Bacteriota and Antibiotic Resistance in Spiders
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Identification of Microbiota
2.3. Antimicrobial Resistance Testing
2.4. Statistical Analyses
3. Results
3.1. Qualitative Analysis of Isolated Microbiota from Spiders
3.2. Isolated Microbial Genera and Microbial Species from Spider Specimens
3.3. Antibiotic Resistance of Isolated Microbial Species of Spiders
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koide, R. Functional complementarity in the arbuscular mycorrhizal symbiosis. New Phytol. 2000, 147, 233–235. [Google Scholar] [CrossRef]
- Selosse, M.-A.; Baudoin, E.; Vandenkoornhuyse, P. Symbiotic microorganisms, a key for ecological success and protection of plants. C. R. Biol. 2004, 327, 639–648. [Google Scholar] [CrossRef]
- Peirano, A. In vivo measurements of the seasonal photosynthetic fluorescence of the Mediterranean coral Cladocora caespitosa (L.). Sci. Mar. 2007, 71, 629–635. [Google Scholar] [CrossRef][Green Version]
- Tripp, E.A.; Zhang, N.; Schneider, H.; Huang, Y.; Mueller, G.M.; Hu, Z.; Häggblom, M.; Bhattacharya, D. Reshaping Darwin’s Tree: Impact of the Symbiome. Trends Ecol. Evol. 2017, 32, 552–555. [Google Scholar] [CrossRef] [PubMed]
- Bredon, M.; Herran, B.; Bertaux, J.; Grève, P.; Moumen, B.; Bouchon, D. Isopod holobionts as promising models for lignocellulose degradation. Biotechnol. Biofuels 2020, 13, 49. [Google Scholar] [CrossRef]
- Suárez, J. The stability of traits conception of the hologenome: An evolutionary account of holobiont individuality. Hist. Philos. Life Sci. 2020, 42, 11. [Google Scholar] [CrossRef]
- Suárez, J.; Triviño, V. What Is a Hologenomic Adaptation? Emergent Individuality and Inter-Identity in Multispecies Systems. Front. Psychol. 2020, 11, 187. [Google Scholar] [CrossRef] [PubMed]
- Bili, M.; Cortesero, A.M.; Mougel, C.; Gauthier, J.P.; Ermel, G.; Simon, J.C.; Outreman, Y.; Terrat, S.; Mahéo, F.; Poinsot, D. Bacterial Community Diversity Harboured by Interacting Species. PLoS ONE 2016, 11, e0155392. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, C.C.; Villegas, L.E.M.; Campolina, T.B.; Pires, A.C.M.A.; Miranda, J.C.; Pimenta, P.F.P.; Secundino, N.F.C. Bacterial diversity of the American sand fly Lutzomyia intermedia using high-throughput metagenomic sequencing. Parasit. Vectors 2016, 9, 480. [Google Scholar] [CrossRef] [PubMed]
- Trout Fryxell, R.T.; DeBruyn, J.M. The Microbiome of Ehrlichia-Infected and Uninfected Lone Star Ticks (Amblyomma americanum). PLoS ONE 2016, 11, e0146651. [Google Scholar] [CrossRef]
- Gotoh, T.; Noda, H.; Ito, S. Cardinium symbionts cause cytoplasmic incompatibility in spider mites. Heredity 2007, 98, 13–20. [Google Scholar] [CrossRef]
- Brownlie, J.C.; Cass, B.N.; Riegler, M.; Witsenburg, J.J.; Iturbe-Ormaetxe, I.; McGraw, E.A.; O’Neill, S.L. Evidence for Metabolic Provisioning by a Common Invertebrate Endosymbiont, Wolbachia pipientis, during Periods of Nutritional Stress. PLoS Pathog. 2009, 5, e1000368. [Google Scholar] [CrossRef] [PubMed]
- Himler, A.G.; Adachi-Hagimori, T.; Bergen, J.E.; Kozuch, A.; Kelly, S.E.; Tabashnik, B.E.; Chiel, E.; Duckworth, V.E.; Dennehy, T.J.; Zchori-Fein, E.; et al. Rapid Spread of a Bacterial Symbiont in an Invasive Whitefly Is Driven by Fitness Benefits and Female Bias. Science 2011, 332, 254–256. [Google Scholar] [CrossRef]
- Hajdamowicz, I.; Rozwałka, R.; Stańska, M.; Rutkowski, T.; Sienkiewicz, P. Xerophilic Alopecosa sulzeri (Pavesi, 1873) (Araneae: Lycosidae)—A new wolf spider species in Poland. Zootaxa 2020, 4899, 175–185. [Google Scholar] [CrossRef]
- Rozwałka, R.; Rutkowski, T.; Sienkiewicz, P.; Renn, K. Occurrence of Talavera aperta (Miller, 1971) (Araneae: Salticidae) in Poland. Biol. Lett. 2015, 52, 3–9. [Google Scholar] [CrossRef][Green Version]
- Busck, M.M.; Settepani, V.; Bechsgaard, J.; Lund, M.B.; Bilde, T.; Schramm, A. Microbiomes and Specific Symbionts of Social Spiders: Compositional Patterns in Host Species, Populations, and Nests. Front. Microbiol. 2020, 11, 1845. [Google Scholar] [CrossRef]
- Goodacre, S.L.; Martin, O.Y.; Thomas, C.F.G.; Hewitt, G.M. Wolbachia and other endosymbiont infections in spiders. Mol. Ecol. 2006, 15, 517–527. [Google Scholar] [CrossRef]
- Duron, O.; Hurst, G.D.D.; Hornett, E.A.; Josling, J.A.; Engelstädter, J. High incidence of the maternally inherited bacterium Cardinium in spiders. Mol. Ecol. 2008, 17, 1427–1437. [Google Scholar] [CrossRef]
- Martin, O.Y.; Goodacre, S.L. Widespread infections by the bacterial endosymbiont Cardinium in Arachnids. J. Arachnol. 2009, 37, 106–108. [Google Scholar] [CrossRef]
- Hu, G.; Zhang, L.; Yun, Y.; Peng, Y. Taking insight into the gut microbiota of three spider species: No characteristic symbiont was found corresponding to the special feeding style of spiders. Ecol. Evol. 2019, 9, 8146–8156. [Google Scholar] [CrossRef]
- Rivera, P.; Stork, R.; Hug, A. A First Look at the Microbial Community of Rabidosa rabida, a Wolf Spider in Searcy, Arkansas. J. Ark. Acad. Sci. 2017, 71, 51–55. [Google Scholar] [CrossRef]
- Foster, T. Staphylococcus. In Medical Microbiology; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996. [Google Scholar]
- Shi, Y.; Lou, K.; Li, C. Growth and photosynthetic efficiency promotion of sugar beet (Beta vulgaris L.) by endophytic bacteria. Photosynth. Res. 2010, 105, 5–13. [Google Scholar] [CrossRef]
- Muturi, E.J.; Ramirez, J.L.; Rooney, A.P.; Kim, C.-H. Comparative analysis of gut microbiota of mosquito communities in central Illinois. PLoS Negl. Trop. Dis. 2017, 11, e0005377. [Google Scholar] [CrossRef] [PubMed]
- Hald, B.; Skovgård, H.; Pedersen, K.; Bunkenborg, H. Influxed Insects as Vectors for Campylobacter jejuni and Campylobacter coli in Danish Broiler Houses. Poult. Sci. 2008, 87, 1428–1434. [Google Scholar] [CrossRef]
- Martínez, J.L. Effect of antibiotics on bacterial populations: A multi-hierarchical selection process. F1000Research 2017, 6, 51. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Chen, Q.-L.; Li, H.; Yang, X.-R.; Christie, P.; Ke, X.; Zhu, Y.-G. Land Use Influences Antibiotic Resistance in the Microbiome of Soil Collembolans Orchesellides sinensis. Environ. Sci. Technol. 2018, 52, 14088–14098. [Google Scholar] [CrossRef]
- Kaňa, J.; Tahovská, K.; Kopáček, J. Response of soil chemistry to forest dieback after bark beetle infestation. Biogeochemistry 2013, 113, 369–383. [Google Scholar] [CrossRef]
- EUCAST The European Committee on Antimicrobial Susceptibility Testing. Available online: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.pdf (accessed on 3 March 2022).
- Zilber-Rosenberg, I.; Rosenberg, E. Role of microorganisms in the evolution of animals and plants: The hologenome theory of evolution. FEMS Microbiol. Rev. 2008, 32, 723–735. [Google Scholar] [CrossRef]
- Ezenwa, V.O.; Gerardo, N.M.; Inouye, D.W.; Medina, M.; Xavier, J.B. Animal Behavior and the Microbiome. Science 2012, 338, 198–199. [Google Scholar] [CrossRef]
- McFall-Ngai, M.; Hadfield, M.G.; Bosch, T.C.G.; Carey, H.V.; Domazet-Lošo, T.; Douglas, A.E.; Dubilier, N.; Eberl, G.; Fukami, T.; Gilbert, S.F.; et al. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. USA 2013, 110, 3229–3236. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Ghosh, A.; Schal, C.; Zurek, L. Insects in confined swine operations carry a large antibiotic resistant and potentially virulent enterococcal community. BMC Microbiol. 2011, 11, 23. [Google Scholar] [CrossRef]
- Voloshyn, V.; Tymchuk, K.; Symochko, L.; Kačániová, M.; Fedoriak, M. Spiders and other Arthropods of Chernivtsi Poultry Farm (Ukraine) and The Preliminary Data About Bacteria Inhabiting Their External Surfaces. Int. J. Ecosyst. Ecol. Sci. 2017, 7, 587–596. [Google Scholar]
- Dillon, R.J.; Dillon, V.M. The gut bacteria of insects: Nonpathogenic interactions. Annu. Rev. Entomol. 2004, 49, 71–92. [Google Scholar] [CrossRef]
- Engel, P.; Moran, N.A. The gut microbiota of insects–diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef] [PubMed]
- Azambuja, P.; Garcia, E.S.; Ratcliffe, N.A. Gut microbiota and parasite transmission by insect vectors. Trends Parasitol. 2005, 21, 568–572. [Google Scholar] [CrossRef]
- Zhang, L.; Yun, Y.; Hu, G.; Peng, Y. Insights into the bacterial symbiont diversity in spiders. Ecol. Evol. 2018, 8, 4899–4906. [Google Scholar] [CrossRef]
- Sheffer, M.M.; Uhl, G.; Prost, S.; Lueders, T.; Urich, T.; Bengtsson, M.M. Tissue- and Population-Level Microbiome Analysis of the Wasp Spider Argiope bruennichi Identified a Novel Dominant Bacterial Symbiont. Microorganisms 2019, 8, 8. [Google Scholar] [CrossRef]
- White, J.A.; Styer, A.; Rosenwald, L.C.; Curry, M.M.; Welch, K.D.; Athey, K.J.; Chapman, E.G. Endosymbiotic Bacteria Are Prevalent and Diverse in Agricultural Spiders. Microb. Ecol. 2020, 79, 472–481. [Google Scholar] [CrossRef] [PubMed]
- Kavanagh, K.; Reeves, E.P. Insect and Mammalian Innate Immune Responses Are Much Alike. Microbe 2007, 2, 596–599. [Google Scholar] [CrossRef]
- Savitzky, A.H.; Mori, A.; Hutchinson, D.A.; Saporito, R.A.; Burghardt, G.M.; Lillywhite, H.B.; Meinwald, J. Sequestered defensive toxins in tetrapod vertebrates: Principles, patterns, and prospects for future studies. Chemoecology 2012, 22, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Baxter, R.H.G.; Contet, A.; Krueger, K. Arthropod Innate Immune Systems and Vector-Borne Diseases. Biochemistry 2017, 56, 907–918. [Google Scholar] [CrossRef] [PubMed]
- Peel, M.M.; Alfredson, D.A.; Gerrard, J.G.; Davis, J.M.; Robson, J.M.; McDougall, R.J.; Scullie, B.L.; Akhurst, R.J. Isolation, Identification, and Molecular Characterization of Strains of Photorhabdus luminescens from Infected Humans in Australia. J. Clin. Microbiol. 1999, 37, 3647–3653. [Google Scholar] [CrossRef]
- Monteiro, C.L.B.; Rubel, R.; Cogo, L.L.; Mangili, O.C.; Gremski, W.; Veiga, S.S. Isolation and identification of Clostridium perfringens in the venom and fangs of Loxosceles intermedia (brown spider): Enhancement of the dermonecrotic lesion in loxoscelism. Toxicon 2002, 40, 409–418. [Google Scholar] [CrossRef]
- Ahrens, B. Bacterial Etiology of Necrotic Arachnidism in Black Widow Spider Bites. J. Clin. Toxicol. 2011, 1, 106. [Google Scholar] [CrossRef]
- Dunbar, J.P.; Khan, N.A.; Abberton, C.L.; Brosnan, P.; Murphy, J.; Afoullouss, S.; O’Flaherty, V.; Dugon, M.M.; Boyd, A. Synanthropic spiders, including the global invasive noble false widow Steatoda nobilis, are reservoirs for medically important and antibiotic resistant bacteria. Sci. Rep. 2020, 10, 20916. [Google Scholar] [CrossRef] [PubMed]
- Giordano, N.; Corallo, C.; Miracco, C.; Papakostas, P.; Montella, A.; Figura, N.; Nuti, R. Erythema nodosum associated with Staphylococcus xylosus septicemia. J. Microbiol. Immunol. Infect. 2016, 49, 134–137. [Google Scholar] [CrossRef]
- Premkrishnan, B.N.V.; Junqueira, A.C.M.; Uchida, A.; Purbojati, R.W.; Houghton, J.N.I.; Chénard, C.; Wong, A.; Kolundžija, S.; Clare, M.E.; Kushwaha, K.K.; et al. Complete Genome Sequence of Staphylococcus haemolyticus Type Strain SGAir0252. Genome Announc. 2018, 6, e00229-18. [Google Scholar] [CrossRef] [PubMed]
- Pain, M.; Hjerde, E.; Klingenberg, C.; Cavanagh, J.P. Comparative Genomic Analysis of Staphylococcus haemolyticus Reveals Key to Hospital Adaptation and Pathogenicity. Front. Microbiol. 2019, 10, 2096. [Google Scholar] [CrossRef]
- Dunbar, J.P.; Sulpice, R.; Dugon, M.M. The kiss of (cell) death: Can venom-induced immune response contribute to dermal necrosis following arthropod envenomations? Clin. Toxicol. 2019, 57, 677–685. [Google Scholar] [CrossRef]
- González-Tokman, D.; Ruch, J.; Pulpitel, T.; Ponton, F. Cuticular Antifungals in Spiders: Density- and Condition Dependence. PLoS ONE 2014, 9, e91785. [Google Scholar] [CrossRef]
- Rosengaus, R.B.; Maxmen, A.B.; Coates, L.E.; Traniello, J.F.A. Disease resistance: A benefit of sociality in the dampwood termite Zootermopsis angusticollis (Isoptera: Termopsidae). Behav. Ecol. Sociobiol. 1998, 44, 125–134. [Google Scholar] [CrossRef]
- Rosengaus, R.B.; Jordan, C.; Lefebvre, M.L.; Traniello, J.F.A. Pathogen Alarm Behavior in a Termite: A New Form of Communication in Social Insects. Naturwissenschaften 1999, 86, 544–548. [Google Scholar] [CrossRef] [PubMed]
- Traniello, J.F.A.; Rosengaus, R.B.; Savoie, K. The development of immunity in a social insect: Evidence for the group facilitation of disease resistance. Proc. Natl. Acad. Sci. USA 2002, 99, 6838–6842. [Google Scholar] [CrossRef] [PubMed]
- Pie, M.R.; Rosengaus, R.B.; Calleri, D.V.; Traniello, J.F.A. Density and disease resistance in group-living insects: Do eusocial species exhibit density-dependent prophylaxis? Ethol. Ecol. Evol. 2005, 17, 41–50. [Google Scholar] [CrossRef]
- Engel, P.; Kwong, W.K.; McFrederick, Q.; Anderson, K.E.; Barribeau, S.M.; Chandler, J.A.; Cornman, R.S.; Dainat, J.; de Miranda, J.R.; Doublet, V.; et al. The Bee Microbiome: Impact on Bee Health and Model for Evolution and Ecology of Host-Microbe Interactions. MBio 2016, 7, e02164-15. [Google Scholar] [CrossRef]


| Location | Spider Species | Gender |
|---|---|---|
| Nitra City, 48°18′ N 18°05′ E, Slaughterhouse | 1. Pholcus alticeps (Spassky, 1932) |  |
| 2. Pholcus alticeps (Spassky, 1932) |  | |
| 3. Pholcus alticeps (Spassky, 1932) |  | |
| 4. Pholcus alticeps (Spassky, 1932) |  | |
| 5. Pholcus alticeps (Spassky, 1932) |  | |
| 6. Steatoda triangulosa (Walckenaer, 1802) |  | |
| 7. Steatoda triangulosa (Walckenaer, 1802) |  | |
| 8. Pholcus alticeps (Spassky, 1932) | juv. | |
| Nové Zámky region, Jatov village, 48°10′ N 18°00′ E, house | 9. Steatoda bipunctata (Linnaeus, 1758) |  |
| 10. Steatoda bipunctata (Linnaeus, 1758) |  | |
| 11. Scytodes thoracica (Latreille, 1802) |  | |
| Nitra City, 48°18′ N 18°05′ E, apartment building | 12. Pholcus phalangioides (Fuesslin, 1775) | juv. |
| 13. Pholcus phalangioides (Fuesslin, 1775) |  | |
| 14. Pholcus phalangioides (Fuesslin, 1775) |  | |
| 15. Pholcus phalangioides (Fuesslin, 1775) | juv. | |
| 16. Pholcus phalangioides (Fuesslin, 1775) |  | |
| 17. Pholcus phalangioides (Fuesslin, 1775) | juv. | |
| 18. Pholcus phalangioides (Fuesslin, 1775) |  | |
| 19. Pholcus phalangioides (Fuesslin, 1775) | juv. | |
| 20. Pholcus phalangioides (Fuesslin, 1775) |  | |
| Nové Zámky region, Jatov village, 48°10′ N 18°00′ E, house | 21. Malthonica ferruginea (Panzer, 1804) | |
| Nitra City, Street, 48°18′ N 18°05′ E, Student dormitory | 22. Steatoda triangulosa (Walckenaer, 1802) | |
| 23. Steatoda triangulosa (Walckenaer, 1802) | ||
| Nitra City, 48°18′ N 18°05′ E, SPU, building | 24. Pholcus phalangioides (Fuesslin, 1775) |  |
| 25. Pholcus phalangioides (Fuesslin, 1775) |  | |
| 26. Pholcus phalangioides (Fuesslin, 1775) | juv. | |
| 27. Pholcus phalangioides (Fuesslin, 1775) |  | |
| 28. Pholcus phalangioides (Fuesslin, 1775) |  | |
| 29. Pholcus phalangioides (Fuesslin, 1775) |  | |
| 30. Steatoda triangulosa (Walckenaer, 1802) |  | |
| 31. Steatoda triangulosa (Walckenaer, 1802) | juv. | |
| Nitra City, 48°18′ N 18°05′ E, apartment building | 32. Pholcus phalangioides (Fuesslin, 1775) | juv. |
| 33. Pholcus phalangioides (Fuesslin, 1775) | juv. | |
| 34. Pholcus phalangioides (Fuesslin, 1775) |  | |
| 35. Pholcus phalangioides (Fuesslin, 1775) |  | |
| 36. Steatoda triangulosa (Walckenaer, 1802) | juv. | |
| 37. Steatoda triangulosa (Walckenaer, 1802) | juv. | |
| Nové Zámky region, Jatov village, 48°10′ N 18°00′ E, house | 38. Steatoda triangulosa (Walckenaer, 1802) | juv. |
| 39. Trochosa robusta (Simon, 1876) |  | |
| Nitra City, 48°18′ N 18°05′ E, Botanical Garden SPU | 40. Pardosa hortensis (Thorell, 1872) |  |
| 41. Pardosa hortensis (Thorell, 1872) |  | |
| 42. Pardosa hortensis (Thorell, 1872) |  | |
| 43. Pardosa hortensis (Thorell, 1872) |  | |
| Veľký Lapáš Bodok, 48°17′24′′ S 18°11′09′′ V, chicken farm | 44. Parasteatoda tepidariorum (C. L. Koch, 1841) |  |
| 45. Parasteatoda tepidariorum (C. L. Koch, 1841) |  | |
| 46. Parasteatoda tepidariorum (C. L. Koch, 1841) | juv. | |
| 47. Pholcus phalangioides (Fuesslin, 1775) |  | |
| 48. Pholcus phalangioides (Fuesslin, 1775) |  | |
| 49. Parasteatoda tepidariorum (C. L. Koch, 1841) | juv. | |
| 50. Steatoda bipunctata (Linnaeus, 1758) |  | |
| 51. Steatoda bipunctata (Linnaeus, 1758) | juv. | |
| 52. Steatoda bipunctata (Linnaeus, 1758) | juv. | |
| 53. Steatoda bipunctata (Linnaeus, 1758) | juv. | |
| 54. Steatoda bipunctata (Linnaeus, 1758) | juv. | |
| 55. Steatoda bipunctata (Linnaeus, 1758) | juv. | |
| 56. Steatoda bipunctata (Linnaeus, 1758) | juv. | |
| 57. Steatoda bipunctata (Linnaeus, 1758) |  | |
| 58. Steatoda bipunctata (Linnaeus, 1758) |  | |
| 59. Steatoda bipunctata (Linnaeus, 1758) | juv. | |
| 60. Steatoda bipunctata (Linnaeus, 1758) | juv. | |
| 61. Pholcus alticeps (Spassky, 1932) |  | |
| 62. Pholcus alticeps (Spassky, 1932) | juv. | |
| 63. Tegenaria domestica (Clerck, 1757) | juv. | |
| 64. Tegenaria domestica (Clerck, 1757) | juv. | |
| 65. Tegenaria domestica (Clerck, 1757) | juv. | |
| 66. Tegenaria domestica (Clerck, 1757) | juv. | |
| 67. Tegenaria domestica (Clerck, 1757) | juv. | |
| 68. Parasteatoda lunata (Clerck, 1757) |  | |
| 69. Parasteatoda tepidariorum (C. L. Koch, 1841) |  | |
| 70. Steatoda triangulosa (Walckenaer, 1802) |  | |
| 71. Steatoda castanea (Clerck, 1757) |  | |
| 72. Salticus scenicus (Clerck, 1757) |  | |
| 73. Nuctenea umbratica (Clerck, 1757) |  | |
| 74. Steatoda triangulosa (Walckenaer, 1802) |  | |
| 75. Pholcus phalangioides (Fuesslin, 1775) |  | |
| 76. Pholcus phalangioides (Fuesslin, 1775) | juv. | |
| 77. Pholcus phalangioides (Fuesslin, 1775) | juv. | |
| 78. Steatoda bipunctata (Linnaeus, 1758) | sub.  | |
| 79. Tegenaria domestica (Clerck, 1757) |  | |
| 80. Steatoda bipunctata (Linnaeus, 1758) |  | |
| 81. Steatoda bipunctata (Linnaeus, 1758) |  | |
| 82. Steatoda triangulosa (Walckenaer, 1802) | juv. | |
| 83. Parasteatoda tepidariorum (C. L. Koch, 1841) |  | |
| 84. Steatoda bipunctata (Linnaeus, 1758) |  | |
| 85. Steatoda bipunctata (Linnaeus, 1758) | juv. | |
| 86. Steatoda bipunctata (Linnaeus, 1758) | juv. | |
| 87. Steatoda bipunctata (Linnaeus, 1758) |  | |
| 88. Steatoda bipunctata (Linnaeus, 1758) |  | |
| 89. Steatoda bipunctata (Linnaeus, 1758) | juv. | |
| 90. Steatoda bipunctata (Linnaeus, 1758) |  | |
| 91. Steatoda bipunctata (Linnaeus, 1758) | juv. | |
| 92. Pholcus phalangioides (Fuesslin, 1775) |  | |
| 93. Pholcus phalangioides (Fuesslin, 1775) | juv. | |
| 94. Pholcus phalangioides (Fuesslin, 1775) | juv. | |
| 95. Pholcus phalangioides (Fuesslin, 1775) | juv. | |
| 96. Pholcus phalangioides (Fuesslin, 1775) | juv. | |
| 97. Pholcus phalangioides (Fuesslin, 1775) | juv. | |
| 98. Pholcus phalangioides (Fuesslin, 1775) | juv. | |
| 99. Pholcus phalangioides (Fuesslin, 1775) |  | |
| 100. Pholcus alticeps (Spassky, 1932) |  | |
| 101. Tegenaria domestica (Clerck, 1757) | juv. | |
| 102. Tegenaria domestica (Clerck, 1757) |  |
 —male;
—male;  —female; juv.—juvenile.
—female; juv.—juvenile.| Spider/Agar | TSA | TSI | BA | AA |
|---|---|---|---|---|
| Malthonica ferruginea | 2.37 ± 0.03 a | 2.81 ± 0.03 b | 2.95 ± 0.02 c | 2.84 ± 0.05 d |
| Nuctenea umbratica | 1.48 ± 0.03 a | 1.26 ± 0.06 b | 1.55 ± 0.10 c | 1.50 ± 0.05 d |
| Parasteatoda lunata | 3.39 ± 0.12 a | 3.26 ± 0.06 b | 2.84 ± 0.06 c | 2.45 ± 0.08 d |
| Parasteatoda tepidariorum | 1.38 ± 0.19 a | 1.43 ± 0.10 b | 1.34 ± 0.12 c | 1.44 ± 0.08 d |
| Pardosa hortensis | 1.83 ± 0.05 a | 1.57 ± 0.09 b | 1.80 ± 0.09 c | 1.53 ± 0.07 d |
| Pholcus alticeps | 1.34 ± 0.09 a | 1.28 ± 0.04 b | 2.58 ± 0.06 c | 1.49 ± 0.04 d |
| Pholcus phalangioides | 1.22 ± 0.06 a | 1.28 ± 0.05 b | 1.19 ± 0.02 c | 1.27 ± 0.09 d |
| Salticus scenicus | 2.30 ± 0.05 a | 2.31 ± 0.16 b | 2.34 ± 0.08 c | 2.27 ± 0.09 d |
| Scytodes thoracica | 1.29 ± 0.06 a | 1.28 ± 0.03 b | 1.18 ± 0.03 c | 1.45 ± 0.08 d |
| Steatoda bipunctata | 1.18 ± 0.07 a | 1.19 ± 0.05 b | 1.22 ± 0.06 c | 1.11 ± 0.06 d |
| Steatoda castanea | 2.64 ± 0.21 a | 2.30 ± 0.06 b | 2.72 ± 0.06 c | 2.28 ± 0.16 d |
| Steatoda triangulosa | 1.43 ± 0.06 a | 1.30 ± 0.06 b | 1.79 ± 0.07 c | 1.43 ± 0.08 d |
| Tegenaria domestica | 1.19 ± 0.05 a | 1.11 ± 0.06 b | 1.27 ± 0.08 c | 1.29 ± 0.12 d |
| Trochosa robusta | 2.46 ± 0.11 a | 2.20 ± 0.03 b | 2.46 ± 0.09 c | 2.21 ± 0.14 d |
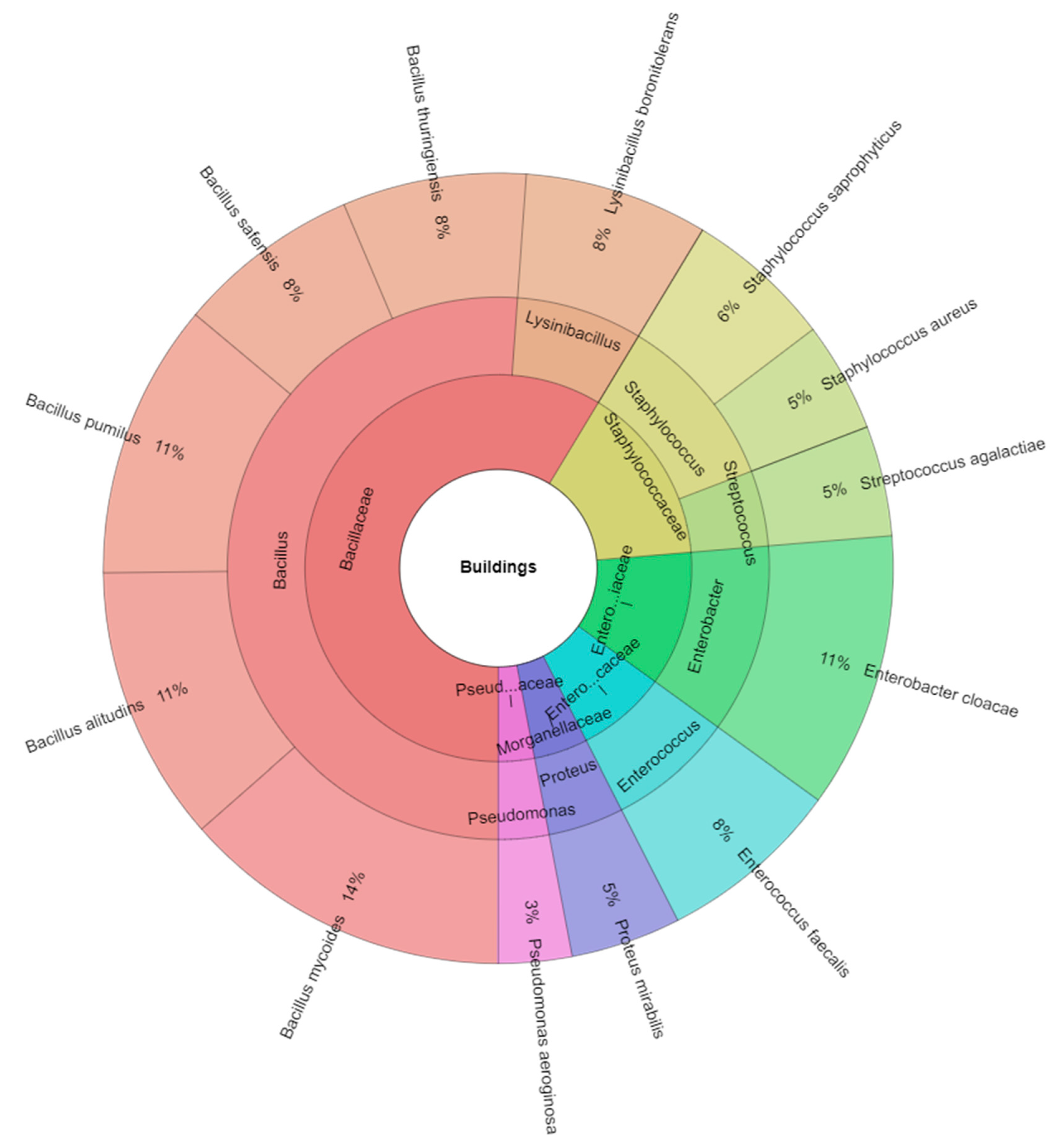
| Phylum | Taxa/Spider Specimens | Slaughterhouse | Chicken Farm | Buildings |
|---|---|---|---|---|
| Proteobacteria | Acinetobacter | |||
| Acinetobacter johnsonii | - | 11 | - | |
| Actinobacteria | Actinomyces | |||
| Actinomyces oris | - | 13 | - | |
| Firmicutes | Aerococcus | |||
| Aerococcus viridans | 6 | - | - | |
| Firmicutes | Bacillus | |||
| Bacillus alitudins | - | - | 15 | |
| Bacillus cereus | 15 | 6 | - | |
| Bacillus licheniformis | 14 | - | - | |
| Bacillus megatherium | - | 10 | - | |
| Bacillus mycoides | - | - | 18 | |
| Bacillus pumilus | 8 | 5 | 15 | |
| Bacillus safensis | 9 | 5 | 10 | |
| Bacillus thuringiensis | 12 | 6 | 10 | |
| Fungi | Candida | |||
| Candida famata | - | 3 | - | |
| Proteobacteria | Capriavidus | |||
| Capriavidus metallidurans | - | 4 | - | |
| Proteobacteria | Citrobacter | |||
| Citrobacter koseri | - | 4 | - | |
| Actinobacteria | Corynebacterium | |||
| Corynebacterium simulans | - | 5 | - | |
| Corynebacterium singulare | - | 6 | - | |
| Corynebacterium xerosis | - | 6 | - | |
| Actinobacteria | Cutibacterium | |||
| Cutibacterium avidum | - | 4 | - | |
| Fungi | Debaryomyces | |||
| Debaryomyces hansenii | - | 3 | - | |
| Proteobacteria | Enterobacter | |||
| Enterobacter cloacae | - | - | 15 | |
| Firmicutes | Enterococcus | |||
| Enterococcus durans | - | 6 | - | |
| Enterococcus faecalis | - | - | 10 | |
| Enterococcus faecium | - | 4 | - | |
| Proteobacteria | Escherichia | |||
| Escherichia coli | 11 | 12 | - | |
| Proteobacteria | Klebsiella | |||
| Klebsiella pneumoniae | - | 11 | - | |
| Actinobacteria | Kocuria | |||
| Kocuria rhizophila | - | 2 | - | |
| Firmicutes | Lactococcus | |||
| Lactococcus lactis | 4 | - | - | |
| Firmicutes | Lysinibacillus | |||
| Lysinibacillus boronitolerans | - | - | 10 | |
| Lysinibacillus fusiformis | - | 5 | - | |
| Lysinibacillus sphaericus | - | 6 | - | |
| Proteobacteria | Moraxella | |||
| Moraxella osloensis | - | 7 | - | |
| Firmicutes | Paenibacillus | |||
| Paenibacillus lautus | 3 | - | - | |
| Paenibacillus polymyxa | 2 | - | - | |
| Actinobacteria | Propionibacterium | |||
| Propionibacterium avidum | - | 4 | - | |
| Proteobacteria | Proteus | |||
| Proteus mirabilis | - | - | 6 | |
| Proteobacteria | Pseudomonas | |||
| Pseudomonas aeroginosa | - | - | 4 | |
| Pseudomonas stutzeri | - | 8 | - | |
| Fungi | Rhodotorula | |||
| Rhodotorula mucilaginosa | 1 | - | - | |
| Proteobacteria | Roseomonas | |||
| Roseomonas muscosa | - | 6 | - | |
| Proteobacteria | Salmonella | |||
| Salmonella spp. | - | 9 | - | |
| Proteobacteria | Sphingomonas | |||
| Sphingomonas parapaucimobilis | - | 6 | - | |
| Sphingomonas vabuuciae | - | 4 | - | |
| Firmicutes | Staphylococcus | |||
| Staphylococcus aureus | - | 5 | 6 | |
| Staphylococcus capitis | 2 | 4 | - | |
| Staphylococcus epidermidis | 8 | 2 | - | |
| Staphylococcus equorum | - | 2 | - | |
| Staphylococcus haemolyticus | 6 | - | - | |
| Staphylococcus hominis | 8 | 6 | - | |
| Staphylococcus oralis | - | 7 | - | |
| Staphylococcus pasteuri | - | 5 | - | |
| Staphylococcus pettenkoferi | 6 | - | - | |
| Staphylococcus saprophyticus | - | - | 8 | |
| Staphylococcus schleiferi | - | 5 | - | |
| Staphylococcus warneri | 4 | - | - | |
| Staphylococcus xylosus | 3 | 5 | - | |
| Firmicutes | Streptococcus | |||
| Streptococcus agalactiae | - | - | 6 | |
| Total isolates | 122 | 222 | 133 |
| Isolated Species | Antibiotic (R/S) | ||||
|---|---|---|---|---|---|
| IPM | MEM | CIP | VA | LZD | |
| Aerococcus viridans | ND | ND | ND | ND | ND |
| Bacillus cereus | 2/13 | 2/13 | 3/12 | 5/10 | 0/15 |
| Bacillus licheniformis | 4//10 | 5/9 | 10/4 | 4/10 | 3/14 |
| Bacillus pumilus | 0/8 | 1/7 | 2/6 | 0/8 | 1/7 |
| Bacillus safensis | 0/9 | 1/8 | 1/8 | 1/8 | 3/6 |
| Bacillus thuringiensis | 2/10 | 3/9 | 2/10 | 1/11 | 0/12 |
| IPM | MEM | CIP | TOB | C | |
| Escherichia coli | 10/1 | 1/10 | 5/6 | 6/5 | 3/8 |
| Lactococcus lactis | ND | ND | ND | ND | ND |
| Paenibacillus lautus | ND | ND | ND | ND | ND |
| Paenibacillus polymyxa | ND | ND | ND | ND | ND |
| Rhodotorula mucilaginosa | ND | ND | ND | ND | ND |
| CIP | NOR | AK | TOB | TGC | |
| Staphylococcus capitis | 0/2 | 0/2 | 0/2 | 0/2 | 1/1 |
| Staphylococcus epidermidis | 2/6 | 1/7 | 2/6 | 3/5 | 4/4 |
| Staphylococcus haemolyticus | 5/1 | 2/4 | 3/3 | 0/6 | 1/5 |
| Staphylococcus hominis | 0/8 | 1/7 | 2/6 | 1/7 | 3/5 |
| Staphylococcus pettenkoferi | 0/6 | 2/4 | 1/5 | 2/4 | 1/5 |
| Staphylococcus warneri | 1/3 | 2/2 | 0/4 | 3/1 | 0/4 |
| Staphylococcus xylosus | 2/1 | 1/2 | 0/3 | 0/3 | 0/3 |
| Total | 28/78 | 22/84 | 31/75 | 26/80 | 20/86 |
| Isolated Species | Antibiotic (R/S) | ||||
|---|---|---|---|---|---|
| IPM | MEM | CIP | VA | LZD | |
| Acinetobacter johnsonii | ND | ND | ND | ND | ND |
| Actinomyces oris | ND | ND | ND | ND | ND |
| Bacillus cereus | 1/5 | 2/4 | 0/6 | 3/3 | 1/5 |
| Priestia megatherium | 0/10 | 1/9 | 0/10 | 1/9 | 2/8 |
| Bacillus pumilus | 0/5 | 0/5 | 0/5 | 0/5 | 1/4 |
| Bacillus safensis | 0/5 | 1/4 | 0/5 | 1/4 | 1/4 |
| Bacillus thuringiensis | 0/6 | 1/5 | 0/6 | 1/5 | 1/5 |
| Candida famata | ND | ND | ND | ND | ND |
| Capriavidus metallidurans | ND | ND | ND | ND | ND |
| IPM | MEM | CIP | TOB | C | |
| Citrobacter koseri | 0/4 | 1/3 | 0/4 | 1/3 | 2/2 |
| Escherichia coli | 2/10 | 3/9 | 2/10 | 5/7 | 2/10 |
| Klebsiella pneumoniae | 1/10 | 2/9 | 2/9 | 0/11 | 1/10 |
| Salmonella spp. | 0/9 | 1/8 | 1/8 | 1/8 | 0/9 |
| CIP | VA | TE | LZD | RD | |
| Corynebacterium simulans | 0/5 | 1/4 | 0/5 | 0//5 | 1/4 |
| Corynebacterium singulare | 1/5 | 0/6 | 2/4 | 0/6 | 1/5 |
| Corynebacterium xerosis | 0/6 | 1/5 | 0/6 | 0/6 | 0/6 |
| MEM | VA | ||||
| Cutibacterium avidum | 0/4 | 0/4 | - | - | - |
| Debaryomyces hansenii | ND | ND | ND | ND | ND |
| IMP | CIP | VA | TGC | LZD | |
| Enterococcus durans | 2/4 | 3/3 | 2/4 | 6/0 | 1/5 |
| Enterococcus faecium | 1/3 | 2/2 | 3/1 | 1/3 | 2/2 |
| Kocuria rhizophila | ND | ND | ND | ND | ND |
| Lysinibacillus fusiformis | ND | ND | ND | ND | ND |
| Lysinibacillus sphaericus | ND | ND | ND | ND | ND |
| Moraxella osloensis | ND | ND | ND | ND | ND |
| IMP | MEM | CIP | TOB | AK | |
| Pseudomonas stutzeri | 1/7 | 2/6 | 0/8 | 0/8 | 0/8 |
| Propionibacterium avidum | ND | ND | ND | ND | ND |
| Roseomonas muscosa | ND | ND | ND | ND | ND |
| Sphingomonas parapaucimobilis | ND | ND | ND | ND | ND |
| Sphingomonas vabuuciae | ND | ND | ND | ND | ND |
| CIP | NOR | AK | TOB | TGC | |
| Staphylococcus aureus | 1/4 | 0/5 | 0/5 | 2/3 | 1/4 |
| Staphylococcus capitis | 1/3 | 1/3 | 0/4 | 0/4 | 1/3 |
| Staphylococcus epidermidis | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 |
| Staphylococcus equorum | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 |
| Staphylococcus hominis | 1/5 | 1/5 | 2/4 | 0/6 | 0/6 |
| Staphylococcus oralis | 2/5 | 0/7 | 1/6 | 2/5 | 1/6 |
| Staphylococcus pasteuri | 1/4 | 0/5 | 0/5 | 1/4 | 1/4 |
| Staphylococcus schleiferi | 0/5 | 0/5 | 1/4 | 1/4 | 1/4 |
| Staphylococcus xylosus | 0/5 | 0/5 | 0/5 | 2/3 | 3/2 |
| Total | 15/133 | 23/125 | 16/128 | 29/115 | 25/119 |
| Isolated Species | Antibiotic (R/S) | ||||
|---|---|---|---|---|---|
| IPM | MEM | CIP | VA | LZD | |
| Bacillus alitudins | 5/10 | 2/13 | 0/15 | 5/10 | 6/9 |
| Bacillus mycoides | 2/16 | 2/16 | 6/12 | 0/18 | 3/15 |
| Bacillus pumilus | 2/8 | 3/7 | 4/6 | 0/10 | 1/9 |
| Bacillus safensis | 2/8 | 2/8 | 0/10 | 5/5 | 6/4 |
| Bacillus thuringiensis | 2/8 | 3/7 | 5/5 | 0/10 | 4/6 |
| IPM | MEM | CIP | TOB | C | |
| Enterobacter cloacae | 0/15 | 0/15 | 0//15 | 0/15 | 0/15 |
| Proteus mirabilis | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 |
| IMP | CIP | VA | TGC | LZD | |
| Enterococcus faecalis | 1/9 | 2/8 | 5/5 | 0/10 | 1/9 |
| Lysinibacillus boronitolerans | ND | ND | ND | ND | ND |
| IMP | MEM | CIP | TOB | AK | |
| Pseudomonas aeroginosa | 0/4 | 1/3 | 0/4 | 0/4 | 0/4 |
| CIP | NOR | AK | TOB | TGC | |
| Staphylococcus aureus | 0/6 | 1/5 | 2/4 | 0/6 | 0/6 |
| Staphylococcus saprophyticus | 1/7 | 1/7 | 0/8 | 0/8 | 2/6 |
| VA | TGC | LZD | C | TE | |
| Streptococcus agalactiae | 5/1 | 1/5 | 2/4 | 0/6 | 1/5 |
| Total | 20/98 | 18/100 | 24/94 | 10/108 | 24/94 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kačániová, M.; Terentjeva, M.; Kowalczewski, P.Ł.; Babošová, M.; Porhajašová, J.I.; Hikal, W.M.; Fedoriak, M. Bacteriota and Antibiotic Resistance in Spiders. Insects 2022, 13, 680. https://doi.org/10.3390/insects13080680
Kačániová M, Terentjeva M, Kowalczewski PŁ, Babošová M, Porhajašová JI, Hikal WM, Fedoriak M. Bacteriota and Antibiotic Resistance in Spiders. Insects. 2022; 13(8):680. https://doi.org/10.3390/insects13080680
Chicago/Turabian StyleKačániová, Miroslava, Margarita Terentjeva, Przemysław Łukasz Kowalczewski, Mária Babošová, Jana Ivanič Porhajašová, Wafaa M. Hikal, and Mariia Fedoriak. 2022. "Bacteriota and Antibiotic Resistance in Spiders" Insects 13, no. 8: 680. https://doi.org/10.3390/insects13080680
APA StyleKačániová, M., Terentjeva, M., Kowalczewski, P. Ł., Babošová, M., Porhajašová, J. I., Hikal, W. M., & Fedoriak, M. (2022). Bacteriota and Antibiotic Resistance in Spiders. Insects, 13(8), 680. https://doi.org/10.3390/insects13080680










