Link between Energy Investment in Biosynthesis and Proteostasis: Testing the Cost–Quality Hypothesis in Insects
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Rearing and Sampling
2.2. Cell Sampling and Culture
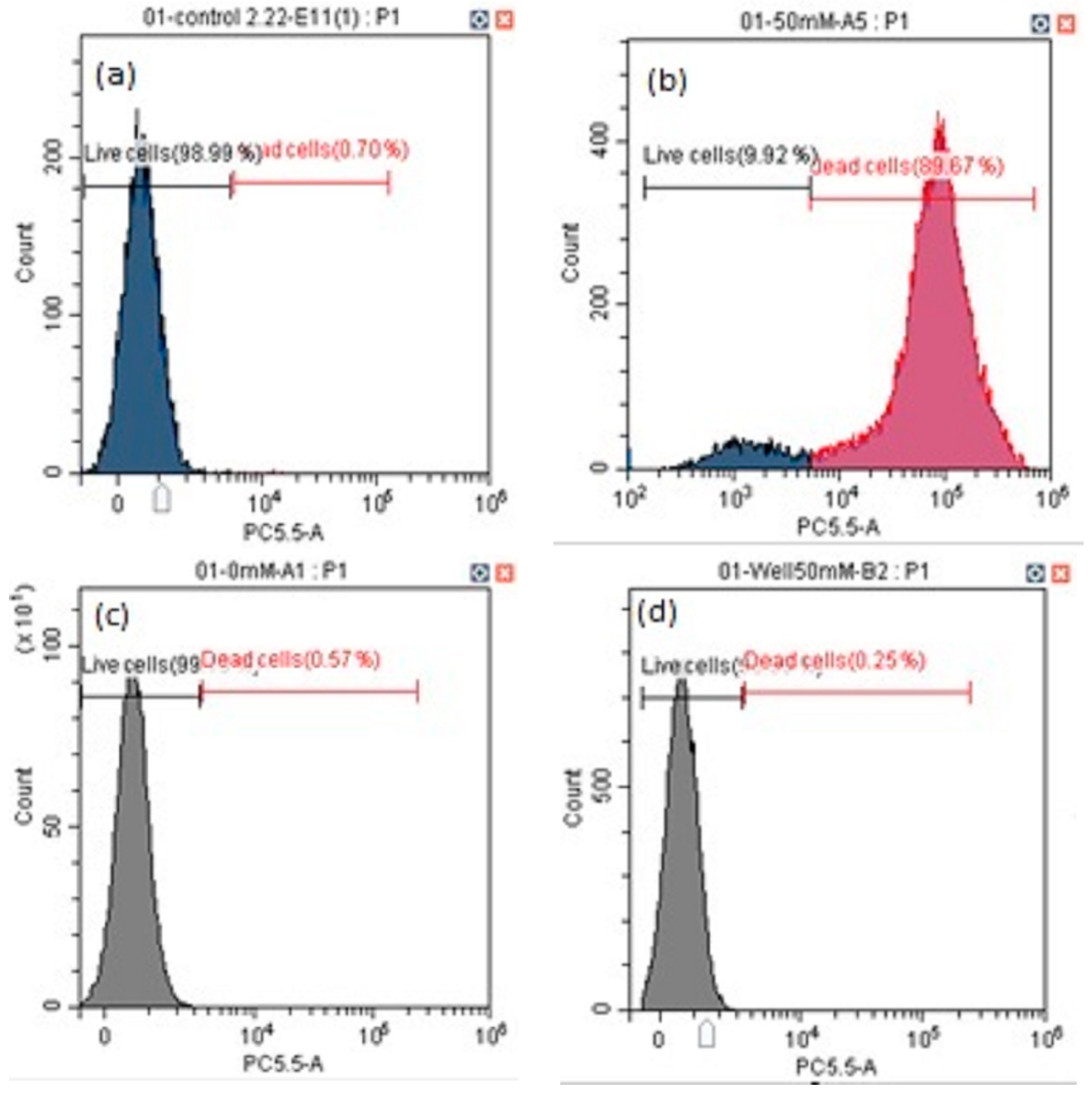
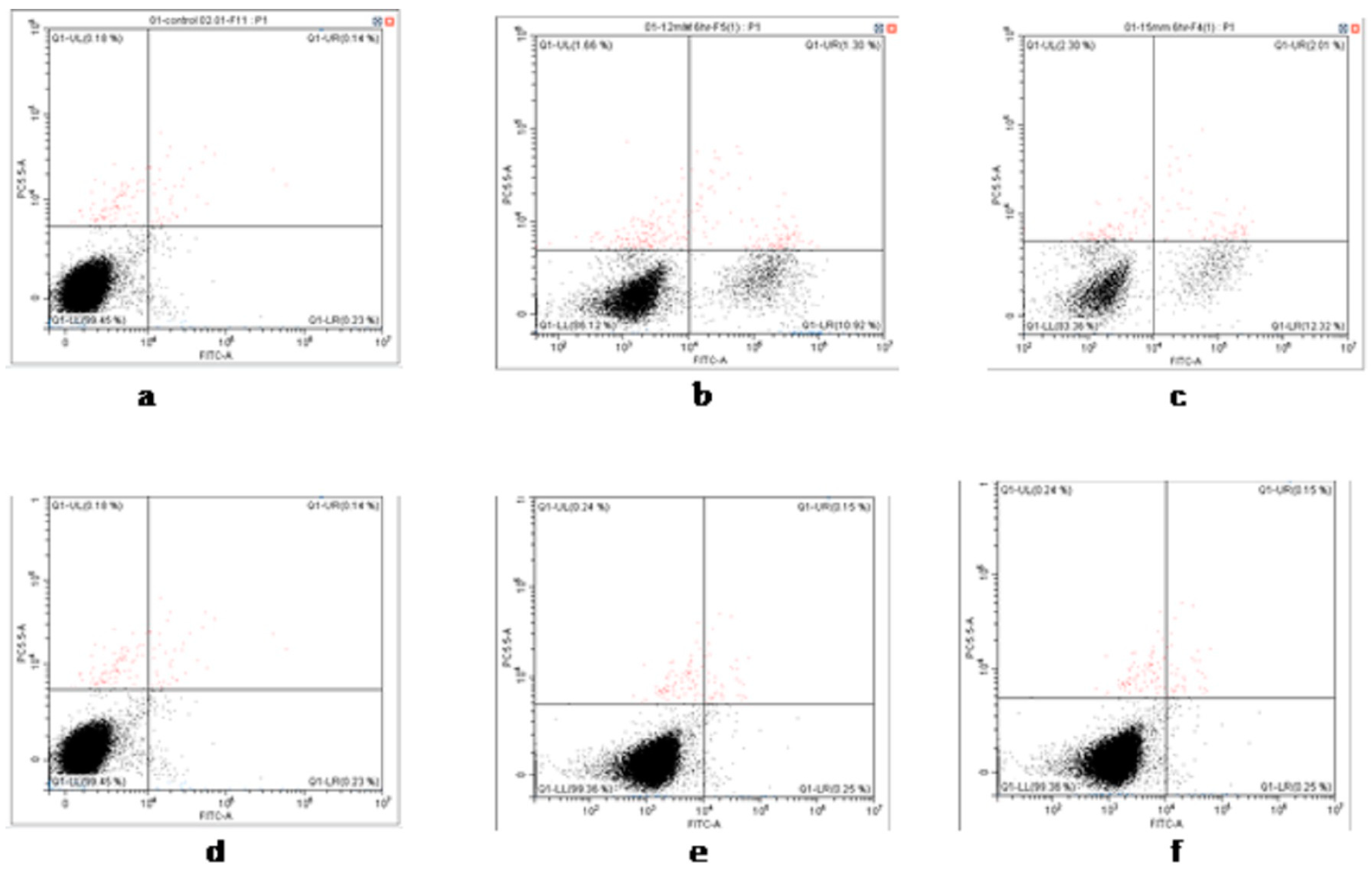
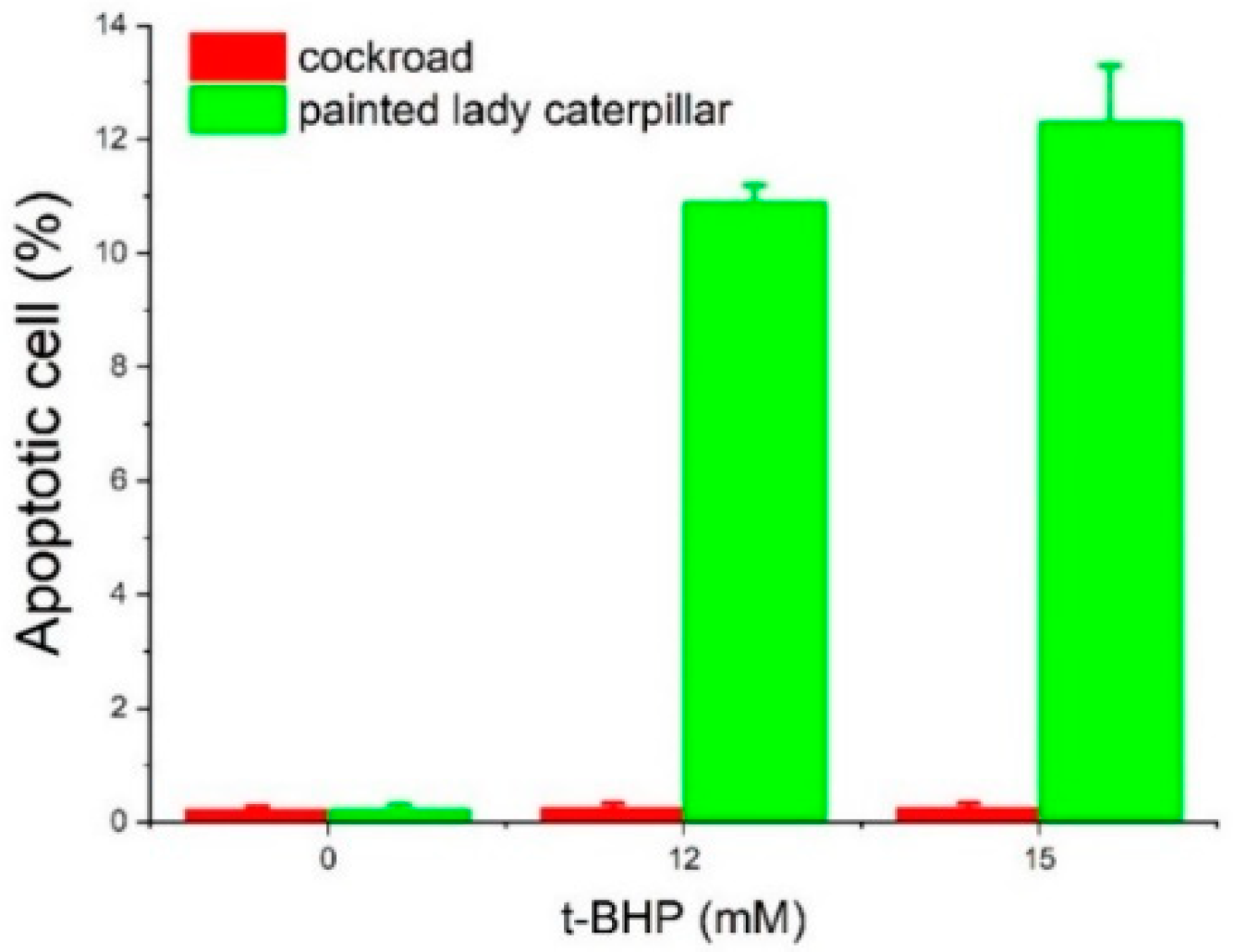
2.3. Cell Viability and Apoptosis
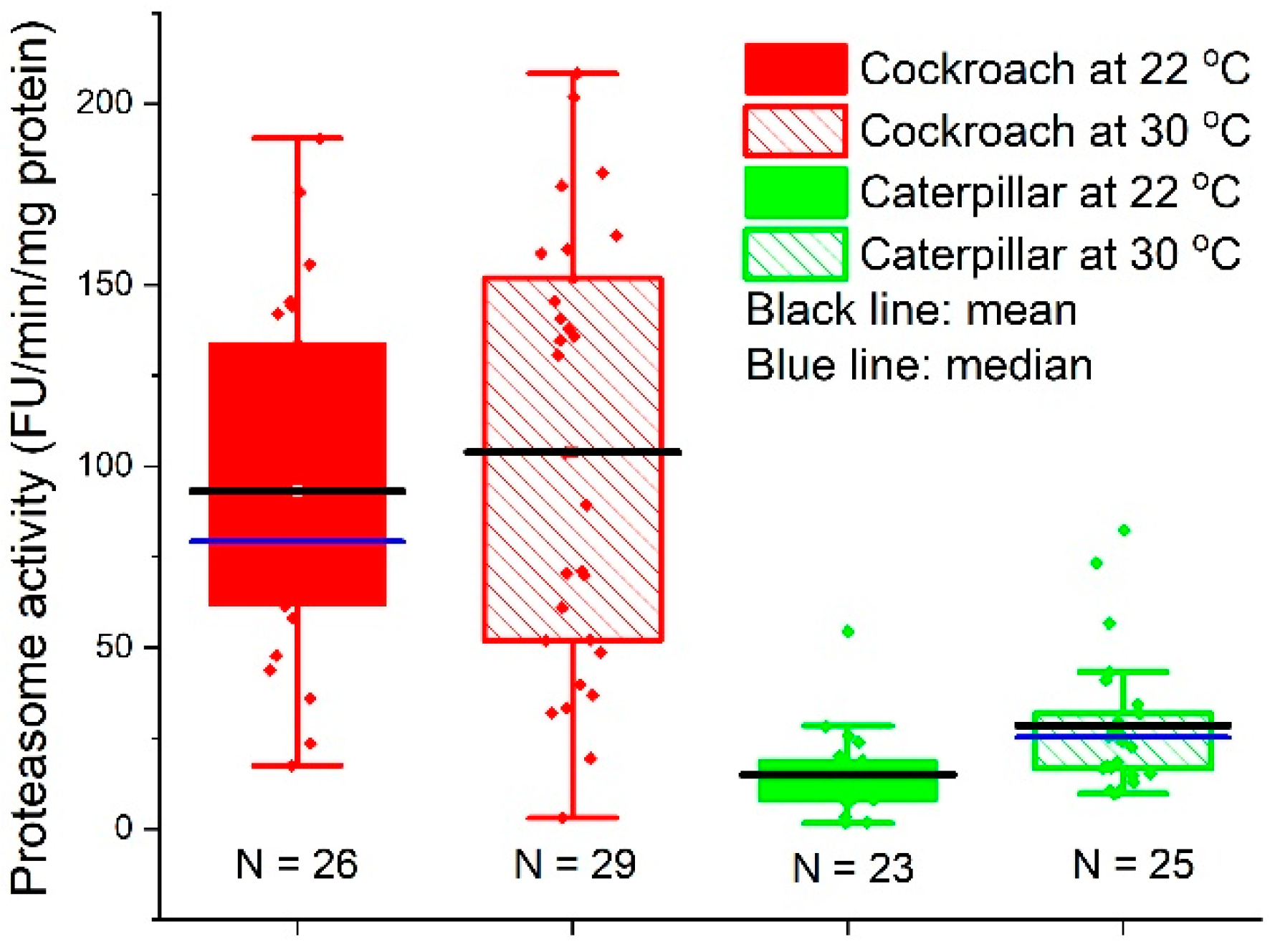
2.4. Proteasome Activity
2.5. Protein Extraction and Quantity
2.6. RNA Extraction and Quantity
2.7. Data Analysis
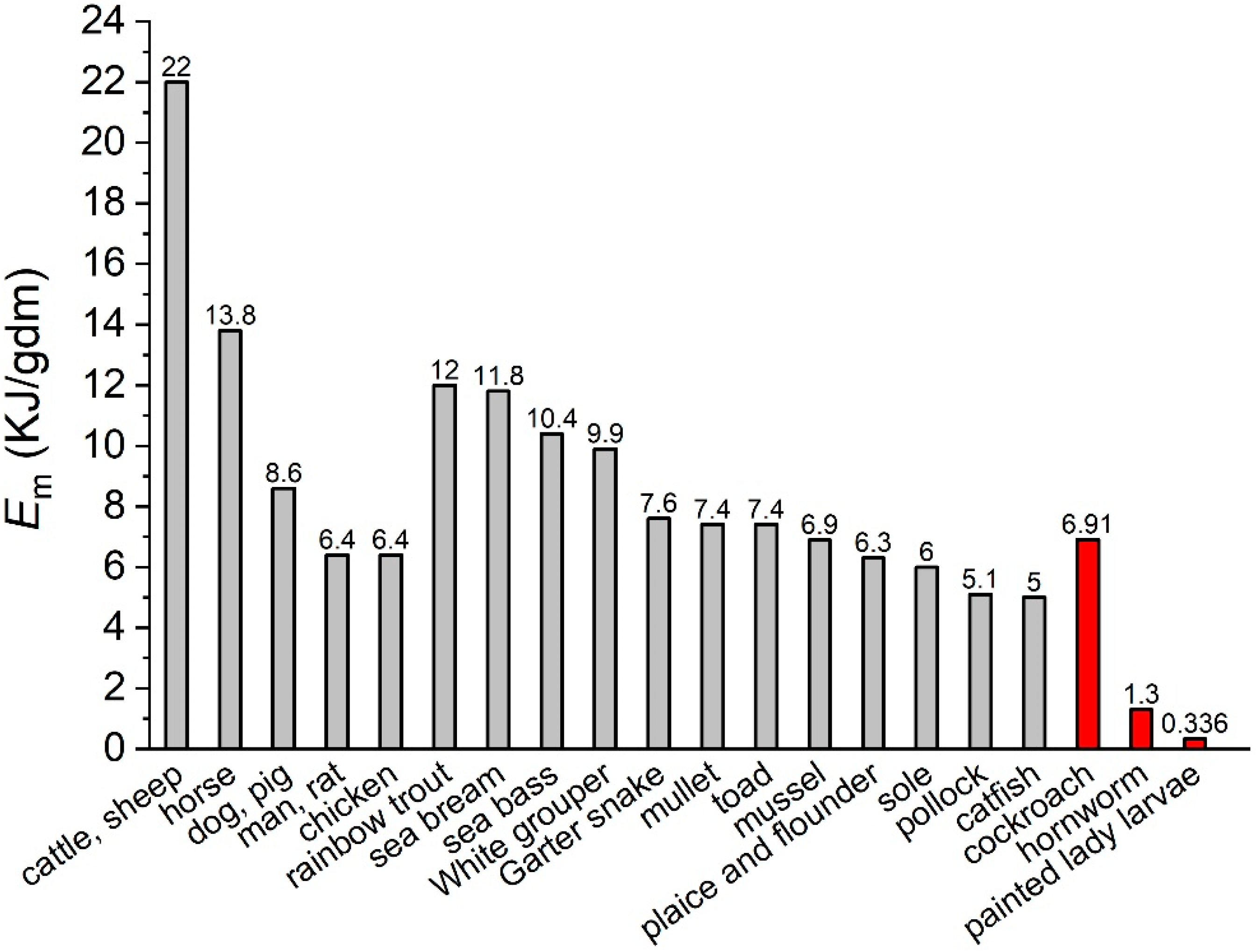
3. Results
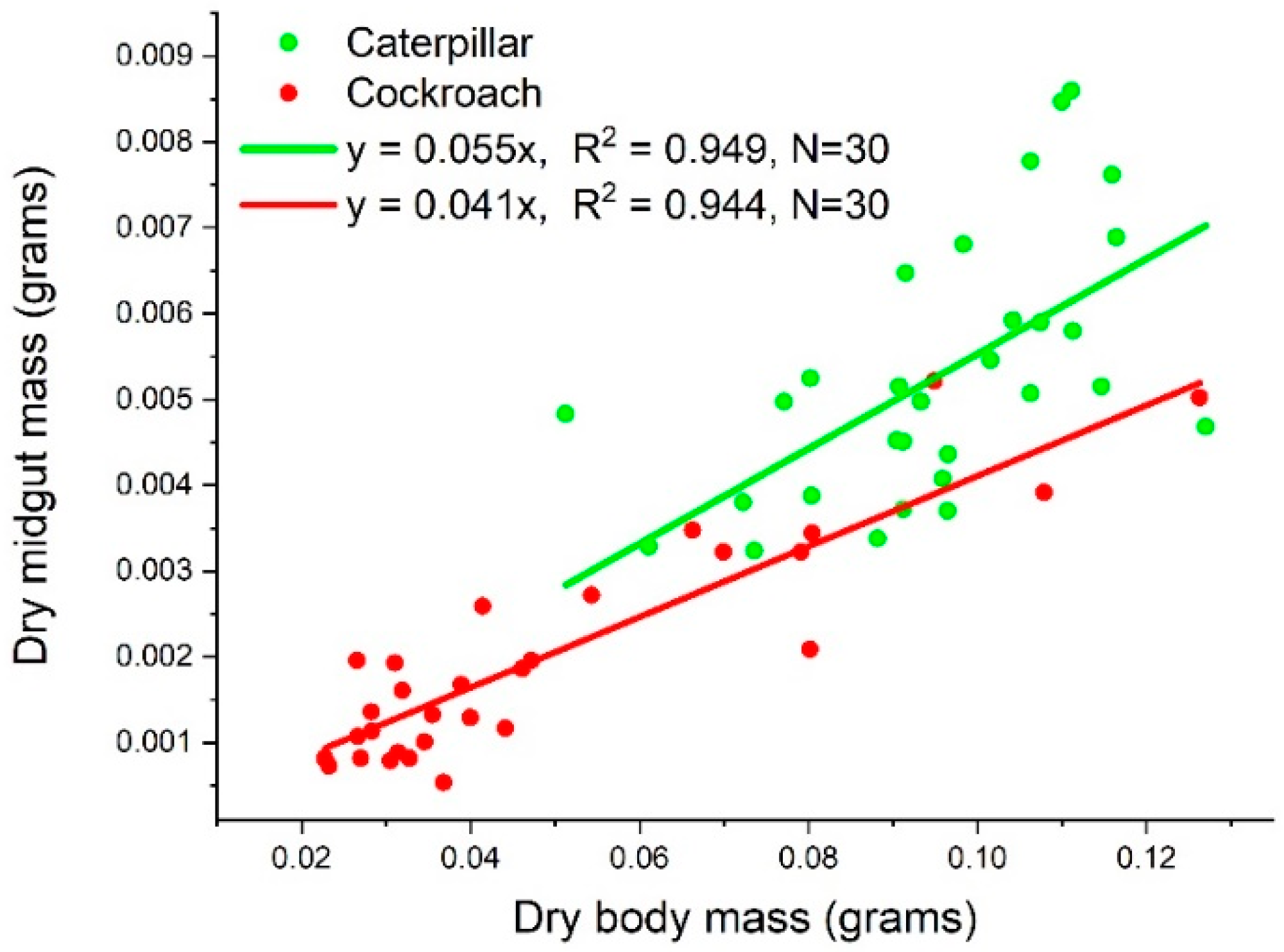
3.1. Dry Midgut Mass
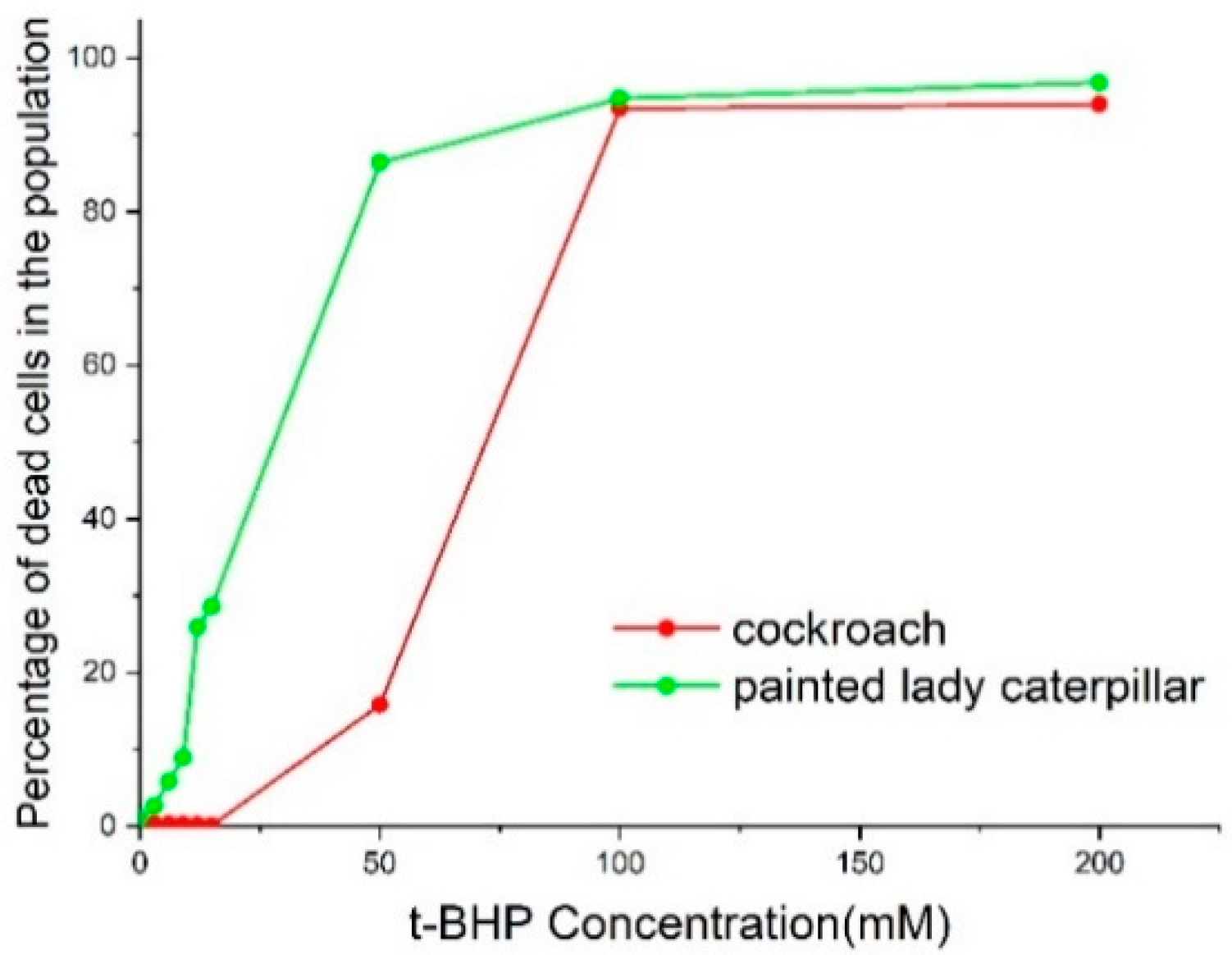
3.2. Cell Viability and Apoptosis
3.3. Proteasome Activity
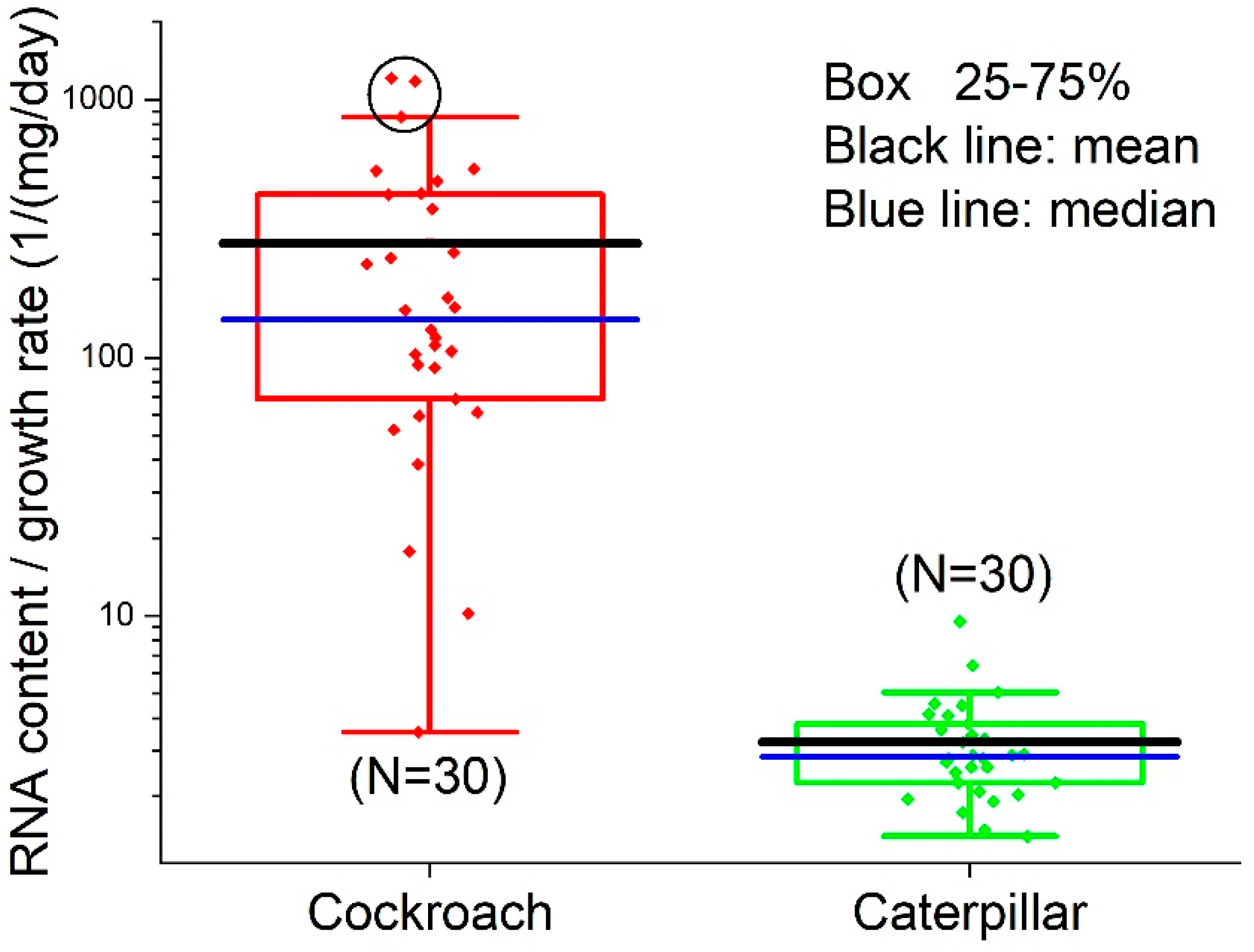
3.4. RNA/Growth Ratio
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stearns, S.C. The Evolution of Life Histories; Oxford University Press: Oxford, UK, 1992. [Google Scholar]
- Lee, W.-S.; Monaghan, P.; Metcalfe, N.B. Experimental demonstration of the growth rate–lifespan trade-off. Proc. R. Soc. B Biol. Sci. 2013, 280, 20122370. [Google Scholar] [CrossRef] [PubMed]
- Monaghan, P.; Metcalfe, N.B.; Torres, R. Oxidative stress as a mediator of life history trade-offs: Mechanisms, measurements and interpretation. Ecol. Lett. 2009, 12, 75–92. [Google Scholar] [CrossRef] [PubMed]
- Kirkwood, T.B.L. The disposable soma theory of aging. In Genetic Effects on Aging II; Harrison, D.E., Ed.; Telford Press: Caldwell, NJ, USA, 1990; pp. 9–19. [Google Scholar]
- Hou, C.; Amunugama, K. On the complex relationship between energy expenditure and longevity: Reconciling the contradictory empirical results with a simple theoretical model. Mech. Ageing Dev. 2015, 149, 50–64. [Google Scholar] [CrossRef] [PubMed]
- Rollo, C.D. Growth negatively impacts the life span of mammals. Evol. Dev. 2002, 4, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Morowitz, H.J. Foundations of Bioenergetics; Academic Press: New York, NJ, USA, 1978. [Google Scholar]
- Hou, C.; Zuo, W.Y.; Moses, M.E.; Woodruff, W.H.; Brown, J.H.; West, G.B. Energy uptake and allocation during ontogeny. Science 2008, 322, 736–739. [Google Scholar] [CrossRef]
- West, G.B.; Brown, J.H.; Enquist, B.J. A general model for ontogenetic growth. Nature 2001, 413, 628–631. [Google Scholar] [CrossRef]
- Moses, M.E.; Hou, C.; Woodruff, W.H.; West, G.B.; Nekola, J.C.; Zuo, W.; Brown, J.H. Revisiting a Model of Ontogenetic Growth: Estimating Model Parameters from Theory and Data. Am. Nat. 2008, 171, 632–645. [Google Scholar] [CrossRef]
- Ferral, N.; Gomez, N.; Holloway, K.; Neeter, H.; Fairfield, M.; Pollman, K.; Huang, Y.W.; Hou, C. The extremely low energy cost of biosynthesis in holometabolous insect larvae. J. Insect Physiol. 2020, 120, 103988. [Google Scholar] [CrossRef]
- Ricklefs, R.E. Energetics of reproduction in birds. In Avian Energetics; Paynter, R.A.J., Ed.; Nuttall Ornithol Club 15: Cambridge, MA, USA, 1974. [Google Scholar]
- Peterson, C.C.; Walton, B.M.; Bennett, A.F. Metabolic costs of growth in free-living Garter Snakes and the energy budgets of ectotherms. Funct. Ecol. 1999, 13, 500–507. [Google Scholar] [CrossRef]
- Wieser, W. Cost of Growth in Cells and Organisms—General Rules and Comparative Aspects. Biol. Rev. Camb. Philos. Soc. 1994, 69, 1–33. [Google Scholar] [CrossRef]
- Calow, P. Conversion Efficiencies in Heterotrophic Organisms. Biol. Rev. 1977, 52, 385–409. [Google Scholar] [CrossRef]
- Brown, J.H.; Gillooly, J.F.; Allen, A.P.; Savage, V.M.; West, G.B. Toward a metabolic theory of ecology. Ecology 2004, 85, 1771–1789. [Google Scholar] [CrossRef]
- van der Meer, J. Metabolic theories in ecology. Trends Ecol. Evol. 2006, 21, 136–140. [Google Scholar] [CrossRef]
- Kooijman, S. Dynamic Energy Budget Theory; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar]
- Bayne, B.L. Physiological Components of Growth Differences between Individual Oysters (Crassostrea gigas) and a Comparison with Saccostrea commercialis. Physiol. Biochem. Zool. 1999, 72, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Lupatsch, I.; Kissil, G.W.; Sklan, D. Comparison of energy and protein efficiency among three fish species gilthead sea bream (Sparus aurata), European sea bass (Dicentrarchus labrax) and white grouper (Epinephelus aeneus): Energy expenditure for protein and lipid deposition. Aquaculture 2003, 225, 175–189. [Google Scholar] [CrossRef]
- Jiao, L.; Amunugama, K.; Hayes, M.; Jennings, M.; Domingo, A.; Hou, C. Food restriction alters energy allocation strategy during growth in tobacco hornworms (Manduca sexta larvae). Sci. Nat. 2015, 102, 40–50. [Google Scholar] [CrossRef]
- Azevedo, P.A.; Young Cho, C.; Leeson, S.; Bureau, D.P. Effects of feeding level and water temperature on growth, nutrient and energy utilization and waste outputs of rainbow trout (Oncorhynchus mykiss). Aquat. Living Resour. 1998, 11, 227–238. [Google Scholar] [CrossRef]
- Rodehutscord, M.; Pfeffer, E. Maintenance requirement for digestible energy and efficiency of utilisation of digestible energy for retention in rainbow trout, Oncorhynchus mykiss. Aquaculture 1999, 179, 95–107. [Google Scholar] [CrossRef]
- Jorgensen, P.; Tyers, M. How Cells Coordinate Growth and Division. Curr. Biol. 2004, 14, R1014–R1027. [Google Scholar] [CrossRef]
- Hawkins, A.; Widdows, J.; Bayne, B. The relevance of whole-body protein metabolism to measured costs of maintenance and growth in Mytilus edulis. Physiol. Zool. 1989, 62, 745–763. [Google Scholar] [CrossRef]
- Sears, K.E.; Kerkhoff, A.J.; Messerman, A.; Itagaki, H. Ontogenetic Scaling of Metabolism, Growth, and Assimilation: Testing Metabolic Scaling Theory with Manduca sexta Larvae. Physiol. Biochem. Zool. 2012, 85, 159–173. [Google Scholar] [CrossRef]
- Blaxter, K.L. Energy Metabolism in Animals and Man; Cambridge University Press: Cambridge, UK, 1989. [Google Scholar]
- Aldaz, S.; Escudero, L.M. Imaginal discs. Curr. Biol. 2010, 20, R429–R431. [Google Scholar] [CrossRef] [PubMed]
- Madhavan, M.M.; Schneiderman, H.A. Histological analysis of the dynamics of growth of imaginal discs and histoblast nests during the larval development of Drosophila melanogaster. Wilhelm Roux’s Arch. Dev. Biol. 1977, 183, 269–305. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.; Garwood, R.J.; Simonsen, T.J.; Bradley, R.S.; Withers, P.J. Metamorphosis revealed: Time-lapse three-dimensional imaging inside a living chrysalis. J. R. Soc. Interface 2013, 10, 20130304. [Google Scholar] [CrossRef] [PubMed]
- Lockshin, R.A. Cell death in metamorphosis. In Cell Death in Biology and Pathology; Bowen, I.D., Lockshin, R.A., Eds.; Springer: Dordrecht, The Netherlands, 1981; pp. 79–121. [Google Scholar]
- Locke, M. Cell Structure during Insect Metamorphosis. In Metamorphosis: A Problem in Developmental Biology; Gilbert, L.I., Frieden, E., Eds.; Springer: Boston, MA, USA, 1981; pp. 75–103. [Google Scholar]
- D’Amico, L.J.; Davidowitz, G.; Nijhout, H.F. The developmental and physiological basis of body size evolution in an insect. Proc. R. Soc. London. Ser. B Biol. Sci. 2001, 268, 1589–1593. [Google Scholar] [CrossRef] [PubMed]
- Davidowitz, G.; D’Amico, L.J.; Nijhout, H.F. Critical weight in the development of insect body size. Evol. Dev. 2003, 5, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Honěk, A. Intraspecific variation in body size and fecundity in insects: A general relationship. Oikos 1993, 66, 483–492. [Google Scholar] [CrossRef]
- Kafri, M.; Metzl-Raz, E.; Jona, G.; Barkai, N. The Cost of Protein Production. Cell Rep. 2016, 14, 22–31. [Google Scholar] [CrossRef]
- Swire, J. Selection on Synthesis Cost Affects Interprotein Amino Acid Usage in All Three Domains of Life. J. Mol. Evol. 2007, 64, 558–571. [Google Scholar] [CrossRef]
- Akashi, H.; Gojobori, T. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2002, 99, 3695–3700. [Google Scholar] [CrossRef]
- Ponnuswamy, P.K.; Muthusamy, R.; Manavalan, P. Amino acid composition and thermal stability of proteins. Int. J. Biol. Macromol. 1982, 4, 186–190. [Google Scholar] [CrossRef]
- Argos, P.; Rossman, M.G.; Grau, U.M.; Zuber, H.; Frank, G.; Tratschin, J.D. Thermal stability and protein structure. Biochemistry 1979, 18, 5698–5703. [Google Scholar] [CrossRef] [PubMed]
- Jakubowski, H. Energy Cost of Translational Proofreading In Vivo. Aminoacylation Transf. RNA Escherichia Coli 1994, 745, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Blomberg, C. Thermodynamic aspects on accuracy in the synthesis of biomolecules. Int. J. Quantum Chem. 1983, 23, 687–707. [Google Scholar] [CrossRef]
- Cochella, L.; Green, R. Fidelity in protein synthesis. Curr. Biol. 2005, 15, R536–R540. [Google Scholar] [CrossRef]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324. [Google Scholar] [CrossRef]
- Chen, B.; Retzlaff, M.; Roos, T.; Frydman, J. Cellular strategies of protein quality control. Cold Spring Harb. Perspect. Biol. 2011, 3, a004374. [Google Scholar] [CrossRef]
- Powers, E.T.; Morimoto, R.I.; Dillin, A.; Kelly, J.W.; Balch, W.E. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 2009, 78, 959–991. [Google Scholar] [CrossRef]
- Richter, K.; Haslbeck, M.; Buchner, J. The Heat Shock Response: Life on the Verge of Death. Mol. Cell 2010, 40, 253–266. [Google Scholar] [CrossRef]
- Amm, I.; Sommer, T.; Wolf, D.H. Protein quality control and elimination of protein waste: The role of the ubiquitin–proteasome system. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Res. 2014, 1843, 182–196. [Google Scholar] [CrossRef]
- Tyedmers, J.; Mogk, A.; Bukau, B. Cellular strategies for controlling protein aggregation. Nat. Rev. Mol. Cell Biol. 2010, 11, 777–788. [Google Scholar] [CrossRef]
- Fredrickson, E.K.; Gardner, R.G. Selective destruction of abnormal proteins by ubiquitin-mediated protein quality control degradation. In Seminars in Cell & Developmental Biology; Academic Press: Cambridge, MA, USA, 2012; pp. 530–537. [Google Scholar]
- Åkerfelt, M.; Morimoto, R.I.; Sistonen, L. Heat shock factors: Integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 2010, 11, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Jung, T.; Grune, T. The proteasome and the degradation of oxidized proteins: Part I—Structure of proteasomes. Redox Biol. 2013, 1, 178–182. [Google Scholar] [CrossRef] [PubMed]
- Jung, T.; Höhn, A.; Grune, T. The proteasome and the degradation of oxidized proteins: Part II–protein oxidation and proteasomal degradation. Redox Biol. 2014, 2, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Santra, M.; Farrell, D.W.; Dill, K.A. Bacterial proteostasis balances energy and chaperone utilization efficiently. Proc. Natl. Acad. Sci. USA 2017, 114, E2654–E2661. [Google Scholar] [CrossRef]
- Salway, K.D.; Gallagher, E.J.; Page, M.M.; Stuart, J.A. Higher levels of heat shock proteins in longer-lived mammals and birds. Mech. Ageing Dev. 2011, 132, 287–297. [Google Scholar] [CrossRef]
- Dahl, J.-U.; Gray, M.J.; Jakob, U. Protein quality control under oxidative stress conditions. J. Mol. Biol. 2015, 427, 1549–1563. [Google Scholar] [CrossRef]
- Reichmann, D.; Voth, W.; Jakob, U. Maintaining a healthy proteome during oxidative stress. Mol. Cell 2018, 69, 203–213. [Google Scholar] [CrossRef]
- Fulda, S.; Gorman, A.M.; Hori, O.; Samali, A. Cellular stress responses: Cell survival and cell death. Int. J. Cell Biol. 2010, 2010, 214074. [Google Scholar] [CrossRef]
- Elser, J.; Acharya, K.; Kyle, M.; Cotner, J.; Makino, W.; Markow, T.; Watts, T.; Hobbie, S.; Fagan, W.; Schade, J. Growth rate–stoichiometry couplings in diverse biota. Ecol. Lett. 2003, 6, 936–943. [Google Scholar] [CrossRef]
- Isanta-Navarro, J.; Prater, C.; Peoples, L.M.; Loladze, I.; Phan, T.; Jeyasingh, P.D.; Church, M.J.; Kuang, Y.; Elser, J.J. Revisiting the growth rate hypothesis: Towards a holistic stoichiometric understanding of growth. Ecol. Lett. 2022, 25, 2324–2339. [Google Scholar] [CrossRef] [PubMed]
- Hakim, R.S.; Caccia, S.; Loeb, M.; Smagghe, G. Primary culture of insect midgut cells. Vitr. Cell. Dev. Biol.-Anim. 2009, 45, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Mandrioli, M.; Monti, M.; Tedeschi, R. A practical guide to insect cell cultures: Establishment and maintenance of primary cell cultures. Halteres 2015, 6, 132–141. [Google Scholar]
- Breusing, N.; Arndt, J.; Voss, P.; Bresgen, N.; Wiswedel, I.; Gardemann, A.; Siems, W.; Grune, T. Inverse correlation of protein oxidation and proteasome activity in liver and lung. Mech. Ageing Dev. 2009, 130, 748–753. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.-Y.; Medhurst, A.; Jackson, M.; Rose, S.; Jenner, P. Proteasomal activity in brain differs between species and brain regions and changes with age. Mech. Ageing Dev. 2005, 126, 760–766. [Google Scholar] [CrossRef] [PubMed]
- Lima, C.F.; Rattan, S.I. Determination of proteasomal activities. In Protein Misfolding and Cellular Stress in Disease and Aging; Springer: Berlin/Heidelberg, Germany, 2010; pp. 183–192. [Google Scholar]
- Friguet, B. Oxidized protein degradation and repair in ageing and oxidative stress. FEBS Lett. 2006, 580, 2910–2916. [Google Scholar] [CrossRef] [PubMed]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The Hallmarks of Aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- Basisty, N.; Meyer, J.G.; Schilling, B. Protein turnover in aging and longevity. Proteomics 2018, 18, 1700108. [Google Scholar] [CrossRef] [PubMed]
- Hipkiss, A.R. Error-protein metabolism and ageing. Biogerontology 2008, 10, 523–529. [Google Scholar] [CrossRef]
- Bozaykut, P.; Ozer, N.K.; Karademir, B. Regulation of protein turnover by heat shock proteins. Free Radic. Biol. Med. 2014, 77, 195–209. [Google Scholar] [CrossRef]
- McClellan, A.J.; Tam, S.; Kaganovich, D.; Frydman, J. Protein quality control: Chaperones culling corrupt conformations. Nat. Cell Biol. 2005, 7, 736–741. [Google Scholar] [CrossRef]
- Kepp, K.P.; Dasmeh, P. A Model of Proteostatic Energy Cost and Its Use in Analysis of Proteome Trends and Sequence Evolution. PLoS ONE 2014, 9, e90504. [Google Scholar] [CrossRef]
- Fraser, K.P.; Rogers, A.D. Protein metabolism in marine animals: The underlying mechanism of growth. Adv. Mar. Biol. 2007, 52, 267–362. [Google Scholar] [PubMed]
- Rosenberger, R.; Carr, A.; Hipkiss, A. Regulation of breakdown of canavanyl proteins in Escherichia coli by growth conditions in Ion+ and Ion− cells. FEMS Microbiol. Lett. 1990, 68, 19–25. [Google Scholar] [CrossRef]
- Hipkiss, A.R. On why decreasing protein synthesis can increase lifespan. Mech. Ageing Dev. 2007, 128, 412–414. [Google Scholar] [CrossRef] [PubMed]
- Chondrogianni, N.; Petropoulos, I.; Grimm, S.; Georgila, K.; Catalgol, B.; Friguet, B.; Grune, T.; Gonos, E.S. Protein damage, repair and proteolysis. Mol. Asp. Med. 2014, 35, 1–71. [Google Scholar] [CrossRef] [PubMed]
- Sterner, R.W.; Elser, J.J. Ecological Stoichiometry; Princeton University Press: Princeton, NJ, USA, 2017. [Google Scholar]
- Elser, J.J.; Dobberfuhl, D.R.; MacKay, N.A.; Schampel, J.H. Organism size, life history, and N: P stoichiometry: Toward a unified view of cellular and ecosystem processes. BioScience 1996, 46, 674–684. [Google Scholar] [CrossRef]
- van der Meer, J. An introduction to Dynamic Energy Budget (DEB) models with special emphasis on parameter estimation. J. Sea Res. 2006, 56, 85–102. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iromini, T.; Tang, X.; Holloway, K.N.; Hou, C. Link between Energy Investment in Biosynthesis and Proteostasis: Testing the Cost–Quality Hypothesis in Insects. Insects 2023, 14, 241. https://doi.org/10.3390/insects14030241
Iromini T, Tang X, Holloway KN, Hou C. Link between Energy Investment in Biosynthesis and Proteostasis: Testing the Cost–Quality Hypothesis in Insects. Insects. 2023; 14(3):241. https://doi.org/10.3390/insects14030241
Chicago/Turabian StyleIromini, Taiwo, Xiaolong Tang, Kyara N. Holloway, and Chen Hou. 2023. "Link between Energy Investment in Biosynthesis and Proteostasis: Testing the Cost–Quality Hypothesis in Insects" Insects 14, no. 3: 241. https://doi.org/10.3390/insects14030241
APA StyleIromini, T., Tang, X., Holloway, K. N., & Hou, C. (2023). Link between Energy Investment in Biosynthesis and Proteostasis: Testing the Cost–Quality Hypothesis in Insects. Insects, 14(3), 241. https://doi.org/10.3390/insects14030241





