Helmet Shape and Phylogeography of the Treehopper Membracis mexicana
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
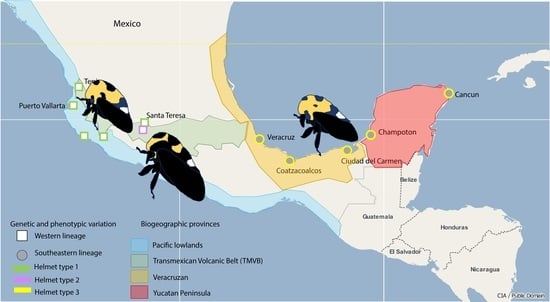
2.1. Study Area
2.2. Insect Sampling
2.3. Geometric Morphometrics
2.4. DNA Extraction, Sequencing, and Processing
2.5. Genetic and Phylogeographic Analyses
3. Results
3.1. Insect Sampling
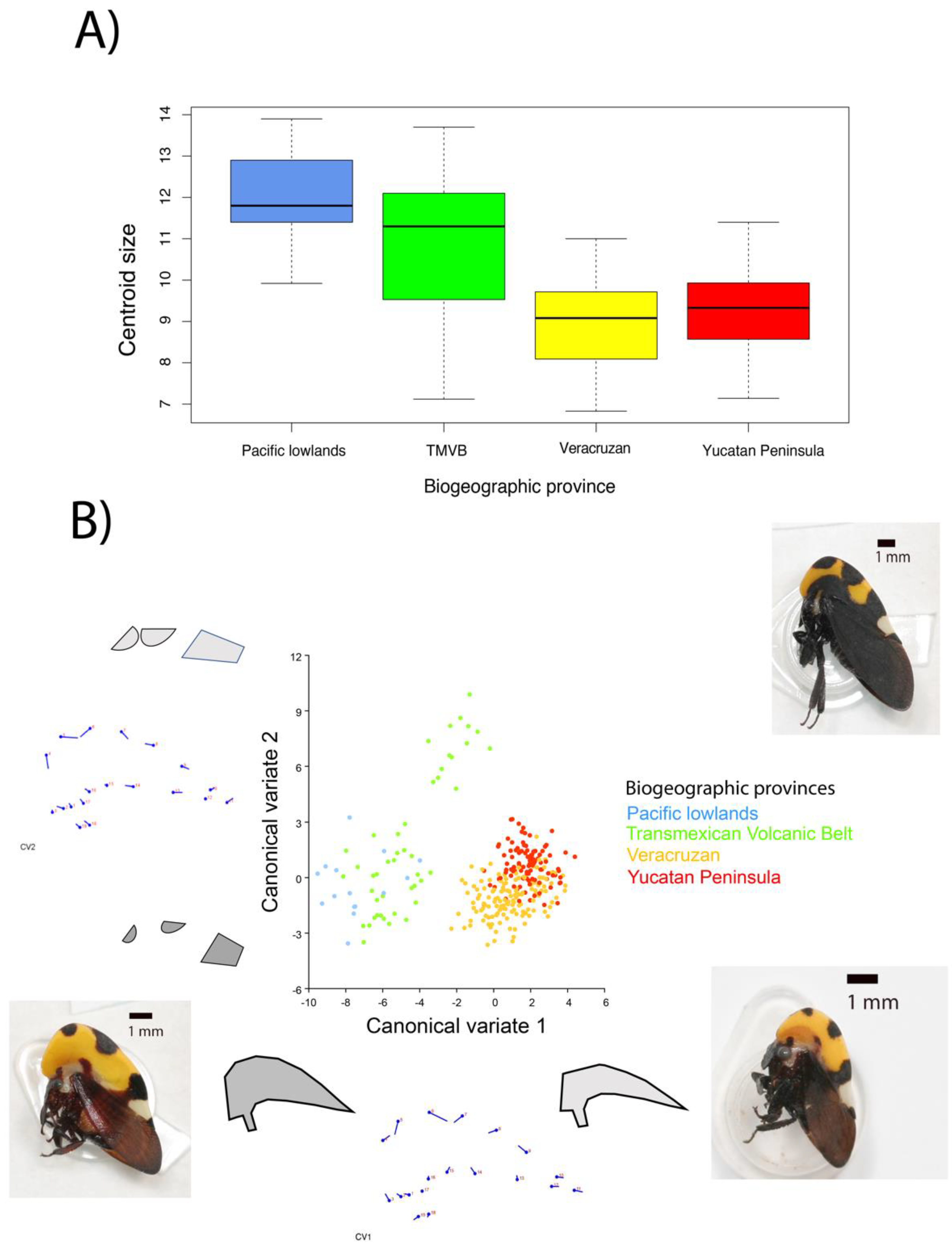
3.2. Geometric Morphometric Analysis
3.2.1. Centroid Size of the Helmet
3.2.2. Principal Components of Helmet Shape
3.2.3. Canonical Variance of Helmet Shape
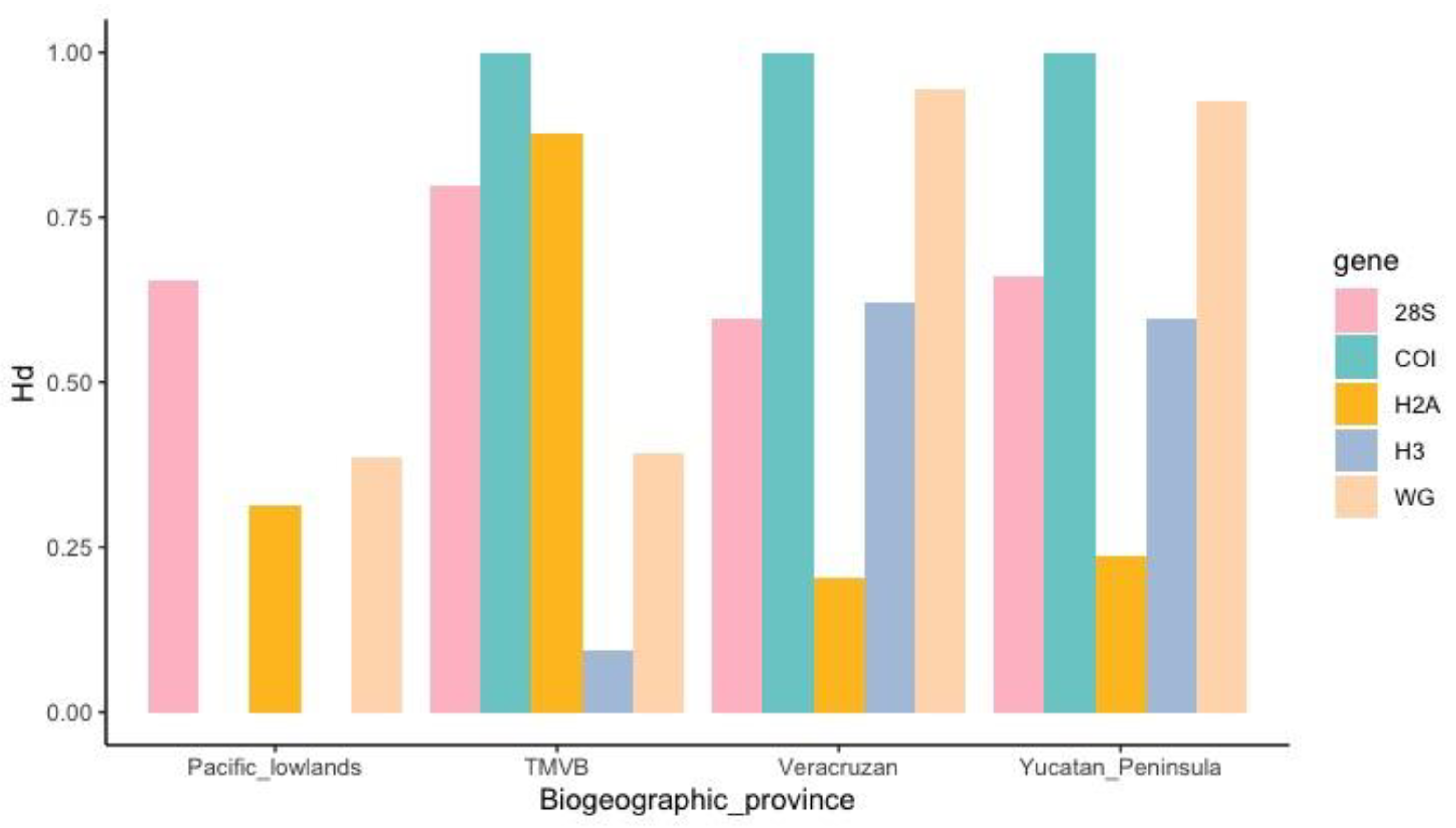
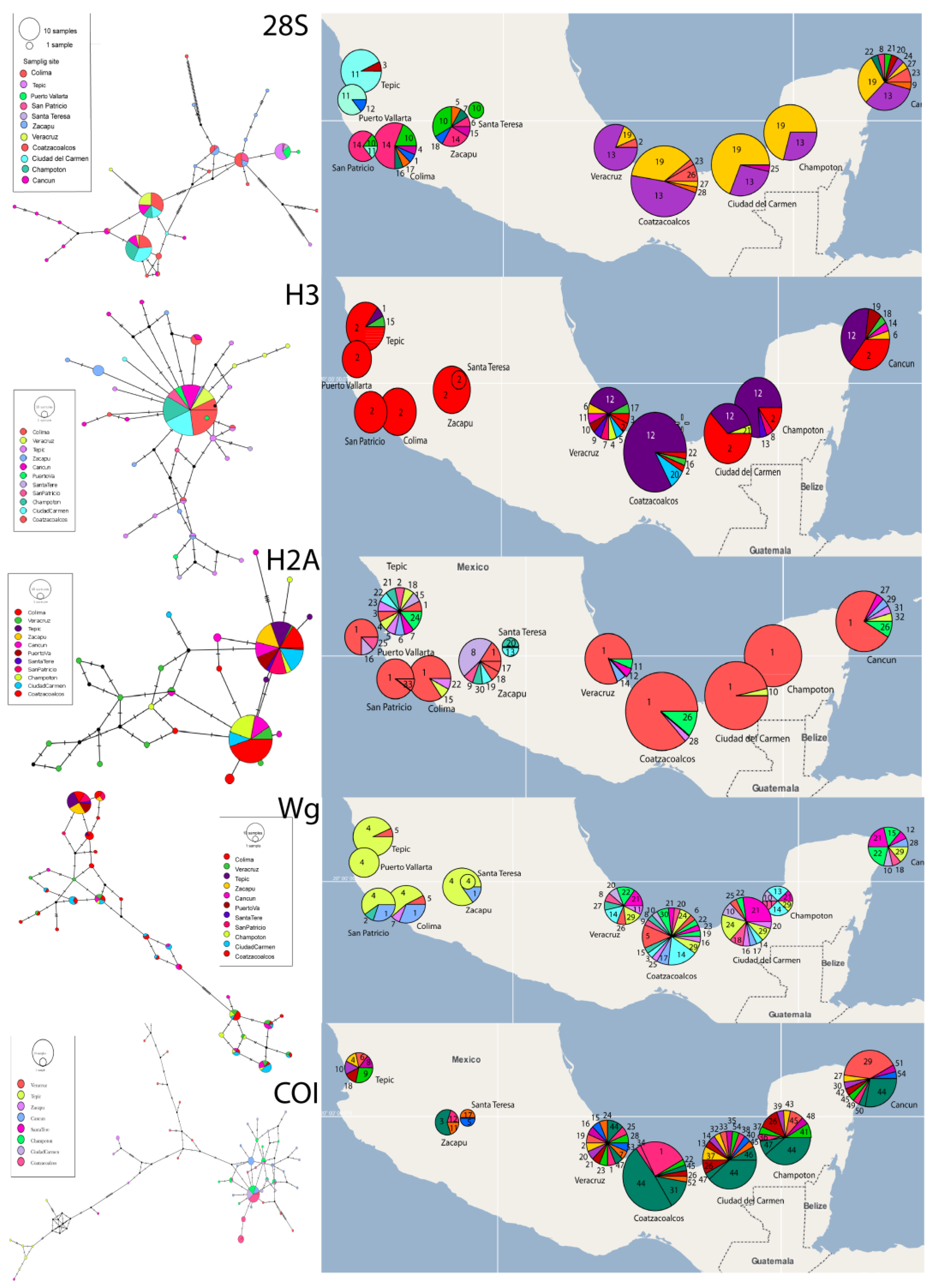
3.3. Genetic and Phylogeographic Analyses
3.3.1. DNA Polymorphism
3.3.2. Haplotype Networks
3.3.3. Analysis of Population Structure
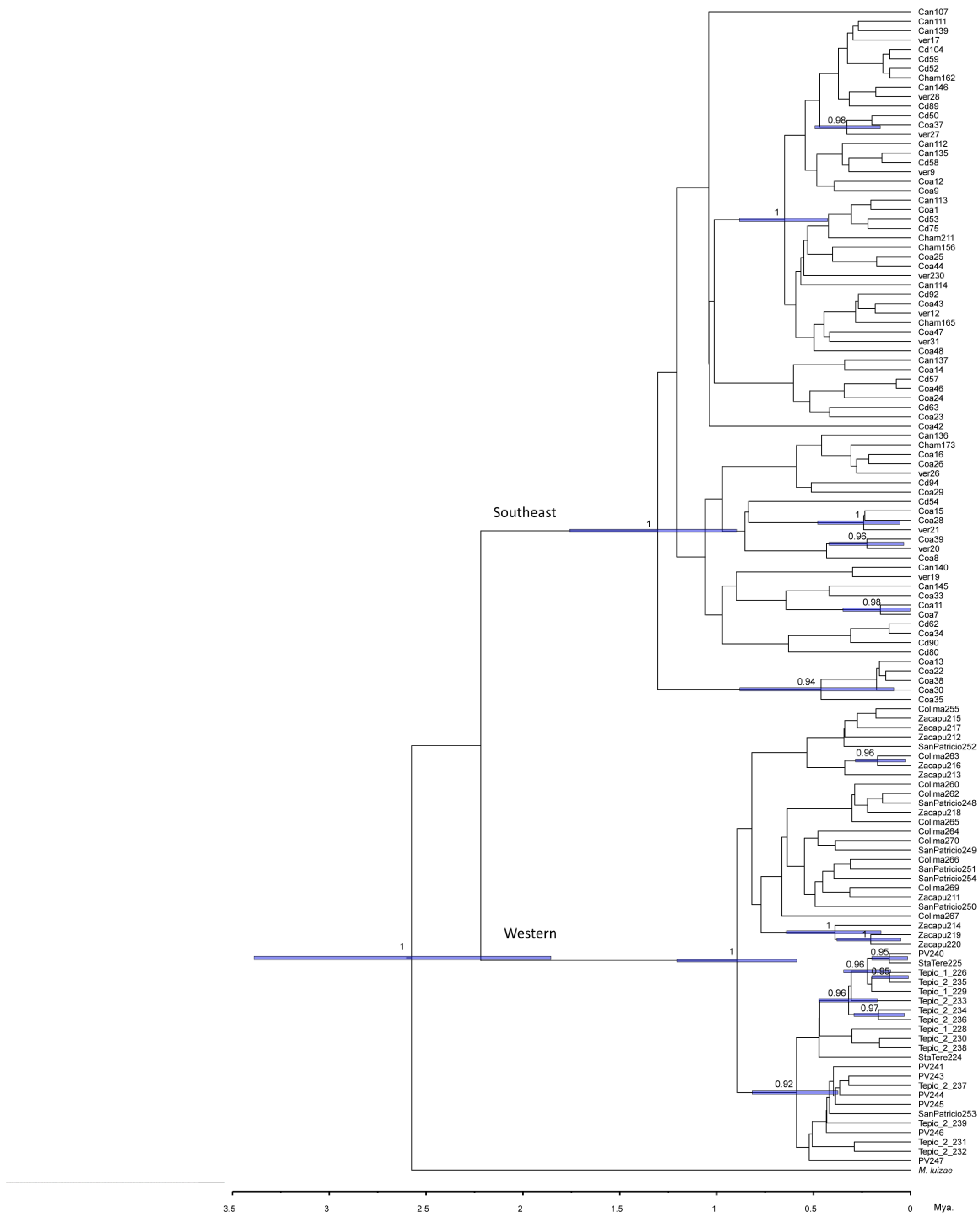
3.3.4. Phylogenetic Analysis
3.3.5. Historical Demography
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferrari, L. Avances en el conocimiento de la Faja Volcánica Transmexicana durante la última década. Boletín Soc. Geológica Mex. 2000, 53, 84–92. [Google Scholar] [CrossRef]
- O’Dea, A.; Lessios, H.A.; Coates, A.G.; Eytan, R.I.; Restrepo-Moreno, S.A.; Cione, A.L.; Stallard, R.F. Formation of the Isthmus of Panama. Sci. Adv. 2016, 2, e1600883. [Google Scholar] [CrossRef] [PubMed]
- Caballero, M.; Lozano-García, S.; Vázquez-Selem, L.; Ortega, B. Evidencias de cambio climático y ambiental en registros glaciales y en cuencas lacustres del centro de México durante el último máximo glacial. Boletín Soc. Geológica Mex. 2010, 62, 359–377. [Google Scholar] [CrossRef]
- Mastretta-Yanes, A.; Moreno-Letelier, A.; Pinero, D.; Jorgensen, T.H.; Emerson, B.C. Biodiversity in the Mexican highlands and the interaction of geology, geography, and climate within the Trans-Mexican Volcanic Belt. J. Biogeogr. 2015, 42, 1586–1600. [Google Scholar] [CrossRef]
- Morrone, J.J. Fundamental biogeographic patterns across the Mexican Transition Zone: An evolutionary approach. Ecography 2010, 33, 355–361. [Google Scholar] [CrossRef]
- Gutiérrez-Ortega, J.S.; Salinas-Rodríguez, M.M.; Martínez, J.F.; Molina-Freaner, F.; Pérez-Farrera, M.A.; Vovides, A.P.; Kajita, T. The phylogeography of the cycad genus Dioon (Zamiaceae) clarifies its Cenozoic expansion and diversification in the Mexican transition zone. Ann. Bot. 2018, 121, 535–548. [Google Scholar] [CrossRef]
- Morrone, J.J.; Espinosa Organista, D.; Llorente Bousquets, J. Mexican biogeographic provinces: Preliminary scheme, general characterizations, and synonymie. Acta Zoológica Mex. 2002, 85, 83–108. [Google Scholar] [CrossRef]
- Halffter, G. Biogeography of the montane entomofauna of Mexico and Central America. Annu. Rev. Entomol. 1987, 32, 95–114. [Google Scholar] [CrossRef]
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; Da Fonseca, G.A.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef]
- Richter, L. Membracidae Colombianae. Revision de las especies Colombianas del genero Membracis. Rev. Acad. Colomb. Cienc. Exactas Físicas Nat. 2017, 41, 222–246. [Google Scholar] [CrossRef]
- Fowler, W.W. 1894–1909. Biologia Centrali-Americana. Insecta Rhynchota. Hemiptera-Homoptera; Biodiversity Heritage Library: Washington, DC, USA, 1996; Volume 11, part I; pp. 1–339. [Google Scholar]
- Rohlf, F. (2007) tpsRelw. Available online: https://www.sbmorphometrics.org/index.html (accessed on 10 January 2022).
- Rohlf, F. (2007) tpsDig. Available online: https://www.sbmorphometrics.org/index.html (accessed on 10 January 2022).
- Bookstein, F.L. Morphometric Tools for Landmark Data; Reprint edition; Cambridge University Press: Cambridge, UK, 1997; ISBN 0521585988. [Google Scholar]
- Klingenberg, C.P. MorphoJ: An integrated software package for geometric morphometrics. Mol. Ecol. Ecol. Resour. 2011, 11, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Klingenberg, C.P.; McIntyre, G.S. Geometric morphometrics of developmental instability: Analyzing patterns of fluctuating asymmetry with Procrustes methods. Evolution 1998, 52, 1363–1375. [Google Scholar] [CrossRef] [PubMed]
- Evangelista, O.; Sakakibara, A.M.; Cryan, J.R.; Urban, J.M. A phylogeny of the treehopper subfamily Heteronotinae reveals convergent pronotal traits (Hemiptera: Auchenorrhyncha: Membracidae). Syst. Entomol. 2017, 42, 410–428. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003, 1, 2–3. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Dupanloup, I.; Schneider, S.; Excoffier, L. A simulated annealing approach to define the genetic structure of populations. Mol. Ecol. 2002, 11, 2571–2581. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.H.; Xie, D.; Drummond, A.J. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Allio, R.; Donega, S.; Galtier, N.; Nabholz, B. Large variation in the ratio of mitochondrial to nuclear mutation rate across animals: Implications for genetic diversity and the use of mitochondrial DNA as a molecular marker. Mol. Biol. Evol. 2017, 34, 2762–2772. [Google Scholar] [CrossRef]
- Montejo-Kovacevich, G.; Smith, J.E.; Meier, J.I.; Bacquet, C.N.; Whiltshire-Romero, E.; Nadeau, N.J.; Jiggins, C.D. Altitude and life-history shape the evolution of Heliconius wings. Evolution 2019, 73, 2436–2450. [Google Scholar] [CrossRef] [PubMed]
- Nolasco-Soto, J.; González-Astorga, J.; de Los Monteros, A.E.; Galante-Patiño, E.; Favila, M.E. Phylogeographic structure of Canthon cyanellus (Coleoptera: Scarabaeidae), a Neotropical dung beetle in the Mexican Transition Zone: Insights on its origin and the impacts of Pleistocene climatic fluctuations on population dynamics. Mol. Phylogenet. Evol. 2017, 109, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Castañeda-Rico, S.; León-Paniagua, L.; Vázquez-Domínguez, E.; Navarro-Sigüenza, A.G. Evolutionary diversification and speciation in rodents of the Mexican lowlands: The Peromyscus melanophrys species group. Mol. Phylogenet. Evol. 2014, 70, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Gómez, F.; Gutiérrez-Rodríguez, C.; Ornelas, J.F. Genetic, phenotypic and ecological divergence with gene flow at the Isthmus of Tehuantepec: The case of the azure-crowned hummingbird (Amazilia cyanocephala). J. Biogeogr. 2013, 40, 1360–1373. [Google Scholar] [CrossRef]
- Suárez-Atilano, M.; Burbrink, F.; Vázquez-Domínguez, E. Phylogeographical structure within Boa constrictor imperator across the lowlands and mountains of Central America and Mexico. J. Biogeogr. 2014, 41, 2371–2384. [Google Scholar] [CrossRef]
- Arteaga, M.C.; Piñero, D.; Eguiarte, L.E.; Gasca, J.; Medellín, R.A. Genetic structure and diversity of the nine-banded armadillo in Mexico. J. Mammal. 2012, 93, 547–559. [Google Scholar] [CrossRef]

| Number | ID | Latitude, Longitude | Host Plant | Sample Size Per Host Plant | State | Country | Sampling Date |
|---|---|---|---|---|---|---|---|
| 1 | Cancun (YP) | 21°10′08.3″ N 86°51′30.9″ W | Terminalia catappa, Ficus carica | 18, 8 | Quintana Roo | Mexico | December 2019 |
| 2 | Champoton (YP) | 19°21′01.0″ N 90°43′40.3″ W | Terminalia catappa | 30 | Campeche | Mexico | December 2019 |
| 3 | Ciudad del Carmen (V) | 18°39′05.7″ N 91°49′25.1″ W | Terminalia catappa | 30 | Campeche | Mexico | December 2019 |
| 4 | Coatzacoalcos (V) | 18°08′53.1″ N 94°27′02.4″ W | Terminalia catappa | 30 | Veracruz | Mexico | December 2019 |
| 5 | Veracruz (V) | 19°11′13.3″ N 96°07′37.2″ W | Terminalia catappa | 25 | Veracruz | Mexico | May 2019 |
| 6 | Tepic (TMVB) | 21°30′08.2″ N 104°54′09.8″ W | Spondias purpurea, Terminalia catappa | 4, 11 | Nayarit | Mexico | May 2021 |
| 7 | Puerto Vallarta (PL) | 20°38′28.5″ N 105°12′49.0″ W | Artocarpus altilis, Terminalia catappa | 3, 5 | Jalisco | Mexico | May 2021 |
| 8 | San Patricio (PL) | 19°13′25.2″ N 104°42′48.0″ W | Terminalia catappa | 7 | Jalisco | Mexico | May 2021 |
| 9 | Colima (TMVB) | 19°14′10.7″ N 103°44′29.3″ W | Cassia fistula, Terminalia catappa | 7, 6 | Colima | Mexico | May 2021 |
| 10 | Santa Teresa (TMVB) | 19°58′18.7″ N 101°38′05.9″ W | Heimia salicifolia | 2 | Michoacan | Mexico | May 2021 |
| 11 | Zacapu (TMVB) | 19°49′22.9″ N 101°47′17.8″ W | Salix babylonica | 13 | Michoacan | Mexico | May 2021 |
| Gene | Clustering | K | FCT | p-Value |
|---|---|---|---|---|
| 28S | (Tepic, Puerto Vallarta) (Colima) (San Patricio) (Santa Teresa) (Zacapu) (Veracruz, Cancun, Champoton, Ciudad del Carmen, Coatzacoalcos) | 6 | 0.8108 | 0.00000 ± 0.00000 |
| H3 | (Puerto Vallarta, Santa Teresa, San Patricio) (Tepic) (Colima) (Zacapu) (Veracruz) (Coatzacoalcos) (Ciudad del Carmen) (Champoton) (Cancun) | 9 | 0.30013 | 0.00782 ± 0.00242 |
| H2A | (Santa Teresa) (Champoton, Tepic, Zacapu, Ciudad del Carmen, Veracruz, Coatzacoalcos, Puerto Vallarta, Cancun, Colima, San Patricio) | 2 | 0.67267 | 0.09677 ± 0.00848 |
| WG | (Colima, San Patricio, Zacapu, Puerto Vallarta, Tepic, Santa Teresa) (Coatzacoalcos, Veracruz, Champoton, Ciudad del Carmen, Cancun) | 2 | 0.58272 | 0.00098 ± 0.00098 |
| COI | (Tepic, Santa Teresa) (Zacapu) (Ciudad del Carmen, Veracruz, Coatzacoalcos, Champoton, Cancun) | 3 | 0.81407 | 0.00293 ± 0.00164 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De-la-Mora, M.; Pinero, D. Helmet Shape and Phylogeography of the Treehopper Membracis mexicana. Insects 2023, 14, 704. https://doi.org/10.3390/insects14080704
De-la-Mora M, Pinero D. Helmet Shape and Phylogeography of the Treehopper Membracis mexicana. Insects. 2023; 14(8):704. https://doi.org/10.3390/insects14080704
Chicago/Turabian StyleDe-la-Mora, Marisol, and Daniel Pinero. 2023. "Helmet Shape and Phylogeography of the Treehopper Membracis mexicana" Insects 14, no. 8: 704. https://doi.org/10.3390/insects14080704
APA StyleDe-la-Mora, M., & Pinero, D. (2023). Helmet Shape and Phylogeography of the Treehopper Membracis mexicana. Insects, 14(8), 704. https://doi.org/10.3390/insects14080704