Comparative Cytogenetics of Lace Bugs (Tingidae, Heteroptera): New Data and a Brief Overview
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
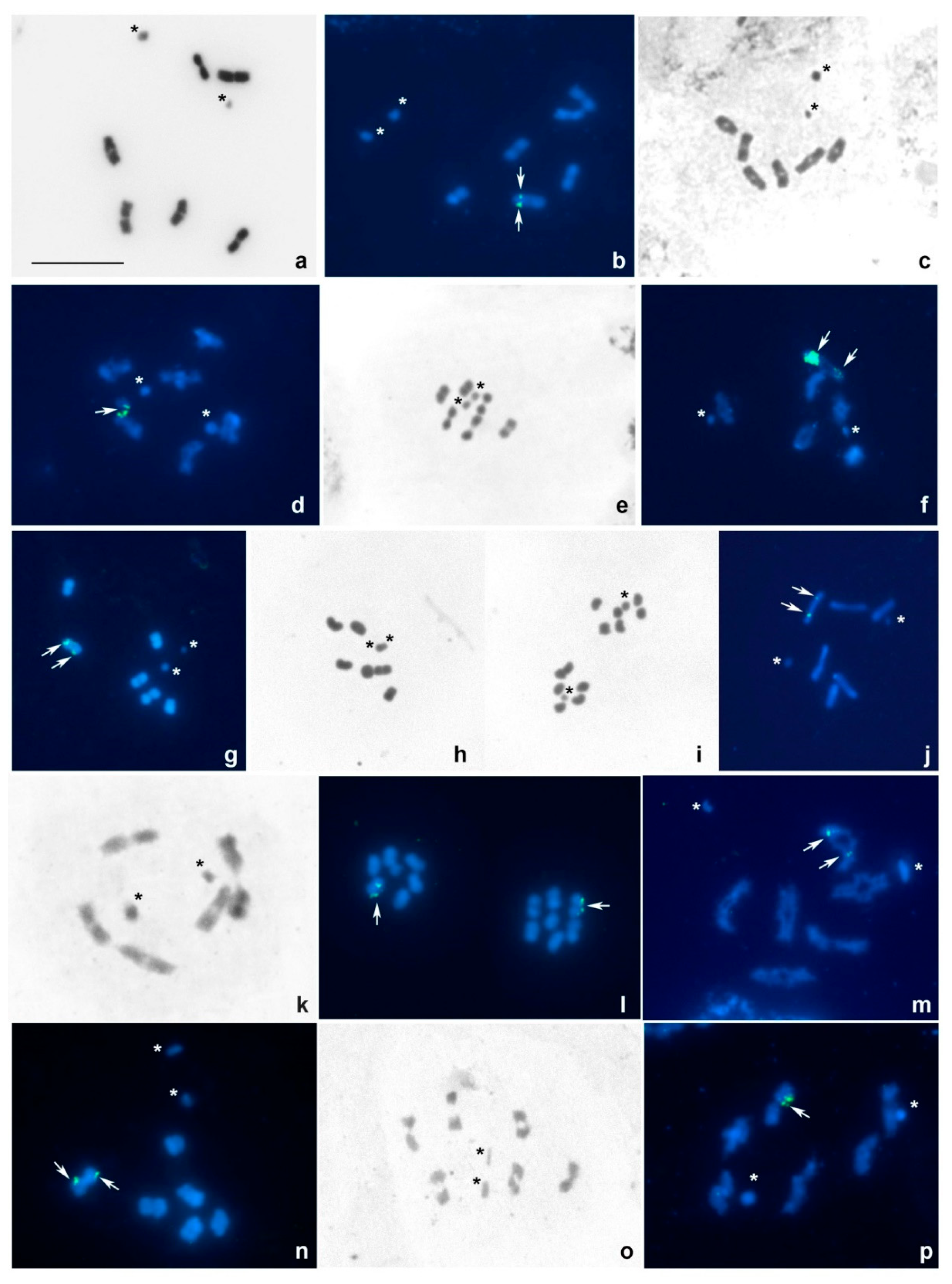
3. Results
3.1. Tribe Tingini
3.2. Tribe Acalyptaini
4. Discussion
4.1. Chromosome Numbers and Sex Chromosome Systems
4.2. Male Meiosis
4.3. Patterns of 18S rDNA Localization
4.4. Lack of the “Insect” Telomere Motif (TTAGG)n
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guilbert, E.; Damgaard, J.; D’Haese, C.A. Phylogeny of the lacebugs (Insecta: Heteroptera: Tingidae) using morphological and molecular data. Syst. Entomol. 2014, 39, 431–441. [Google Scholar] [CrossRef]
- Scudder, G.G.E. The female genitalia of the Heteroptera: Morphology and bearing on classification. Trans. R. Entomol. Soc. Lond. 1959, 111, 405–467. [Google Scholar] [CrossRef]
- Štys, P.; Kerzhner, I.M. The rank and nomenclature of higher taxa in recent Heteroptera. Acta Entomol. Bohemoslov. 1975, 72, 64–79. [Google Scholar]
- Péricart, J.; Golub, V.B. Superfamily Tingoidea Laporte, 1832. In Catalogue of the Heteroptera of the Palaearctic Region II; Aukema, B., Rieger, C., Eds.; The Netherlands Entomological Society (NEV): Amsterdam, The Netherlands, 1996; pp. 3–78. [Google Scholar]
- Golub, V.B.; Popov, Y.A. Historical development and problems of classification of the heteropteran insects of the superfamily Tingoidea (Hemiptera: Heteroptera, Cimicomorpha). Meet. Mem. NA Cholodkovsky 2016, 66, 1–93. (In Russian) [Google Scholar]
- Drake, C.; Davis, N. The morphology, phylogeny, and higher classification of the family Tingidae, including the description of a new genus and species of the subfamily Vianaidinae (Hemiptera: Heteroptera). Entomol. Am. 1960, 39, 100. [Google Scholar]
- Schuh, R.T.; Štys, P. Phylogenetic analysis of cimicomorphan family relationships (Heteroptera). J. N. Y. Entomol. Soc. 1991, 99, 298–350. [Google Scholar]
- Schuh, R.T.; Weirauch, C. True Bugs of the World (Hemiptera: Heteroptera). Classification and Natural History, 2nd ed.; Monograph Series 8; Siri Scientific Press: Rochdale, UK, 2020; 767p. [Google Scholar]
- Schuh, R.T.; Weirauch, C.; Wheeler, W.C. Phylogenetic relationships within the Cimicomorpha (Hemiptera: Heteroptera): A total-evidence analysis. Syst. Entomol. 2009, 34, 15–48. [Google Scholar] [CrossRef]
- Weirauch, C.; Schuch, R.T.; Cassis, G.; Wheeler, W.C. Revisiting habitat and lifestyle transitions in Heteroptera (Insecta: Hemiptera): Insights from a combined morphological and molecular phylogeny. Cladistics 2019, 35, 67–105. [Google Scholar] [CrossRef]
- Lis, B.; Zielinska, A.; Lis, J.A. The King’s Lace Bug Recaredus rex Distant, 1909 (Hemiptera: Heteroptera: Tingidae): Systematic Position, First Palaearctic and Afrotropical Records, and Ecological Niche Modelling. Insects 2022, 13, 558. [Google Scholar] [CrossRef]
- Lis, B. Phylogeny and classification of Cantacaderini [= Cantacaderidae stat. nov.] (Hemiptera: Tingoidea). Ann. Zool. 1999, 49, 157–196. [Google Scholar]
- Schuh, R.T.; Cassis, G.; Guilbert, E. Description of the first recent macropterus species of Vianaidinae (Heteroptera: Tingidae) with comments on the phylogenetic relationships of the family within the Cimicomorpha. J. N. Y. Entomol. Soc. 2006, 114, 33–53. [Google Scholar] [CrossRef]
- Golub, V.B.; Golub, N.V. On the status of the genera complex Acalypta, Dictyonota, Kalama and Derephysia (Heteroptera: Tingidae: Tinginae) having common morphological and karyological features. Zoosyst. Ross. 2019, 28, 228–237. [Google Scholar] [CrossRef]
- Montgomery, T.H. A study of the chromosomes of the germ cells of Metazoa. Trans. Am. Philos. Soc. 1901, 20, 154–236. [Google Scholar] [CrossRef]
- Montgomery, T.H. Chromosomes in the spermatogenesis of the Hemiptera-Heteroptera. Trans. Am. Philos. Soc. 1906, 21, 97–173. [Google Scholar] [CrossRef]
- Toshioka, S. On the chromosomes in certain Heterotpera. Oyo-Dobutsugaku-Zasshi 1934, 6, 109–115. [Google Scholar]
- Southwood, T.R.E.; Leston, D. Land and Water Bugs of the British Isles; Frederick Warne and Co., Ltd.: London, UK; New York, NY, USA, 1959; 436p. [Google Scholar]
- Jande, S.S. Pre-reductional sex-chromosomes in the family Tingidae (Gymnocerata—Heteroptera). Experientia 1960, 16, 440. [Google Scholar] [CrossRef]
- Takenouchi, Y.; Muramoto, N. A survey of the chromosomes in twenty species of Heteroptera insects. Hokkaido Univ. Educ. 1967, 18, 1–15. (In Japanese) [Google Scholar]
- Harley, K.L.S.; Kassulke, R.C. Tingidae for biological control of Lantana camara (Verbenaceae). Entomophaga 1971, 16, 384–410. [Google Scholar] [CrossRef]
- Muramoto, N. A list of the chromosome numbers of Heteropteran insects of Japan. Chromosome Inf. Serv. 1973, 14, 29–31. [Google Scholar]
- Ueshima, N. Animal Cytogenetics. Insecta 6. Hemiptera II: Heteroptera; Gebrüder Borntraeger: Berlin, Germany; Stuttgart, Germany, 1979; Volume 117, 528p. [Google Scholar]
- Nokkala, S.; Nokkala, C. Occurrence of the XO sex chromosomes system in Dictyonota tricornis (Schr.) (Tingidae, Hemiptera) and its significance for concepts of sex chromosome evolution in Heteroptera. Hereditas 1984, 100, 299–301. [Google Scholar] [CrossRef]
- Grozeva, S.; Nokkala, S. Chromosome numbers, sex determining systems, and patterns of the C-heterochromatin distribution in 13 species of lace bugs (Heteroptera, Tingidae). Fol. Biol. 2001, 49, 29–41. [Google Scholar]
- Golub, N.V.; Golub, V.B.; Kuznetsova, V.G. Variability of 18rDNA loci in four lace bug species (Hemiptera, Tingidae) with the same chromosome number. Comp. Cytogenet. 2015, 9, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Golub, N.V.; Golub, V.B.; Kuznetsova, V.G. Further evidence for the variability of the 18S rDNA loci in the family Tingidae (Hemiptera, Heteroptera). Comp. Cytogenet. 2016, 10, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Golub, N.V.; Golub, V.B.; Kuznetsova, V.G. Distribution of the major rDNA loci among four Hemipteran species of the family Tingidae (Heteroptera, Cimicomorpha). Fol. Biol. 2017, 65, 155–158. [Google Scholar] [CrossRef]
- Golub, N.V.; Golub, V.B.; Kuznetsova, V.G. New data on karyotypes of lace bugs (Tingidae, Cimicomorpha, Hemiptera) with analysis of the 18S rDNA clusters distribution. Comp. Cytogenet. 2018, 12, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Grozeva, S.; Nokkala, S. Chromosomes and their meiotic behavior in two families of the primitive infraorder Dipsocoromorpha (Heteroptera). Hereditas 1996, 125, 189–192. [Google Scholar] [CrossRef]
- Grozeva, S.; Anokhin, B.; Kuznetsova, V.G. Bed bugs (Hemiptera). In Protocols for Cytogenetic Mapping of Arthropod Genomes; Sharachov, I., Ed.; CRC Press, Taylor and Francis: Boca Raton, FL, USA, 2015; pp. 285–326. [Google Scholar] [CrossRef]
- Anokhin, B.; Hemmrich-Stanisak, G.; Bosch, T.C.G. Karyotyping and single-gene detection using fluorescence in situ hybridization on chromosomes of Hydra magnipapillata. Comp. Cytogenet. 2010, 4, 97–110. [Google Scholar] [CrossRef][Green Version]
- Grozeva, S.; Kuznetsova, V.G.; Anokhin, B.A. Karyotypes, male meiosis and comparative FISH mapping of 18S ribosomal DNA and telomeric (TTAGG)n repeat in eight species of true bugs (Hemiptera, Heteroptera). Comp. Cytogenet. 2011, 5, 355–374. [Google Scholar] [CrossRef]
- Golub, N.; Anokhin, B.; Kuznetsova, V. Comparative FISH mapping of ribosomal DNA clusters and TTAGG telomeric sequences to holokinetic chromosomes of eight species of the insect order Psocoptera. Comp. Cytogenet. 2019, 13, 403–410. [Google Scholar] [CrossRef]
- Kuznetsova, V.G.; Grozeva, S.M.; Nokkala, S.; Nokkala, C. Cytogenetics of the true bug infraorder Cimicomorpha (Hemiptera, Heteroptera): A review. ZooKeys 2011, 154, 31–70. [Google Scholar] [CrossRef]
- Papeschi, A.G.; Bressa, M.J. Evolutionary cytogenetics in Heteroptera. J. Biol. Res. 2006, 5, 3–21. [Google Scholar]
- Nokkala, S.; Nokkala, C. Achiasmatic male meiosis in two species of Saldula (Saldidae, Hemiptera). Hereditas 1983, 99, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Nokkala, S.; Kuznetsova, V.; Maryańska-Nadachowska, A. Achiasmate segregation of a B-chromosome from the X-chromosome in two species of psyllids (Psylloidea: Homoptera). Genetica 2000, 108, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Nokkala, S.; Grozeva, S.; Kuznetsova, V.; Maryańska-Nadachowska, A. The origin of the achiasmatic XY sex chromosome system in Cacopsylla peregrine (Frst.) (Psylloidea, Homoptera). Genetica 2003, 119, 327–332. [Google Scholar] [CrossRef] [PubMed]
- White, M.J.D. Animal Cytology and Evolution; Cambridge University Press: Cambridge, UK; London, UK; New York, NY, USA, 1973; 961p. [Google Scholar]
- Grozeva, S.; Nokkala, S.; Simov, N. First evidence of sex chromosomes pre-reduction in male meiosis in the Miridae bugs (Heteroptera). Fol. Biol. 2006, 54, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Grozeva, S.; Simov, N.; Josifov, M. Karyotaxonomy of some European Macrolophus species (Heteroptera, Miridae). Mainzer Naturwiss. Arch. 2007, 31, 81–87. [Google Scholar]
- Grozeva, S.; Kuznetsova, V.G.; Simov, N.; Langourov, M.; Dalakchieva, S. Sex chromosome pre-reduction in male meiosis of Lethocerus patruelis (Stal, 1854) (Heteroptera, Belostomatidae) with some notes on the distribution of the species. ZooKeys 2013, 319, 119–135. [Google Scholar] [CrossRef]
- Toscani, M.A.; Pigozzi, M.I.; Papeschi, A.G.; Bressa, M.J. Histone H3 methylation and autosomal vs. sex chromosome segregation during male meiosis in Heteroptera. Front. Ecol. Evol. 2022, 10, 836786. [Google Scholar] [CrossRef]
- Bressa, M.J.; Papeschi, A.G.; Vítková, M.; Kubícková, S.; Fuková, I.; Pigozzi, M.I.; Marec, F. Sex chromosome evolution in cotton stainers of the genus Dysdercus (Heteroptera: Pyrrhocoridae). Cytogenet. Genome Res. 2009, 125, 292–305. [Google Scholar] [CrossRef]
- Toscani, M.A.; Pigozzi, M.I.; Bressa, M.J.; Papeschi, A.G. Synapsis with and without recombination in the male meiosis of the leaf-footed bug Holhymenia rubiginosa (Coreidae, Heteroptera). Genetica 2008, 132, 173–178. [Google Scholar] [CrossRef]
- Bressa, M.J.; Franco, M.J.; Toscani, M.A.; Papeschi, A.G. Heterochromatin heteromorphism in Holhymenia rubiginosa (Heteroptera: Coreidae). Eur. J. Entomol. 2008, 105, 65–72. [Google Scholar] [CrossRef]
- Panzera, F.; Pita, S.; Lorite, P. Chromosome structure and evolution of Triatominae: A review. In Triatominae—The Biology of Chagas Disease Vectors. Entomology in Focus; Guarneri, A., Lorenzo, M., Eds.; Springer: New York, NY, USA, 2021; Volume 5, pp. 65–99. [Google Scholar] [CrossRef]
- Pita, S.; Panzera, F.; Mora, P.; Vela, J.; Palomeque, T.; Lorite, P. The presence of the ancestral insect telomeric motif in kissing bugs (Triatominae) rules out the hypothesis of its loss in evolutionarily advanced Heteroptera (Cimicomorpha). Comp. Cytogenet. 2016, 10, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Poggio, M.G.; Provecho, Y.M.; Papeschi, A.G.; Bressa, M.J. Possible origin of polymorphism for chromosome number in the assassin bug Zelurus femoralis longispinis (Reduviidae: Reduviinae). Biol. J. Linn. Soc. Lond. 2013, 110, 757–764. [Google Scholar] [CrossRef]
- Bardella, V.B.; Fernandes, T.; Vanzela, A.L.L. The conservation of number and location of 18S sites indicates the relative stability of rDNA in species of Pentatomomorpha (Heteroptera). Genome 2013, 56, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Bardella, V.B.; Fernandes, J.A.M.; Cabral-de-Mello, D.C. Chromosomal evolutionary dynamics of four multigene families in Coreidae and Pentatomidae (Heteroptera) true bugs. Mol. Genet. Genomics 2016, 291, 1919–1925. [Google Scholar] [CrossRef] [PubMed]
- Gapon, D.A.; Kuznetsova, V.G.; Maryańska-Nadachowska, A. A new species of the genus Rhaphidosoma Amyot et Serville, 1843 (Heteroptera, Reduviidae), with data on its chromosome complement. Comp. Cytogenet. 2021, 15, 467–505. [Google Scholar] [CrossRef] [PubMed]
- Raskina, O.; Barber, J.C.; Nevo, E.; Belyayev, A. Repetitive DNA and chromosomal rearrangements: Speciation-related events in plant genomes. Cytogenet. Genome Res. 2008, 120, 351–357. [Google Scholar] [CrossRef]
- Cabrero, J.; Camacho, J.P.M. Location and expression of ribosomal RNA genes in grasshoppers: Abundance of silent and cryptic loci. Chromosome Res. 2008, 16, 595–607. [Google Scholar] [CrossRef]
- Nakajima, R.T.; Cabral-de-Mello, D.C.; Valente, G.T.; Venere, P.C.; Martins, C. Evolutionary dynamics of rRNA gene clusters in cichlid fish. BMC Evol. Biol. 2012, 12, 198. [Google Scholar] [CrossRef]
- Jakubczak, J.L.; Burke, W.D.; Eickbush, T.H. Retrotransposable Elements RI and R2 interrupt the rRNA genes of most insects. Proc. Natl. Acad. Sci. USA 1991, 88, 3295–3299. [Google Scholar] [CrossRef]
- Eickbush, T.H.; Eickbush, D.G. Finely orchestrated movements: Evolution of the ribosomal RNA genes. Genetics 2007, 175, 477–485. [Google Scholar] [CrossRef]
- Raskina, O.; Belyayev, A.; Nevo, E. Activity of the En/Spm-like transposons in meiosis as a base for chromosome repatterning in a small, isolated, peripheral population of Aegilops speltoides Tausch. Chromosome Res. 2004, 12, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Eickbush, M.T.; Eickbush, T.H. Role of recombination in the long-term retention of transposable elements in rRNA gene loci. Genetics 2008, 180, 1617–1626. [Google Scholar] [CrossRef] [PubMed]
- Cabral-de-Mello, D.C.; Oliveira, S.G.; de Moura, R.C.; Martins, C. Chromosomal organization of the 18S and 5S rRNAs and histone H3 genes in Scarabaeinae coleopterans: Insights into the evolutionary dynamics of multigene families and heterochro-matin. BMC Genet. 2011, 12, 88. [Google Scholar] [CrossRef] [PubMed]
- Proenҫa, S.J.R.; Serrano, A.R.M.; Serrano, J.; Galián, J. Patterns of rDNA chromosomal localization in Palearctic Cephalota and Cylindera (Coleoptera: Carabidae: Cicindelini) with different numbers of X-chromosomes. Comp. Cytogen. 2011, 5, 47–59. [Google Scholar] [CrossRef]
- Symonová, R.; Majtanova, Z.; Sember, A.; Staaks, G.B.O.; Bohlen, J.; Freyhof, J.; Rabova, M.; Rab, P. Genome differentiation in a species pair of coregonine fishes: An extremely rapid speciation driven by stress-activated retrotransposons mediating exten-sive ribosomal DNA multiplications. BMC Evol. Biol. 2013, 13, 42. [Google Scholar] [CrossRef]
- Sochorová, J.; Garcia, S.; Gálvez, F.; Symonová, R.; Kovařík, A. Evolutionary trends in animal ribosomal DNA loci: Introduction to a new online database. Chromosoma 2018, 127, 141–150. [Google Scholar] [CrossRef]
- Sochorová, J.; Gálvez, F.; Matyášek, R.; Garcia, S.; Kovařík, A. Analyses of the Updated “Animal rDNA Loci Database” with an Emphasis on Its New Features. Int. J. Mol. Sci. 2021, 22, 11403. [Google Scholar] [CrossRef]
- Frydrychová, R.; Grossmann, P.; Trubac, P.; Vitková, M.; Marec, F.E. Phylogenetic distribution of TTAGG telomeric repeats in insects. Genome 2004, 47, 16–178. [Google Scholar] [CrossRef]
- Grozeva, S.; Anokhin, B.A.; Simov, N.; Kuznetsova, V.G. New evidence for the presence of the telomere motif (TTAGG)n in the family Reduviidae and its absence in the families Nabidae and Miridae (Hemiptera, Cimicomorpha). Comp. Cytogenet. 2019, 13, 285–295. [Google Scholar] [CrossRef]
- Lukhtanov, V.A. Diversity and evolution of telomere and subtelomere DNA sequences in insects. bioRxiv 2022. [Google Scholar] [CrossRef]
- Kuznetsova, V.; Grozeva, S.; Gokhman, V. Telomere structure in insects: A review. J. Zool. Syst. Evol. Res. 2020, 58, 127–158. [Google Scholar] [CrossRef]
- Prušáková, D.; Peska, V.; Pekár, S.; Bubeník, M.; Čížek, L.; Bezděk, A.; Frydrychová, R.C. Telomeric DNA sequences in beetle taxa vary with species richness. Sci. Rep. 2021, 11, 13319. [Google Scholar] [CrossRef] [PubMed]

| Species | Number of Males Examined | Host Plant, Date, and Locality of Collection |
|---|---|---|
| Tribe Tingini | ||
| Agramma blandulum (Horváth, 1905) | 2 | Carex sp., 1 June 2017, Nizhny Baskunchak vic., Astrakhan Prov., Russia |
| A. minutum Horváth, 1874 | 7 | Carex sp., 4 August 2020, Karadag Nature Reserve, Crimea, Russia |
| Copium adumbratum Horváth, 1832 | 9 | Teucrium sp., 4 June 2018, Erevan vic., Armenia |
| C. brevicorne (Jakovlev, 1879) | 5 | Teucrium sp., 4 June 2018, Erevan vic., Armenia |
| C. clavicorne (Linnaeus, 1758) | 3 | Teucrium sp., 28 July 2021, Teberda Nature Reserve, Teberda vic., North Caucasus, Russia |
| Corythucha arcuata (Say, 1832) | 15 | Quercus sp., 2 June 2021, Goryachy Klyuch vic., Krasnodar Krai, Russia |
| C. ciliata (Say, 1832) | 7 | Platanus orientalis Linnaeus, 1753, 21 July 2021, Maykop Prov., Republic of Adygea, Russia |
| Galeatus affinis (Herrich-Schaeffer, 1835) | 10 | Helichrysum arenarium Moench, 1794, and Artemisia marschalliana Sprengel, 1826, 1–15 July 2021, Voronezh Prov., Russia |
| Physatocheilaputshkovi Golub, 1976 | 7 | Padus avium, 27 July 2019, vill. Artybash, Altai Republic, Russia |
| Ph.smreczynskii China, 1952 | 12 | Padus avium Linnaeus, 1753, 1–15 July 2021, Voronezh Prov., Russia |
| Stephanitisoschanini Vasiliev, 1935 | 4 | Malus sp., 30 May 2018, Khosrov Forest State Reserve, Ararat Prov., Armenia |
| Tingis (Tingis) brevicornis (Horváth, 1902) | 2 | Grass community, 30 May 2018, Khosrov Forest State Reserve, Goravan desert, Ararat Prov., Armenia |
| Tribe Acalyptaini | ||
| Acalypta gracilis (Fieber, 1844) | 11 | Moss community, 20 June 2020, Voronezh Prov., Russia |
| A. hellenica (Reuter, 1888) | 2 | Moss community, 7 August 2020, Karadag Nature Reserve, Crimea, Russia |
| Derephysia. (Paraderephysia) cristata (Panzer, 1806) | 9 | Roots of Artemisia marschalliana, 1–15 July 2020, Voronezh Prov., Russia |
| Kalama beckeri (Jakovlev, 1871) | 3 | Moss community, 7 August 2020, Karadag Nature Reserve, Crimea, Russia |
| Species | Meioformula and Pattern of rDNA Localization (in Curly Brackets), References |
|---|---|
| Tribe Tingini | |
| 1. Agramma atricapillum (Spinola, 1837) | 12 + XY {AA 1}, [29] |
| 2. A. blandulum (Horváth, 1905) | 12 + XY, present study |
| 3. A. femorale Thomson, 1871 | 12 + XY {X}, [26] |
| 4. A. fallax (Horvath, 1906) | 12 + XY {X and Y}, [28] |
| 5. A. nexile (Drake, 1948) | 12 + XY, [22] |
| 6. A. minutum Horváth, 1874 | 12 + XY {AA}, present study |
| 7. Bredenbachius consanguensis Distant, 1903 | 12 + XY, [19] |
| 8. Cochlochila lewisi (Scott, 1880) | 12 + XY, [20] |
| 9. Catoplathus carthusianus (Goeze, 1778) | 12 + XY {AA}, [28] |
| 10. Copium adumbratum (Horváth, 1891) | 12 + XY, present study |
| 11. C. clavicorne (Linnaeus, 1758) | 12 + XY, [25] 12 + XY {AA}, present study |
| 12. C. brevicorne (Jakovlev, 1879) | 12 + XY {AA}, present study |
| 13. C. teucrii (Host, 1788) | 12 + XY {AA}, [28] |
| 14. Corythucha arcuata (Say, 1832) | 12 + XY {AA}, present study |
| 15. C. ciliata (Say, 1832) | 12 + XY, [25] 12 + XY {AA}, present study |
| 16. Dasytingis rudis Drake and Poor, 1936 | 12 + XY, [19] |
| 17. Dictyla echii (Schrank, 1782) | 12 + XY, [25] 12 + XY {X and Y}, [27] |
| 18. D. humuli (Fabricius, 1794) | 12 + XY, [18] 12 + XY {AA}, [28] |
| 19. D. platyoma (Fieber, 1861) | 12 + XY {AA}, [29] |
| 20. D. rotundata (Herrich-Schaeffer, 1835) | 12 + XY {AA}, [27] |
| 21. Elasmotropis testacea (Herrich-Schaeffer, 1830) | 12 + XY, [25] 12 + XY {AA}, [26] |
| 22. Galeatus affinis (Herrich-Schaeffer, 1835) | 12 + XY {AA}, present study |
| 23. G. sinuatus (Herrich-Schaeffer, 1838) | 12 + XY {AA}, [28] |
| 24. Lasiacantha capucina (Germar, 1837) | 12 + XY, [25] 12 + XY {AA}, [27] |
| 25. L. hermani Vasarhelyi 1977 | 12 + XY {AA}, [29] |
| 26. Leptobyrsa decora Drake, 1922 | 12 + XY, [21] |
| 27. Melanorhopala clavata Stål, 1873 | 12 + XY, [15,16] |
| 28. Oncochila simplex (Herrich-Schaeffer, 1830) | 12 + XY {AA}, [29] |
| 29. Physatocheila confinis (Horváth, 1906) | 12 + XY {AA}, [27] |
| 30. Ph. smreczynskii China, 1952 | 12 + XY, [25] 12 + XY {AA}, present study |
| 31. Ph. putshkovi Golub, 1976 | 12 + XY {AA}, present study |
| 32. Stephanitis caucasica Kiritshenko, 1939 | 12 + XY {AA}, [27] |
| 33. S. nashi Esaki and Takeya 1931 | 12 + XY, [17] |
| 34. S. oberti (Kolenati, 1856) | 12 + XY, [25] |
| 35. S. pyri (Fabricius, 1775) | 12 + XY {AA}, [27] |
| 36. S. oschanini Vasiliev, 1935 | 12 + XY, present study |
| 37. S. takeyai Drake and Maa, 1955 | 12 + XY, [19] |
| 38. Teleonemia elata Drake, 1935 | 12 + XY, [21] |
| 39. T. scrupulosa Stål, 1873 | 12 + XY, [21] |
| 40. Tingis (Neolasiotropis) pilosa Hummel, 1825 | 12 + XY {AA}, [29] |
| 41. T. (Tropidocheila) caucasica (Jakovlev, 1880) | 12 + XY, [25] |
| 42. T. (Tr.) reticulata Herrich-Schaeffer, 1835 | 12 + XY {AA}, [29] |
| 43. T. (Tr.) sideritis Štusák, 1973 | 12 + XY, [25] |
| 44. T. (Tingis) ampilata (Herrich-Schaeffer, 1838) | 12 + XY, [18] |
| 45. T. (T.) brevicornis (Horváth, 1902) | 12 + XY {AA}, present study |
| 46. T. (T.) cardui (Linnaeus, 1758) | 12 + XY, [18] 12 + XY {AA}, [27] |
| 47. T. (T.) crispata (Herrich-Schaeffer, 1838) | 12 + XY {X and Y}, [26] |
| 48. T. (T.) lasiocera Matsumura, 1907 | 12 + XY, [22] |
| Tribe Acalyptaini | |
| 49. Acalypta carinata (Panzer, 1806) | 12 + X(0), [25] 12 + X(0) {AA}, [29] |
| 50. A. gracilis (Fieber, 1844) | 12 + X(0) {AA}, present study |
| 51. A. hellenica Reuter, 1888 | 12 + X(0) {AA}, present study |
| 52. A. marginata (Wolff, 1804) | 12 + X(0) {AA}, [29] |
| 53. A. nigrina (Fallén, 1807) | 12 + X(0) [25] |
| 54. A. parvula (Fallen, 1807) | 10 + XY, [18] 12 + X(0), [25] |
| 55. Derephysia (Paraderephysia) longispina Golub, 1974 | 12 + X(0) {AA and X}, [29] |
| 56. D. (P.) cristata (Panzer, 1806) | 12 + X(0) {AA}, present study |
| 57. Dictyonota fuliginosa Costa, 1855 | 12 + XY, [18] |
| 58. D. strichnocera Fieber, 1844 | 12 + X(0) {AA}, [29] |
| 59. Kalama beckeri (Jakovlev, 1871) | 12 + X(0) {AA}, present study |
| 60. K. tricornis (Schrank, 1801) | 12 + X(0), [24,25] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Golub, N.V.; Golub, V.B.; Anokhin, B.A.; Kuznetsova, V.G. Comparative Cytogenetics of Lace Bugs (Tingidae, Heteroptera): New Data and a Brief Overview. Insects 2022, 13, 608. https://doi.org/10.3390/insects13070608
Golub NV, Golub VB, Anokhin BA, Kuznetsova VG. Comparative Cytogenetics of Lace Bugs (Tingidae, Heteroptera): New Data and a Brief Overview. Insects. 2022; 13(7):608. https://doi.org/10.3390/insects13070608
Chicago/Turabian StyleGolub, Natalia V., Viktor B. Golub, Boris A. Anokhin, and Valentina G. Kuznetsova. 2022. "Comparative Cytogenetics of Lace Bugs (Tingidae, Heteroptera): New Data and a Brief Overview" Insects 13, no. 7: 608. https://doi.org/10.3390/insects13070608
APA StyleGolub, N. V., Golub, V. B., Anokhin, B. A., & Kuznetsova, V. G. (2022). Comparative Cytogenetics of Lace Bugs (Tingidae, Heteroptera): New Data and a Brief Overview. Insects, 13(7), 608. https://doi.org/10.3390/insects13070608








