SMRT Sequencing Technology Was Used to Construct the Batocera horsfieldi (Hope) Transcriptome and Reveal Its Features
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Preparation
2.2. Library Preparation and SMRT Sequencing
2.3. SMRT Sequencing Data Processing
2.4. Functional Annotation of Transcripts
2.5. CDS Prediction
2.6. SSR Analysis
2.7. Transcription Factor (TF) Analysis
2.8. lncRNAs Analysis
3. Results
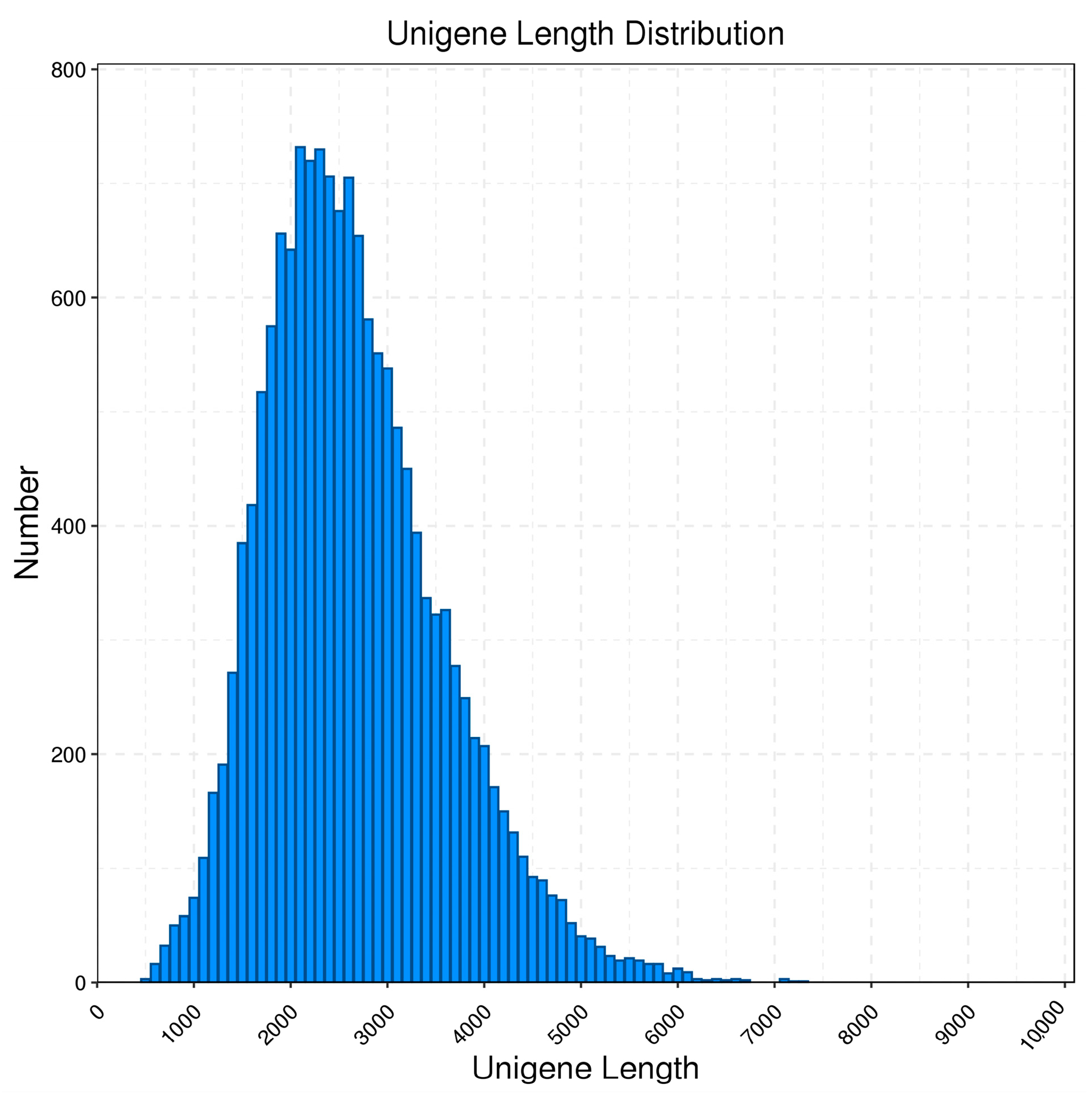
3.1. Transcriptome Analysis Was Performed using Pacbio Sequencing
3.2. Functional Annotation of B. horsfieldi
3.3. CDS Predictions
3.4. Identification of Transcription Factors
3.5. SSR Analysis
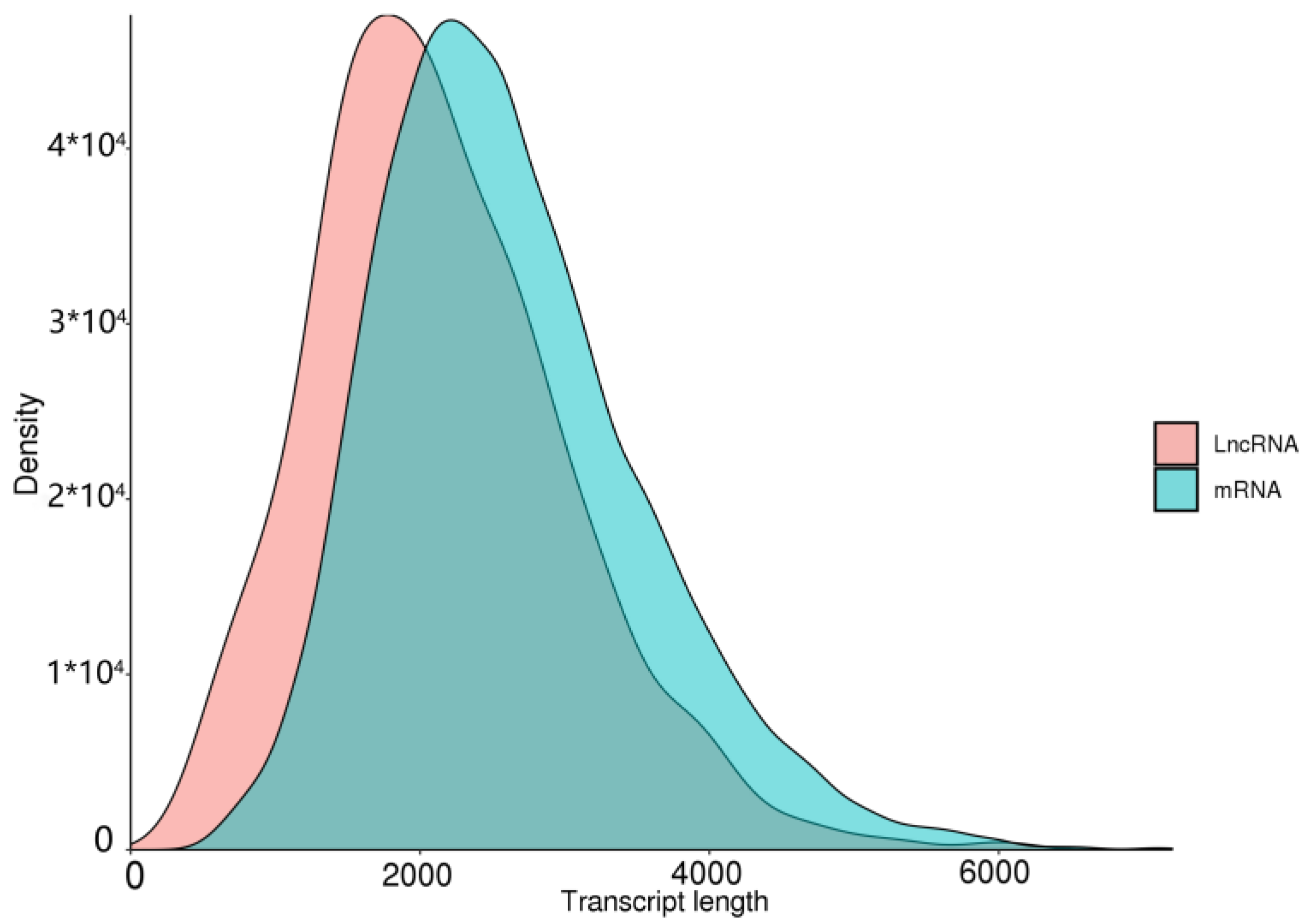
3.6. lncRNA Forecasts
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, J. Biocontrol of Batocera horsfieldi (Coleoptera: Cerambycidae) with Parasitoid Dastarcus helophoroides (Coleoptera: Bothrideridae). Ph.D. Thesis, Northwest A&F University, Xianyang, China, 2009. [Google Scholar]
- Yang, H.; Yang, M.; Yang, W.; Yang, C.; Dong, J.; Shen, Y. A Study on the Spatial Distribution Pattern and the Living-inhabiting Tunnel of the Larvae of Batocera horsfieldi (Hope). J. Sichuan Agr. Univ. 2010, 28, 148–152. [Google Scholar]
- Li, A.; Wang, J.; Wang, R.; Yang, H.; Yang, W.; Yang, C.; Jin, Z. MaxEnt modeling to predict current and future distributions of Batocera lineolata (Coleoptera: Cerambycidae) under climate change in China. Écoscience 2020, 27, 23–31. [Google Scholar] [CrossRef]
- Wang, S.; Wang, H.; Xia, M.; Dong, S. Regularity of occurrence and control of Batocera horsfieldi in walnut trees. China Fruits 2004, 2, 11–13. [Google Scholar]
- Mei, A.; Chen, J.; Wu, G.; Du, X.; Luo, F. Investigation on Yang Tree Pests, Their Occurrence Reasons, and Main Pest Control Measures in the Jianghan Plain. For. Pest Dis. 1998, 02, 36–38. [Google Scholar]
- Yang, H.; Cai, Y.; Zhuo, Z.; Yang, W.; Yang, C.; Zhang, J.; Yang, Y.; Wang, B.; Guan, F. Transcriptome analysis in different developmental stages of Batocera horsfieldi (Coleoptera: Cerambycidae) and comparison of candidate olfactory genes. PLoS ONE 2018, 13, e192730. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y. Theory and Techniques of Ecological Regulation of Poplar Longhorned Beetle Disaster in Shelter-Forest. Ph.D. Thesis, Beijing Forestry University, Beijing, China, 2005. [Google Scholar]
- Zheng, K.; Wu, S.; Zhang, D.; Wu, J.; Du, Y.; Fan, J. Differences in feeding and oviposition behavior of different populations of Batocera horsfieldi. J. Zhejiang AF Univ. 2022, 39, 159–165. [Google Scholar]
- Wu, Y. Identification and Binding Characteristics of Odorant-Binding Proteins in Batocera horsfieldi. Master’s Thesis, Huazhong Agricultural University, Wuhan, China, 2022. [Google Scholar]
- Yang, H.; Yang, W.; Yang, C.; Zhu, T.; Huang, Q.; Han, S.; Xiao, J. Electrophysiological and behavioral responses of the white-striped longhorned beetle, Batocera lineolata, to the diurnal rhythm of host plant volatiles of holly, Viburnum awabuki. J. Insect Sci. 2013, 13, 85. [Google Scholar] [CrossRef]
- Zhuge, P.P.; Luo, S.L.; Wang, M.Q.; Zhang, G. Electrophysiological responses of Batocera horsfieldi (Hope) adults to plant volatiles. J. Appl. Entomol. 2010, 134, 600–607. [Google Scholar] [CrossRef]
- Zhuo, Z.; Jin, Y.; Xu, D.; Liao, W. Electroantennogram responses of Batocera horsfieldi (Hope) to the selected volatile components of host plants, Rosa cymosa Tratt. and Rosa multiflora Thunb. Glob. Ecol. Conserv. 2022, 33, e1986. [Google Scholar] [CrossRef]
- Hu, J.; Xu, D.; Zhuo, Z.; Yang, W.; Yang, H.; Zheng, Y. Analysis of antennal transcriptome and olfaction-related genes of adult Batocera horsfieldi (Hope). Chin. J. Appl. Entomol. 2019, 56, 1037–1047. [Google Scholar]
- Hittinger, C.T.; Johnston, M.; Tossberg, J.T.; Rokas, A. Leveraging skewed transcript abundance by RNA-Seq to increase the genomic depth of the tree of life. Proc. Natl. Acad. Sci. USA 2010, 107, 1476–1481. [Google Scholar] [CrossRef] [PubMed]
- Jia, D.; Wang, Y.; Liu, Y.; Hu, J.; Guo, Y.; Gao, L.; Ma, R. SMRT sequencing of full-length transcriptome of flea beetle Agasicles hygrophila (Selman and Vogt). Sci. Rep. 2018, 8, 2197. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Xu, D.; Zhuo, Z.; Hu, J.; Lu, B. SMRT sequencing of the full-length transcriptome of the Rhynchophorus ferrugineus (Coleoptera: Curculionidae). PeerJ 2020, 8, e9133. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Ekblom, R.; Galindo, J. Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity 2011, 107, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Won, H.I.; Schulze, T.T.; Clement, E.J.; Watson, G.F.; Watson, S.M.; Warner, R.C.; Ramler, E.A.M.; Witte, E.J.; Schoenbeck, M.A.; Rauter, C.M.; et al. De novo Assembly of the Burying Beetle Nicrophorus orbicollis (Coleoptera: Silphidae) Transcriptome Across Developmental Stages with Identification of Key Immune Transcripts. J. Genom. 2018, 6, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Lü, J.; Bai, C.; Gu, C.; Guo, C. Transcriptome analysis reveals adaptation mechanism of Tribolium castaneum (Herbst) (Coleoptera: Tenebrionidae) adults to benzoquinone stress. J. Stored Prod. Res. 2023, 101, 102083. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, H.; Zhuang, G.; Zheng, H.; Zhang, X. Comparative transcriptome analysis of Callosobruchus chinensis (L.) (Coleoptera: Chrysomelidae-Bruchinae) after heat and cold stress exposure. J. Therm. Biol. 2023, 112, 103479. [Google Scholar] [CrossRef]
- Wang, G.; Chang, Y.; Guo, J.; Xi, J.; Liang, T.; Zhang, S.; Yang, M.; Hu, L.; Mu, W.; Song, J. Identification and Expression Profiles of Putative Soluble Chemoreception Proteins from Lasioderma serricorne (Coleoptera: Anobiidae) Antennal Transcriptome. Environ. Entomol. 2022, 51, 700–709. [Google Scholar] [CrossRef]
- Coghlan, A.; Fiedler, T.; Mckay, S.; Flicek, P.; Harris, T.; Blasiar, D.; Stein, L. nGASP-the nematode genome annotation assessment project. BMC Bioinform. 2008, 9, 549. [Google Scholar] [CrossRef]
- Li, Y.; Fang, C.; Fu, Y.; Hu, A.; Li, C.; Zou, C.; Li, X.; Zhao, S.; Zhang, C.; Li, C. A survey of transcriptome complexity in Sus scrofa using single-molecule long-read sequencing. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2018, 25, 421–437. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Lin, X.; Zhu, J.; Yu, X.; Xia, X.; Yao, F.; Yang, G.; You, M. Characterization and expression profiling of serine protease inhibitors in the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). BMC Genom. 2017, 18, 162. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Yang, Q.; Wang, Q.; An, X.; Liu, Y.; Li, L. Application of single molecule real time sequencing in environmental microorganisms research. Microbiol. China 2019, 46, 3140–3147. [Google Scholar]
- Koren, S.; Schatz, M.C.; Walenz, B.P.; Martin, J.; Howard, J.T.; Ganapathy, G.; Wang, Z.; Rasko, D.A.; Mccombie, W.R.; Jarvis, E.D.; et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat. Biotechnol. 2012, 30, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tang, X.; Wang, Z.; Tang, F. The transcriptomic response of Hyphantria cunea (Drury) to the infection of Serratia marcescens Bizio based on full-length SMRT transcriptome sequencing. Front. Cell. Infect. Microbiol. 2023, 13, 1093432. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, H.; Wang, X.; Zheng, X.; Lu, W.; Qin, F.; Chen, C. Full-Length SMRT Transcriptome Sequencing and SSR Analysis of Bactrocera dorsalis (Hendel). Insects 2021, 12, 938. [Google Scholar] [CrossRef]
- Kai, F.; Xiaoyu, L.; Jian, L.; Fang, T. SMRT sequencing of the full-length transcriptome of Odontotermes formosanus (Shiraki) under Serratia marcescens treatment. Sci. Rep. 2020, 10, 15909. [Google Scholar]
- Chen, J.; Yu, Y.; Kang, K.; Zhang, D. SMRT sequencing of the full-length transcriptome of the white-backed planthopper Sogatella furcifera. PeerJ 2020, 8, e9320. [Google Scholar] [CrossRef]
- Wang, X.; Xu, X.; Ullah, F.; Ding, Q.; Gao, X.; Desneux, N.; Song, D. Comparison of full-length transcriptomes of different imidacloprid-resistant strains of Rhopalosiphum padi (L.). Entomol. Gen. 2020, 41, 289–304. [Google Scholar] [CrossRef]
- Chin, C.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E.; et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef]
- Salmela, L.; Rivals, E. LoRDEC: Accurate and efficient long read error correction. Bioinformatics 2014, 30, 3506–3514. [Google Scholar] [CrossRef]
- Pirooznia, M.; Perkins, E.; Deng, Y. Batch Blast Extractor: An automated blastx parser application. BMC Genom. 2008, 9 (Suppl. 2), S10. [Google Scholar] [CrossRef]
- Li, W.; Jaroszewski, L.; Godzik, A. Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics 2002, 18, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.; Fedorova, N.; Jackson, J.; Jacobs, A.; Kiryutin, B.; Koonin, E.; Krylov, D.; Mazumder, R.; Mekhedov, S.; Nikolskaya, A.; et al. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Bairoch, A.; Apweiler, R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000, 28, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Shimizu, K.; Adachi, J.; Muraoka, Y. ANGLE: A sequencing errors resistant program for predicting protein coding regions in unfinished cDNA. J. Bioinform. Comput. Biol. 2006, 4, 649–664. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, T.; Liu, C.; Song, S.; Zhang, X.; Liu, W.; Jia, H.; Xue, Y.; Guo, A. AnimalTFDB 2.0: A resource for expression, prediction and functional study of animal transcription factors. Nucleic. Nucleic Acids Res. 2015, 43, D76–D81. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Yang, D.; Kong, L.; Hou, M.; Meng, Y.; Wei, L.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Zhang, J.; Zhou, Z. PLEK: A tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinform. 2014, 15, 311. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Shen, W.; Chen, S.; Gan, Z.; Zhang, Y.; Yue, T.; Chen, M.; Xue, Y.; Hu, H.; Guo, A. AnimalTFDB 4.0: A comprehensive animal transcription factor database updated with variation and expression annotations. Nucleic Acids Res. 2022, 51, D39–D45. [Google Scholar] [CrossRef]
- Schadt, E.E.; Turner, S.; Kasarskis, A. A window into third-generation sequencing. Hum. Mol. Genet. 2010, 19, R227–R240. [Google Scholar] [CrossRef]
- Yang, F.; Chen, T.; Mi, L.; Ma, L.; Xie, Y.; Li, J.; Liu, Z. Current status andprospect of biological research on transcriptome sequencing. Anim. Husb. Vet. Med. 2019, 51, 133–138. [Google Scholar]
- Zhao, L. Analysis of Full-Length Transcriptome and Mitochondrial Transcriptome of Three Orthoptera Insects. Ph.D. Thesis, Shaanxi Normal Univiversity, Xi’an, China, 2018. [Google Scholar]
- Jing-E, M.; Hai-Ying, J.; Lin-Miao, L.; Xiu-Juan, Z.; Hui-Ming, L.; Guan-Yu, L.; Da-Ying, M.; Jin-Ping, C. SMRT sequencing of the full-length transcriptome of the Sunda pangolin (Manis javanica). Gene 2019, 692, 208–216. [Google Scholar]
- Chaisson, M.J.; Tesler, G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): Application and theory. BMC Bioinform. 2012, 13, 1. [Google Scholar] [CrossRef]
- Lv, Y. Variation Analysis of Single-Molecule Real-Time Sequencing Data Based on Deep Learning. Master’s Thesis, Beijing University of Chemical Technology, Beijing, China, 2021. [Google Scholar]
- Yang, H.; Hu, J.; Wang, Z.; Xu, D.; Zhuo, Z. Using PacBio Iso-Seq to determine the transcriptome of Rhynchophorus ferrugineus. Chin. J. Appl. Entomol. 2021, 58, 655–663. [Google Scholar]
- Dong, L.; Liu, H.; Zhang, J.; Yang, S.; Kong, G.; Chu, J.S.C.; Chen, N.; Wang, D. Single-molecule real-time transcript sequencing facilitates common wheat genome annotation and grain transcriptome research. BMC Genom. 2015, 16, 1039. [Google Scholar] [CrossRef] [PubMed]
- Noriega, D.D.; Arias, P.L.; Barbosa, H.R.; Arraes, F.B.M.; Ossa, G.A.; Villegas, B.; Coelho, R.R.; Albuquerque, E.V.S.; Togawa, R.C.; Grynberg, P.; et al. Transcriptome and gene expression analysis of three developmental stages of the coffee berry borer, Hypothenemus hampei. Sci. Rep. 2019, 9, 12804. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhao, X.; Qiao, H.; He, X.; Tan, J.; Hao, D. Comparative Transcriptome Analysis of the Heat Stress Response in Monochamus alternatus Hope (Coleoptera: Cerambycidae). Front. Physiol. 2020, 10, 1568. [Google Scholar] [CrossRef]
- Jian-Kun, Y.; Shuang, Y.; Shang, W.; Jun, W.; Xin-Xin, Z.; Yan, L.; Jing-Hui, X. Identification of candidate chemosensory receptors in the antennal transcriptome of the large black chafer Holotrichia parallela Motschulsky (Coleoptera: Scarabaeidae). Comp. Biochem. Physiol. Part D Genom. Proteom. 2018, 28, 63–71. [Google Scholar]
- Wei, G.; Jing, L.; Mujuan, G.; Shimin, C.; Baoli, Q.; Wen, S.; Chunxiao, Y.; Youjun, Z.; Huipeng, P. De Novo Transcriptome Analysis Reveals Abundant Gonad-specific Genes in the Ovary and Testis of Henosepilachna vigintioctopunctata. Int. J. Mol. Sci. 2019, 20, 4084. [Google Scholar]
- Liang, X. Preference of Batocera horsfieldi (Hope) for Host of Supplementary Feeding. Master’s Thesis, Sichuan Agricultural University, Chengdu, China, 2007. [Google Scholar]
- Zhuge, P. The Semiochemicals in Host Location of Longhorn Beetle Batocera horsfieldi (Hope). Master’s Thesis, Huazhong Agricultural University, Wuhan, China, 2009. [Google Scholar]
- Fang, F.; Yao, Y.; Ma, Z. Exploration of the Long Noncoding RNAs Involved in the Crosstalk between M2 Macrophages and Tumor Metabolism in Lung Cancer. Genet. Res. 2023, 2023, 4512820. [Google Scholar] [CrossRef]
- Pedro, J.B.; Howard, Y.C. Long Noncoding RNAs: Cellular Address Codes in Development and Disease. Cell 2013, 152, 1298–1307. [Google Scholar]
- Lou, F.; Han, Z. Full-length transcripts facilitates Portunus trituberculatus genome structure annotation. J. Oceanol. Limnol. 2022, 40, 2042–2051. [Google Scholar] [CrossRef]









| Data Type | Total Bases (bp) | Total Number | Mean Length | Min_Length | Max_Length | N50 |
|---|---|---|---|---|---|---|
| polymerase read | 47.83 G | 462,134 | 103,503 | - | - | 165,195 |
| subread | 46.31 G | 20,356,793 | 2275 | - | - | 2465 |
| CCS | - | 432,091 | 2527 | 62 | 18,215 | 2674 |
| FLNC | - | 395,851 | 2407 | 86 | 14,739 | 2555 |
| Sample | Library Name | Raw Reads | Clean Reads | Raw Base (G) | Clean Base (G) | Effective (%) | Error (%) | Q20 (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| BHM | FRAS220000762-4r | 48699628 | 45453468 | 7.3 | 6.82 | 93.33 | 0.03 | 97.65 | 93.02 | 36.65 |
| BHM1 | FRAS220000763-4r | 56680480 | 52076758 | 8.5 | 7.81 | 91.88 | 0.03 | 97.74 | 93.17 | 36.37 |
| BHM2 | FRAS220000764-3r | 39332558 | 36671048 | 5.9 | 5.5 | 93.23 | 0.03 | 97.46 | 92.46 | 32.88 |
| BHF | FRAS220000759-4r | 43125796 | 40408496 | 6.47 | 6.06 | 93.7 | 0.03 | 97.87 | 93.26 | 34.54 |
| BHF1 | FRAS220000760-5r | 42298740 | 39126102 | 6.34 | 5.87 | 92.5 | 0.03 | 97.77 | 93.14 | 35.24 |
| BHF2 | FRAS220000761-4r | 44239794 | 41348904 | 6.64 | 6.2 | 93.47 | 0.03 | 97.76 | 93.13 | 34.36 |
| Transcript Length Interval | <500 bp | 500–1 kbp | 1 k–2 kbp | 2 k–3 kbp | >3 kbp | Total |
|---|---|---|---|---|---|---|
| Number of transcripts | 21 | 800 | 13,054 | 16,657 | 9380 | 39,912 |
| Number of genes | 3 | 230 | 3930 | 6593 | 4477 | 15,233 |
| Isoform number | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| Unigene number | 10,226 | 2059 | 887 | 523 | 313 | 223 | 159 | 137 | 86 | 620 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, X.; Xu, D.; Liu, Z.; Liu, Q.; Zhuo, Z. SMRT Sequencing Technology Was Used to Construct the Batocera horsfieldi (Hope) Transcriptome and Reveal Its Features. Insects 2023, 14, 625. https://doi.org/10.3390/insects14070625
Wei X, Xu D, Liu Z, Liu Q, Zhuo Z. SMRT Sequencing Technology Was Used to Construct the Batocera horsfieldi (Hope) Transcriptome and Reveal Its Features. Insects. 2023; 14(7):625. https://doi.org/10.3390/insects14070625
Chicago/Turabian StyleWei, Xinju, Danping Xu, Zhiqian Liu, Quanwei Liu, and Zhihang Zhuo. 2023. "SMRT Sequencing Technology Was Used to Construct the Batocera horsfieldi (Hope) Transcriptome and Reveal Its Features" Insects 14, no. 7: 625. https://doi.org/10.3390/insects14070625
APA StyleWei, X., Xu, D., Liu, Z., Liu, Q., & Zhuo, Z. (2023). SMRT Sequencing Technology Was Used to Construct the Batocera horsfieldi (Hope) Transcriptome and Reveal Its Features. Insects, 14(7), 625. https://doi.org/10.3390/insects14070625






