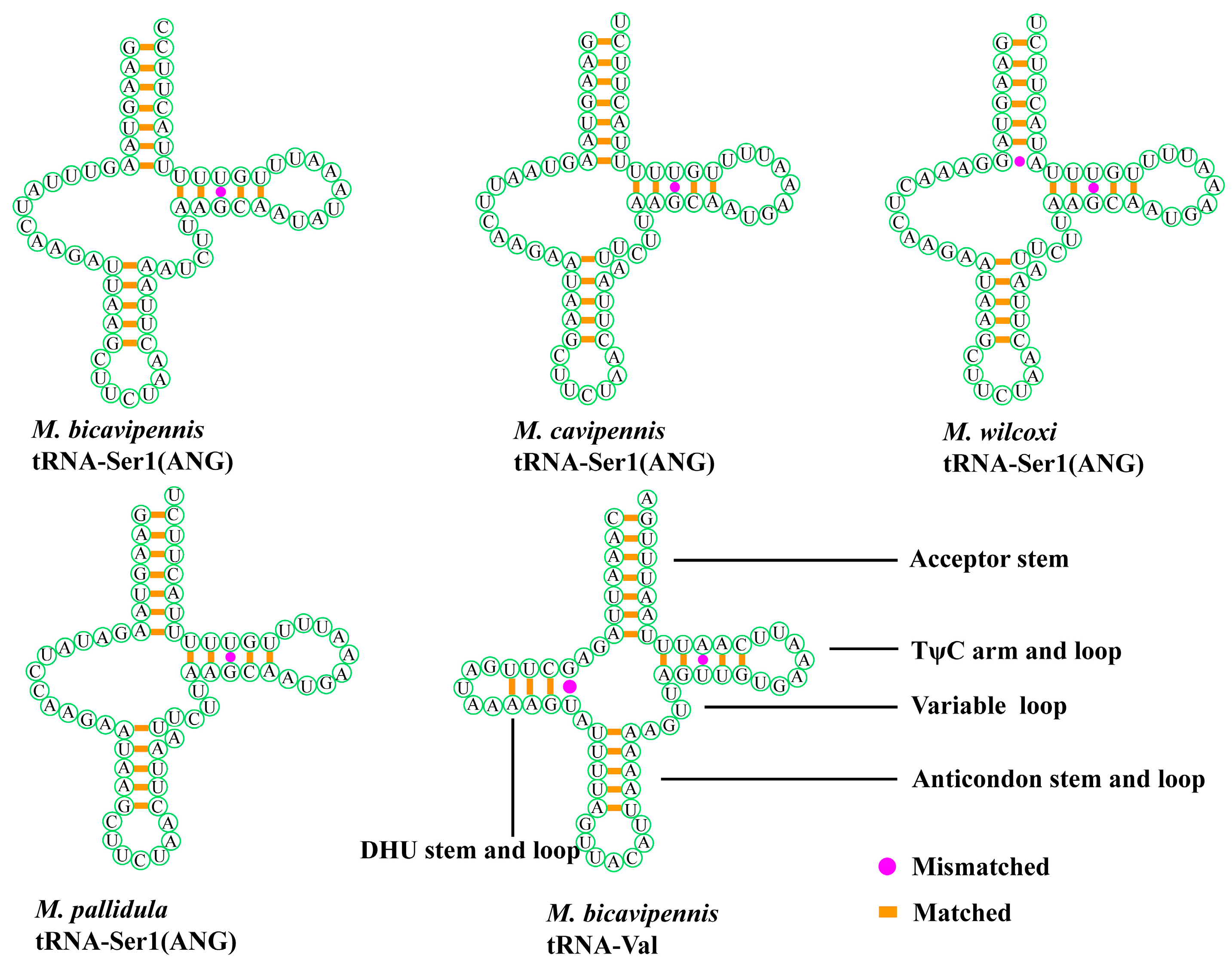
3.1. Sequence Data, Mitogenomic Organization, and Composition in Monolepta
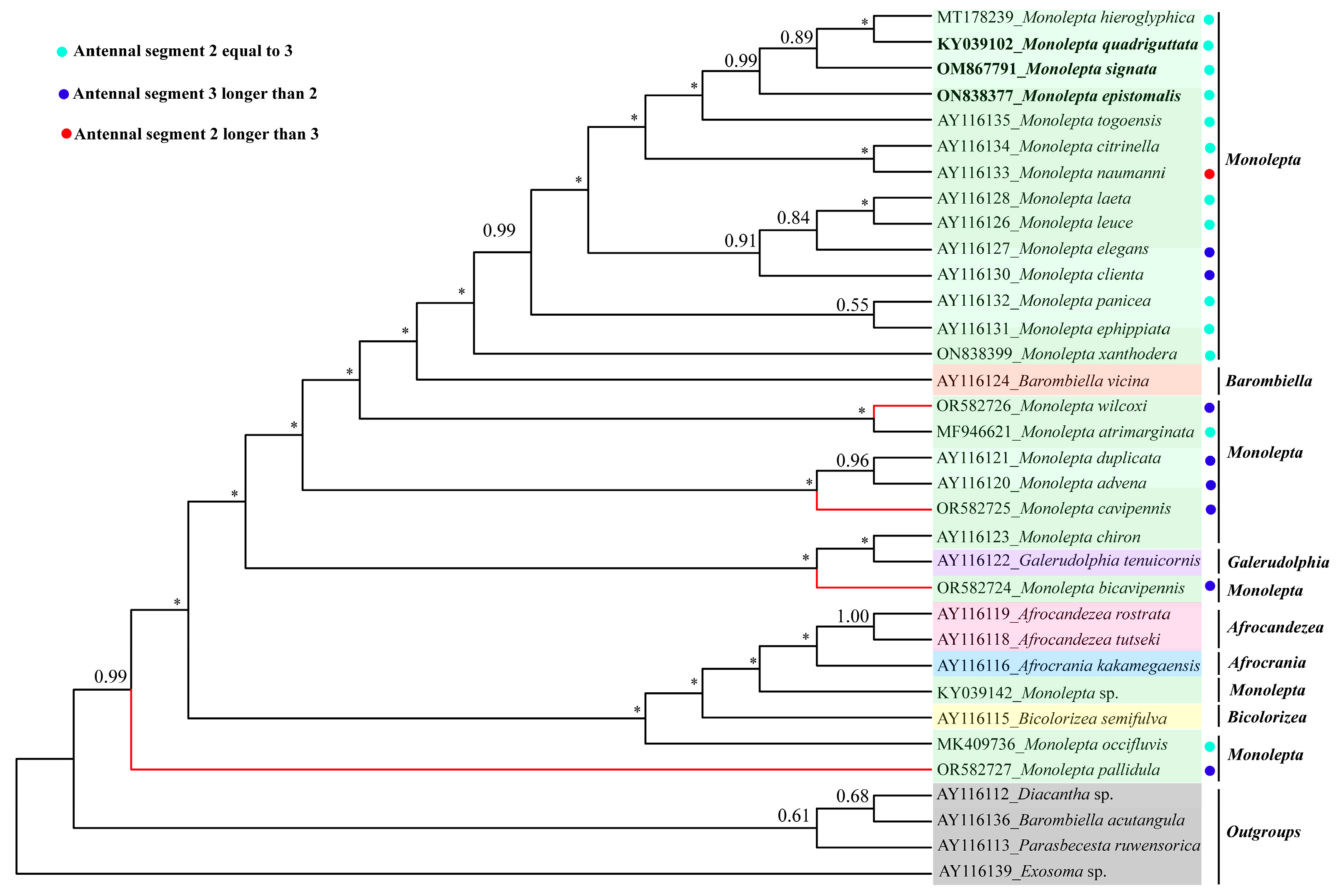
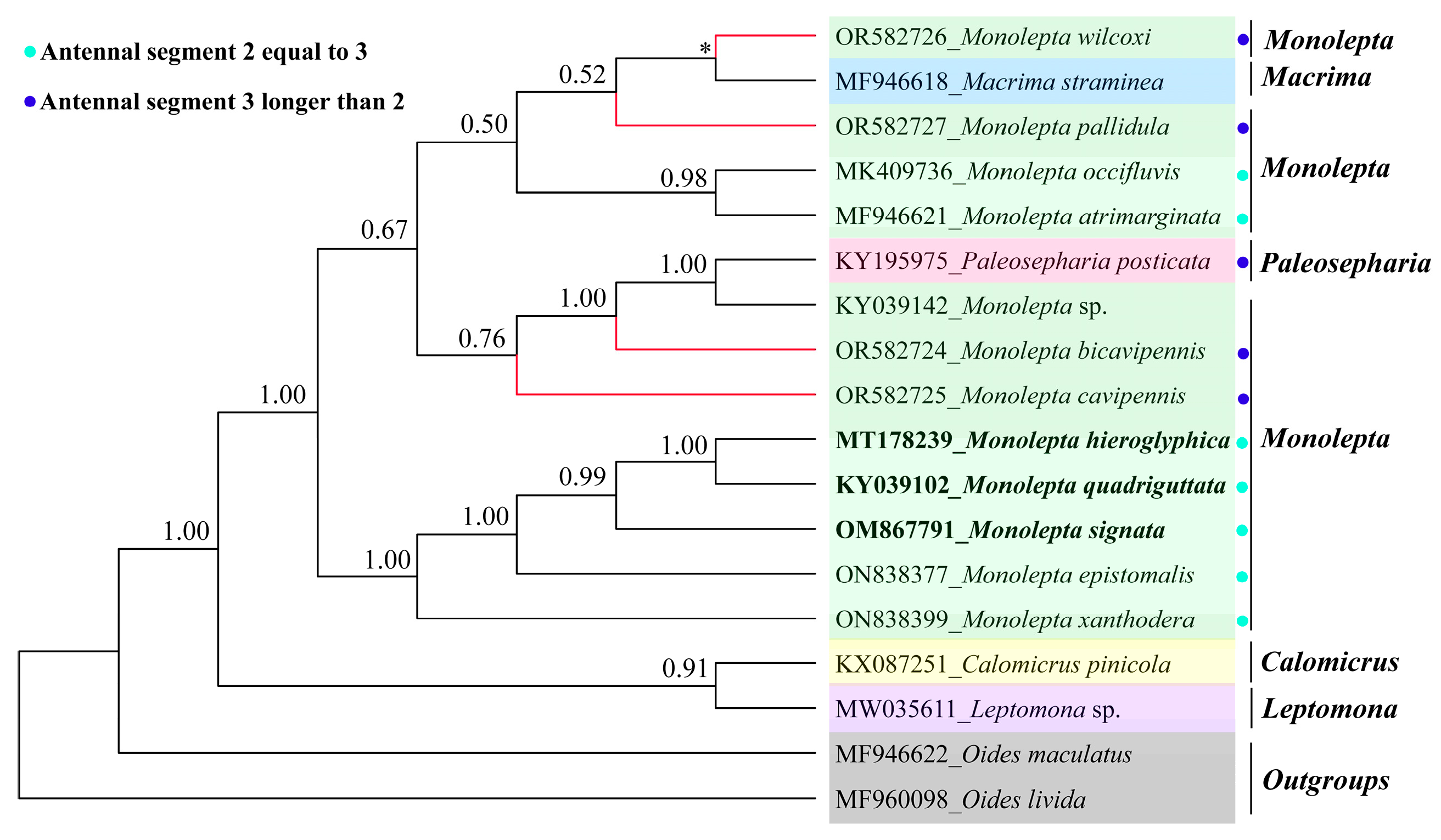
3.2. The Phylogeny of Monolepta
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chevrolat, L.A.A. Monolepta. In Catalogues Des Coléopterès De La Collection De M. LeComte Dejean, Troisième éd.; revue, corrigée et augmentée, livraison, Dejean, P.F.M.A., Ed.; Méquignon-Marvis Père et Fils: Paris, France, 1837; Volume 5, pp. 385–503. [Google Scholar]
- Nie, R.E.; Bezděk, J.; Yang, X.K. How many genera and species of Galerucinaes. str. do we know? Updated statistics (Coleoptera, Chrysomelidae). Zookeys 2017, 720, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.K. Genus Monolepta Chevrolat, 1837. In Chinese Leaf Beetles; Yang, X.K., Ge, S.Q., Nie, R.E., Ruan, Y.Y., Li, W.Z., Eds.; Science Press: Beijing, China, 2015; pp. 261–274. [Google Scholar]
- Zhang, C.; Yuan, Z.H.; Wang, Z.Y.; He, K.L.; Bai, S.X. Population dynamics of Monolepta hieroglyphica (Motschulsky) in cornfields. J. Appl. Entomol. 2014, 51, 668–675. [Google Scholar]
- Chapuis, F. Famille des Phytophages. In Histoire Naturelle Des Insects; Lacordaire, T., Chapuis, F., Eds.; Genera des Coleopteres: Paris, France, 1875; Volume 11, pp. 1–420. [Google Scholar]
- Wilcox, J.A. Chrysomelidae: Galerucinae: Luperini: Luperina. In Coleopterorum Catalogus Supplementa. Pars 78 (3); Wilcox, J., Ed.; BIBLIO: Gravenhage, The Netherlands, 1973; pp. 433–664. [Google Scholar]
- Bolz, H.; Wagner, T. Neobarombiella, a diverse, newly described genus of Afrotropical Galerucinae (Coleoptera, Chrysomelidae). Zootaxa 2012, 3463, 1–112. [Google Scholar] [CrossRef]
- Stapel, H.; Misof, B.; Wagner, T. A molecular and morphological phylogenetic analysis of Afrotropical Monolepta species and related Galerucinae (Coleoptera: Chrysomelidae). Arthropod Syst. Phylogeny 2008, 66, 3–17. [Google Scholar] [CrossRef]
- Wagner, T. Monolepta Chevrolat, 1837, the most speciose galerucine taxon: Redescription of the type species Monolepta bioculata (Fabricius, 1781) and key to related genera from Africa (Chrysomelidae, Coleoptera). J. Nat. Hist. 2007, 41, 81–100. [Google Scholar] [CrossRef]
- Wagner, T. Revision of afrotropical Monolepta species (Coleoptera, Chrysomelidae, Galerucinae). Part I: Species with red and black coloured elytra, pronotum and head, with description of new species. Entomol. Z. 2000, 110, 226–237. [Google Scholar]
- Wagner, T. Revision of Afrotropical Monolepta Chevrolat, 1837 (Coleoptera, Chrysomelidae, Galerucinae). Part II: Species with red elytra, pronotum and elytra, with descriptions of new species. Bonn. Zool. Beitr. 2001, 50, 49–65. [Google Scholar]
- Wagner, T. Revision of Afrotropical Monolepta species (Coleoptera, Chrysomelidae, Galerucinae). Part III: Species with red elytra and yellow prothorax, including description of new species. Dtsch. Entomol. Z. 2002, 49, 27–45. [Google Scholar] [CrossRef]
- Steiner, I.; Wagner, T. Afronaumannia gen. nov., a new monophyletic group of leaf beetles from Africa (Coleoptera: Chrysomelidae, Galerucinae). Entomol. Z. 2005, 115, 15–24. [Google Scholar]
- Wagner, T. Description of Monoleptoides gen.nov. from the Afrotropical Region, including the revision of nine species (Coleoptera: Chrysomelidae: Galerucinae). Bonn Zool. Bull. 2011, 60, 169–199. [Google Scholar]
- Wagner, T.; Bieneck, S. Galerucine type material described by Victor Motschu in 1858 and 1866 from the Zoological Museum Moscow (Coleoptera: Chrysomelidae, Galerucinae). Entomol. Z. 2012, 122, 205–216. [Google Scholar]
- Hazmi, I.R.; Wagner, T. Revision of Neolepta Jacoby, 1884 and related Galerucines from the Oriental region, including descriptions of two new genera (Coleoptera: Chrysomelidae: Galerucinae). Raffles Bull. Zool. 2013, 61, 73–95. [Google Scholar]
- Heunemann, L.O.; Dalstein, V.; Schulze, M.; Wagner, T. Bicolorizea gen. nov. from tropical Africa (Coleoptera: Chrysomelidae, Galerucinae). Mitt. Mus. Naturkunde Berl. Dtsch. Entomol. Z. 2015, 125, 235–246. [Google Scholar]
- Wagner, T. Doeberllepta gen. nov., a new monotypic genus for a widely distributed galerucine species from Africa (Coleoptera: Chrysomelidae). Entomol. Blätter 2017, 113, 245–253. [Google Scholar]
- Laboissière, V. Observations sur les Galerucini asiatiques principalement du Tonkin et du Yunnan et descriptions de nouveaux genres et espèces (5e partie). Ann. Soc. Entomol. Fr. 1936, 105, 239–261. [Google Scholar] [CrossRef]
- Chûjô, M.H. Sauter’s. Formosa-Ausbeute: Subfamily Galerucinae (Coleoptera: Chrysomelidae). Arb. Morph. Taxon. Ent. Berlin-Dahlem 1935, 2, 160–174. [Google Scholar]
- Chûjô, M. A taxonomic study on the Chrysomelidae (Insecta: Coleoptera) from Formosa XI. Subfamily Galerucinae. Philipp. J. Sci. 1962, 91, 1–239. [Google Scholar]
- Kimoto, S. Notes on the Chryosmelidae from Taiwan, China. II. Esakia 1969, 7, 1–68. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, S. Notes on the Chrysomelidae from Taiwan, China, XIII. Entomol. Rev. Jpn. 1996, 51, 27–51. [Google Scholar]
- Gressitt, J.L.; Kimoto, S. The Chrysomelidae (Coleopt.) of China and Korea. Part II. Pac. Insects Monogr. 1963, 1B, 301–1026. [Google Scholar]
- Lee, C.F. Revision of Taiwanese species of Atrachya Chevrolat, 1836 (Coleoptera, Chrysomelidae, Galerucinae): Descriptions of three new genera, two new species, and designations of three new synonyms. Zookeys 2020, 940, 117–159. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.F. A taxonomic revision of Monolepta pallidula species group in Taiwan (Coleoptera: Chrysomelidae: Galerucinae). Zootaxa 2009, 2170, 15–27. [Google Scholar] [CrossRef]
- Lei, Q.L.; Xu, S.Y.; Yang, X.K.; Nie, R.E. Five new species of the leaf-beetle genus Monolepta Chevrolat (Coleoptera, Chrysomelidae, Galerucinae) from China. Zookeys 2021, 1056, 35–57. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.C.; Shi, C.M.; Bai, M.; Cao, Z.Y.; Cao, L.P.; Wang, Z.Y.; Dong, J.A.; Wang, Y.Y. Molecular data confirm Monolepta hieroglyphica (Motschulsky, 1858) and M. quadriguttata (Motschulsky, 1860) being synonyms of M. signata (Oliver, 1808). Insect Syst. Evol. 2023, 54, 402–412. [Google Scholar] [CrossRef]
- Bolz, H.; Wagner, T. Revision of Galerudolphia Hincks, 1949 (Coleoptera: Chrysomelidae, Galerucinae). Insect Syst. Evol. 2004, 35, 361–400. [Google Scholar] [CrossRef]
- Nie, R.E.; Breeschoten, T.; Timmermans, M.J.T.N.; Nadein, K.; Xue, H.J.; Bai, M.; Huang, Y.; Yang, X.K.; Vogler, A.P. The phylogeny of Galerucinae (Coleoptera: Chrysomelidae) and the performance of mitochondrial genomes in phylogenetic inference compared to nuclear rRNA genes. Cladistics 2018, 34, 113–130. [Google Scholar] [CrossRef] [PubMed]
- Sheffield, N.C.; Song, H.; Cameron, S.L.; Whiting, M.F. A comparative analysis of mitochondrial genomes in Coleoptera (Arthropoda: Insecta) and genome descriptions of six new beetles. Mol. Biol. Evol. 2008, 25, 2499–2509. [Google Scholar] [CrossRef]
- Sheffield, N.C.; Song, H.; Cameron, S.L.; Whiting, M.F. Nonstationary evolution and compositional heterogeneity in beetle mitochondrial phylogenomics. Syst. Biol. 2009, 58, 381–394. [Google Scholar] [CrossRef]
- Pons, J.; Ribera, I.; Bertranpetit, J.; Balke, M. Nucleotide substitution rates for the full set of mitochondrial protein-coding genes in Coleoptera. Mol. Phylogenet. Evol. 2010, 56, 796–807. [Google Scholar] [CrossRef]
- Yuan, M.L.; Zhang, Q.L.; Zhang, L.; Guo, Z.L.; Liu, Y.J.; Shen, Y.Y.; Shao, R. High-level phylogeny of the Coleoptera inferred with mitochondrial genome sequences. Mol. Phylogenet. Evol. 2016, 104, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Timmermans, M.J.T.N.; Barton, C.; Haran, J.; Ahrens, D.; Culverwell, C.L.; Ollikainen, A.; Dodsworth, S.; Foster, P.G.; Bocak, L.; Vogler, A.P. Family-level sampling of mitochondrial genomes in Coleoptera: Compositional heterogeneity and phylogenetics. Genome Biol. Evol. 2016, 8, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Nie, R.E.; Vogler, A.P.; Yang, X.K.; Lin, M.Y. Higher-level phylogeny of longhorn beetles (Coleoptera: Chrysomeloidea) inferred from mitochondrial genomes. Syst. Entomol. 2021, 46, 56–70. [Google Scholar] [CrossRef]
- Nie, R.E.; Andújar, C.; Gómez-Rodríguez, C.; Bai, M.; Xue, H.J.; Tang, M.; Yang, C.T.; Tang, P.; Yang, X.K.; Vogler, A.P. The phylogeny of leaf beetles (Chrysomelidae) inferred from mitochondrial genomes. Syst. Entomol. 2020, 45, 188–204. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; dePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Xiang, C.Y.; Gao, F.L.; Jakovlić, L.; Lei, H.P.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.T.; Zhang, D. Using PhyloSuite for molecular phylogeny and tree-based analyses. iMeta 2023, 2, e87. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11 Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.H. DAMBE7: New and Improved Tools for Data Analysis in Molecular Biology and Evolution. Mol. Biol. Evol. 2018, 35, 1550–1552. [Google Scholar] [CrossRef]
- Bininda-Emonds, O.R. transAlign: Using amino acids to facilitate the multiple alignment of protein-coding DNA sequences. BMC Bioinform. 2005, 6, 156. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Patrick, P.S.; Hammersley, J.; Loizou, L.; Kettunen, M.I.; Rodrigues, T.B.; Hu, D.E.; Tee, S.S.; Hesketh, R.; Lyons, S.K.; Soloviev, D.; et al. Dual-modality gene reporter for in vivo imaging. Proc. Natl. Acad. Sci. USA 2014, 111, 415–420. [Google Scholar] [CrossRef]
- Lartillot, N.; Rodrigue, N.; Stubbs, D.; Richer, J. Phylobayes MPI: Phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 2013, 62, 611–615. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.L.; Wang, D.M.; Wang, B.F.; Song, X.Y. Genome survey and mitochondrial genome analysis of Monolepta hieroglyphica. J. Environ. Entomol. 2019, 41, 1287–1296. [Google Scholar]
- Song, N.; Yin, X.M.; Zhao, X.C.; Chen, J.H.; Yin, J. Reconstruction of mitogenomes by NGS and phylogenetic implications for leaf beetles. Mitochondrial DNA Part A 2018, 29, 1041–1050. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.X. Galerucinae nouveaux de la faune Chinoise. Notes D’entomologie Chin. 1942, 9, 9–67. [Google Scholar]
- Baly, J.S. Descriptions of the phytophagous Coleoptera collected by the late Dr. F. Stoliczka during Forsyth’s expedition to Kashgar in 1873–1874. Cistula Entomol. 1878, 2, 369–383. [Google Scholar]
- Baly, J.S. Catalogue of the phytophagous Coleoptera of Japan, with descriptions of the species new to science. Trans. Ent. Soc. Lond. 1874, 22, 161–217. [Google Scholar] [CrossRef]
- Gressitt, J.L.; Kimoto, S. Second supplement to ‘The Chrysomelidae (Coleopt.) of China and Korea. Pac. Insects 1965, 7, 799–806. [Google Scholar]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef]
- Hebert, P.D.; Ratnasingham, S.; deWaard, J.R. Barcoding animal life: Cytochrome c oxidase subunit I divergences among closely related species. Proc. Biol. Sci. 2003, 270 (Suppl. 1), S96–S99. [Google Scholar] [CrossRef] [PubMed]
- Wilcox, J.A. Coleopterum Catalogus Supplementa (Chrysomelidae: Galerucinae, Oidini, Galerucini, Metacyclini, Sermylini), 2nd ed.; BIBLIO: Gravenhage, The Netherlands, 1971; pp. 1–770. [Google Scholar]
- Lee, C.F. The genus Paleosepharia Laboissiere, 1936 in Taiwan: Review and nomenclatural changes (Coleoptera, Chrysomelidae, Galerucinae). Zookeys 2018, 744, 19–41. [Google Scholar] [CrossRef] [PubMed]
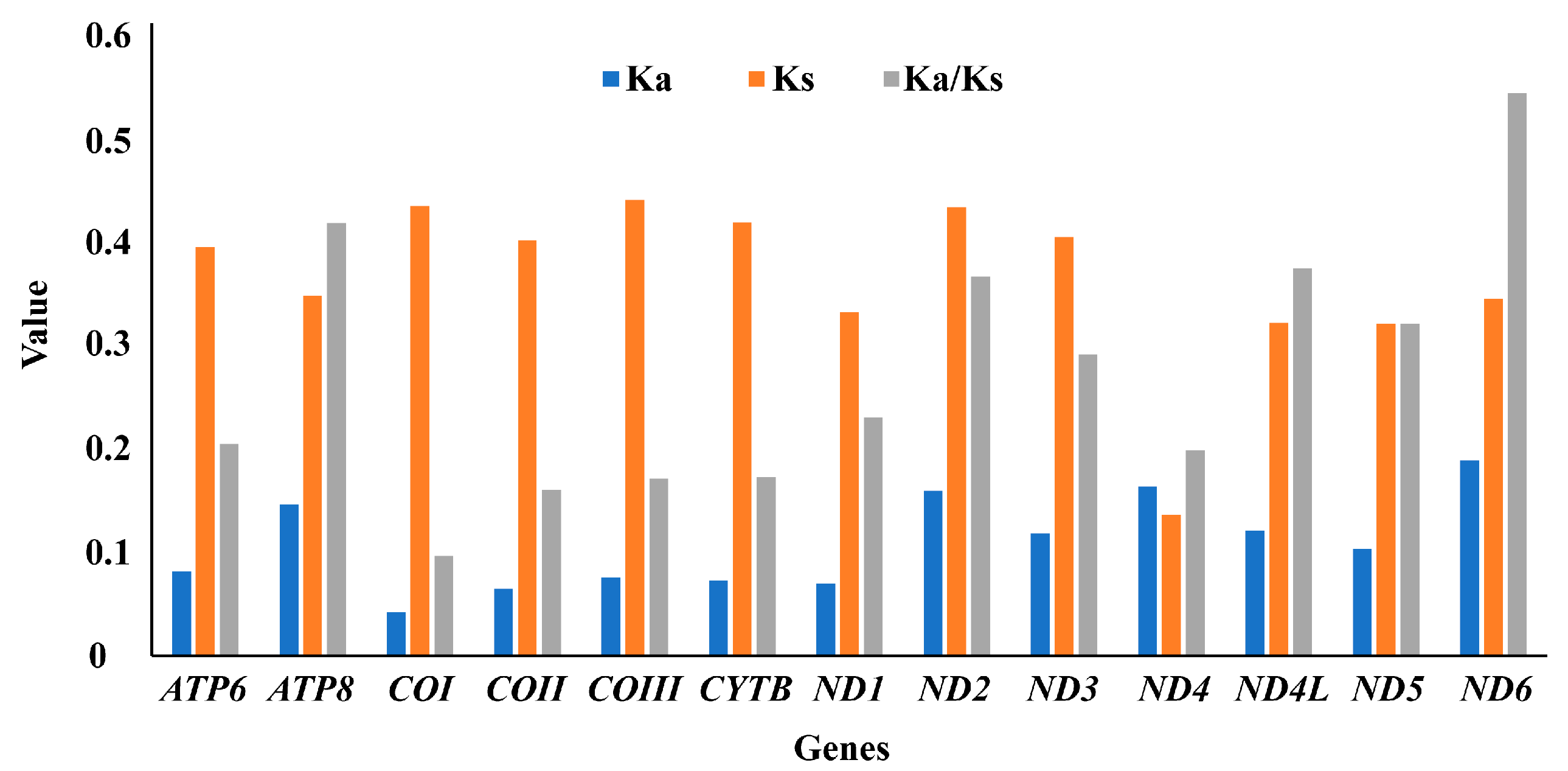

| Subfamily | Tribe | Species | Length (bp) | Acc. No. | References |
|---|---|---|---|---|---|
| Galerucinae | Luperini | Calomicrus pinicola | 15,436 | KX087251 | unpublished |
| Galerucinae | Luperini | Leptomona sp. | 16,697 | MW035611 | unpublished |
| Galerucinae | Luperini | Macrima straminea | 15,567 | MF946618 | [30] |
| Galerucinae | Luperini | Monolepta atrimarginata | 15,143 | MF946621 | [30] |
| Galerucinae | Luperini | Monolepta bicavipennis | 16,672 | OR582724 | this study |
| Galerucinae | Luperini | Monolepta cavipennis | 16,965 | OR582725 | this study |
| Galerucinae | Luperini | Monolepta epistomalis | 15,161 | ON838377 | unpublished |
| Galerucinae | Luperini | * Monolepta hieroglyphica | 16,299 | MT178239 | [58] |
| Galerucinae | Luperini | Monolepta occifluvis | 15,998 | MK409736 | unpublished |
| Galerucinae | Luperini | * Monolepta quadriguttata | 16,130 | KY039102 | [59] |
| Galerucinae | Luperini | Monolepta pallidula | 15,866 | OR582727 | this study |
| Galerucinae | Luperini | * Monolepta signata | 16,329 | OM867791 | unpublished |
| Galerucinae | Luperini | Monolepta sp. | 15,792 | KY039142 | [59] |
| Galerucinae | Luperini | Monolepta wilcoxi | 16,012 | OR582726 | this study |
| Galerucinae | Luperini | Monolepta xanthodera | 14,782 | ON838399 | unpublished |
| Galerucinae | Luperini | Paleosepharia posticata | 15,729 | KY195975 | unpublished |
| Galerucinae | Oidini | Oides livida | 16,127 | MF960098 | [30] |
| Galerucinae | Oidini | Oides maculatus | 15,089 | MF946622 | [30] |
| Species | Whole Mitogenome | Protein-Coding Genes | 12S rRNA Genes | 16S rRNA Genes | Control Region | ||
|---|---|---|---|---|---|---|---|
| A + T% | A + T% | AT-Skew | GC-Skew | A + T% | A + T% | A + T% | |
| M. bicavipennis | 79.1 | 77.6 | −0.145 | 0.003 | 81.2 | 83.0 | 83.4 |
| M. cavipennis | 79.3 | 77.6 | −0.142 | 0.017 | 81.7 | 82.0 | 83.1 |
| M. pallidula | 78.8 | 77.3 | −0.143 | 0.015 | 80.7 | 83.0 | 85.9 |
| M. wilcoxi | 79.8 | 78.4 | −0.143 | 0.018 | 83.3 | 83.1 | 82.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, R.-R.; Lei, Q.-L.; Jin, X.; Zafar, I.; Yang, X.-K.; Su, C.-Y.; Hao, J.-S.; Nie, R.-E. Characterization of Four Complete Mitogenomes of Monolepta Species and Their Related Phylogenetic Implications. Insects 2024, 15, 50. https://doi.org/10.3390/insects15010050
Gao R-R, Lei Q-L, Jin X, Zafar I, Yang X-K, Su C-Y, Hao J-S, Nie R-E. Characterization of Four Complete Mitogenomes of Monolepta Species and Their Related Phylogenetic Implications. Insects. 2024; 15(1):50. https://doi.org/10.3390/insects15010050
Chicago/Turabian StyleGao, Rong-Rong, Qi-Long Lei, Xu Jin, Iqbal Zafar, Xing-Ke Yang, Cheng-Yong Su, Jia-Sheng Hao, and Rui-E Nie. 2024. "Characterization of Four Complete Mitogenomes of Monolepta Species and Their Related Phylogenetic Implications" Insects 15, no. 1: 50. https://doi.org/10.3390/insects15010050
APA StyleGao, R.-R., Lei, Q.-L., Jin, X., Zafar, I., Yang, X.-K., Su, C.-Y., Hao, J.-S., & Nie, R.-E. (2024). Characterization of Four Complete Mitogenomes of Monolepta Species and Their Related Phylogenetic Implications. Insects, 15(1), 50. https://doi.org/10.3390/insects15010050









