First Comprehensive Analysis of Both Mitochondrial Characteristics and Mitogenome-Based Phylogenetics in the Subfamily Eumeninae (Hymenoptera: Vespidae)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Whole-Genome Sequencing and Assembling
2.3. Mitogenome Annotation and Sequence Analysis
2.4. Phylogenetic Analyses
3. Results and Discussion
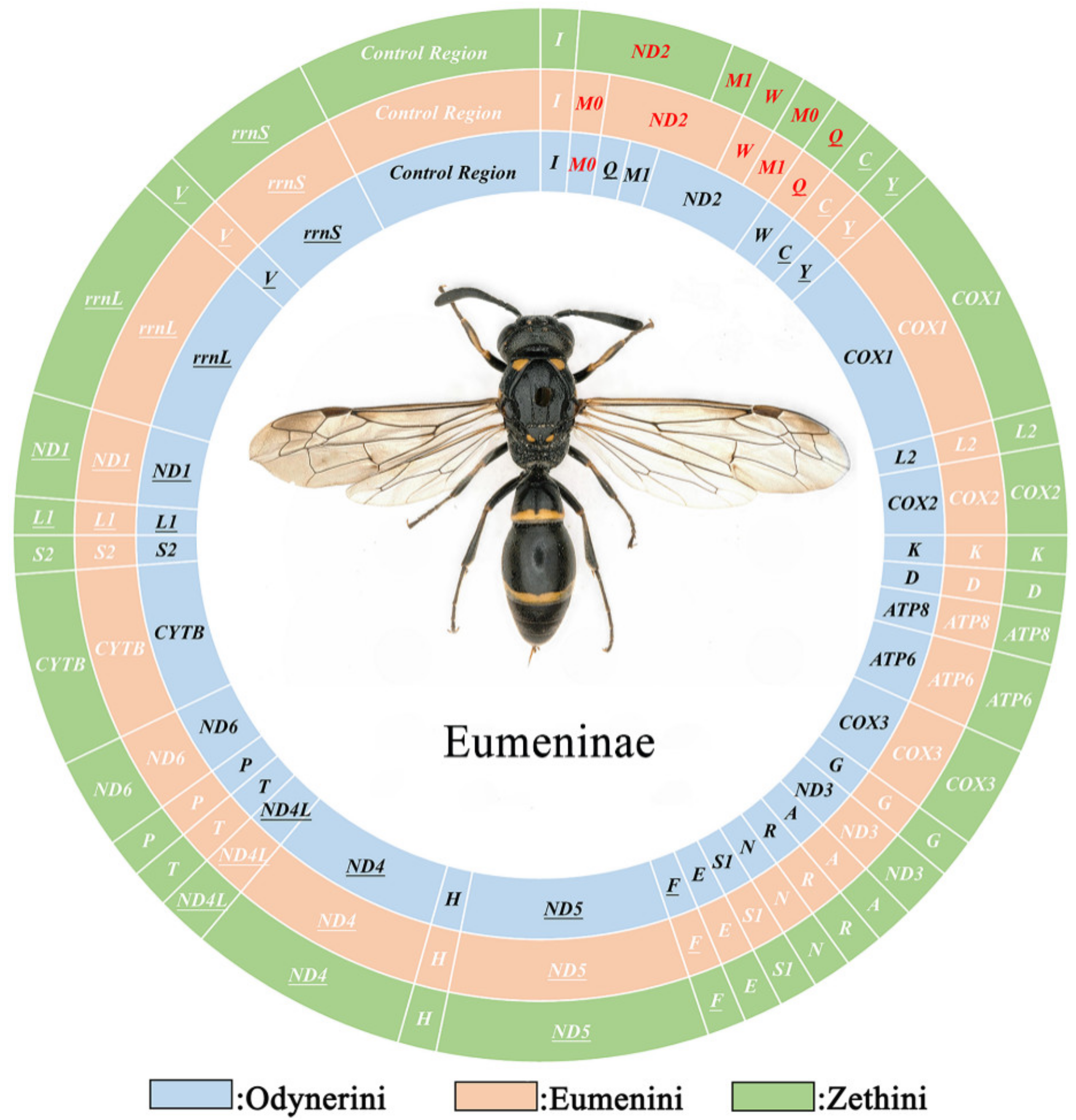
3.1. Mitochondrial Genome Organization
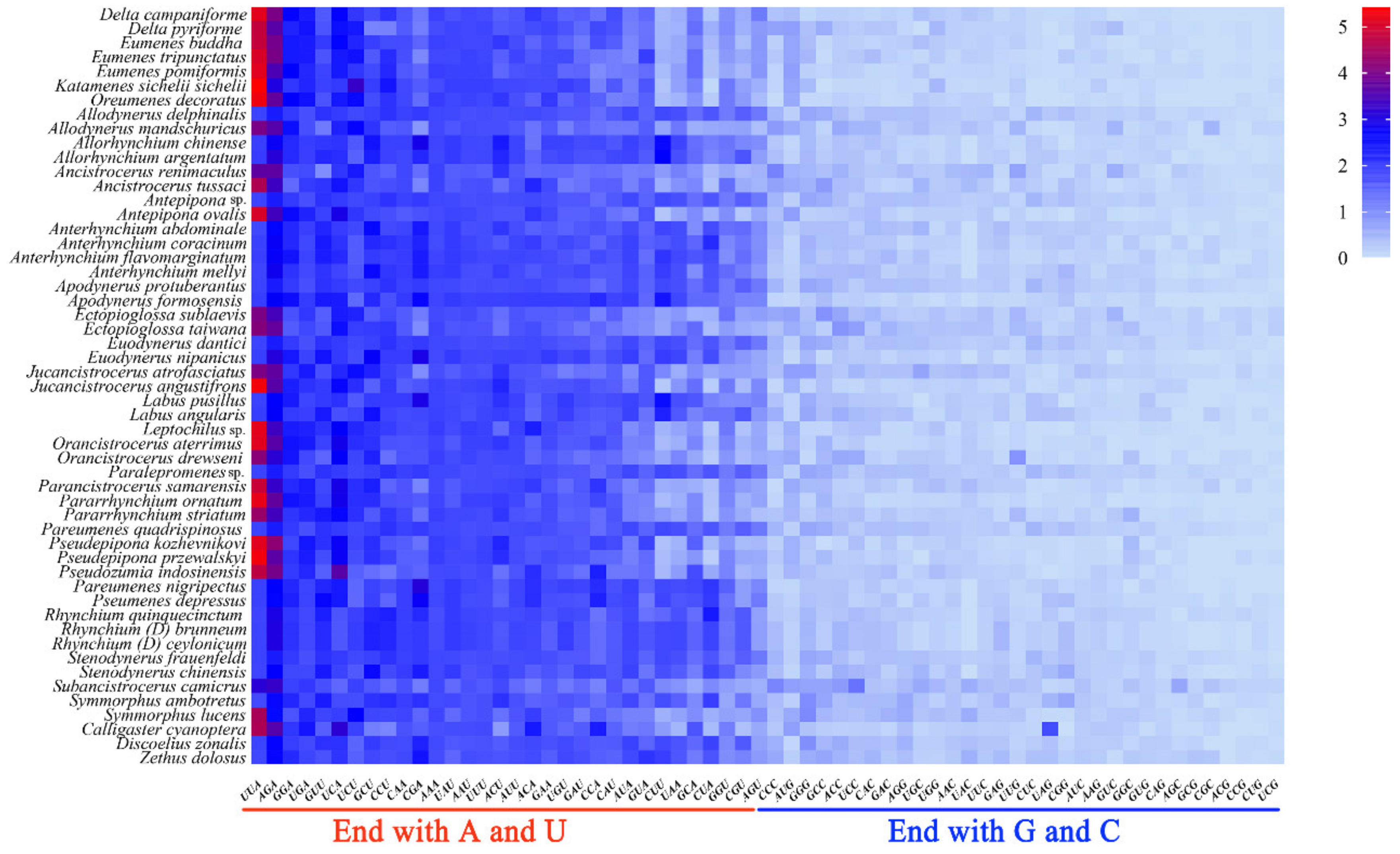
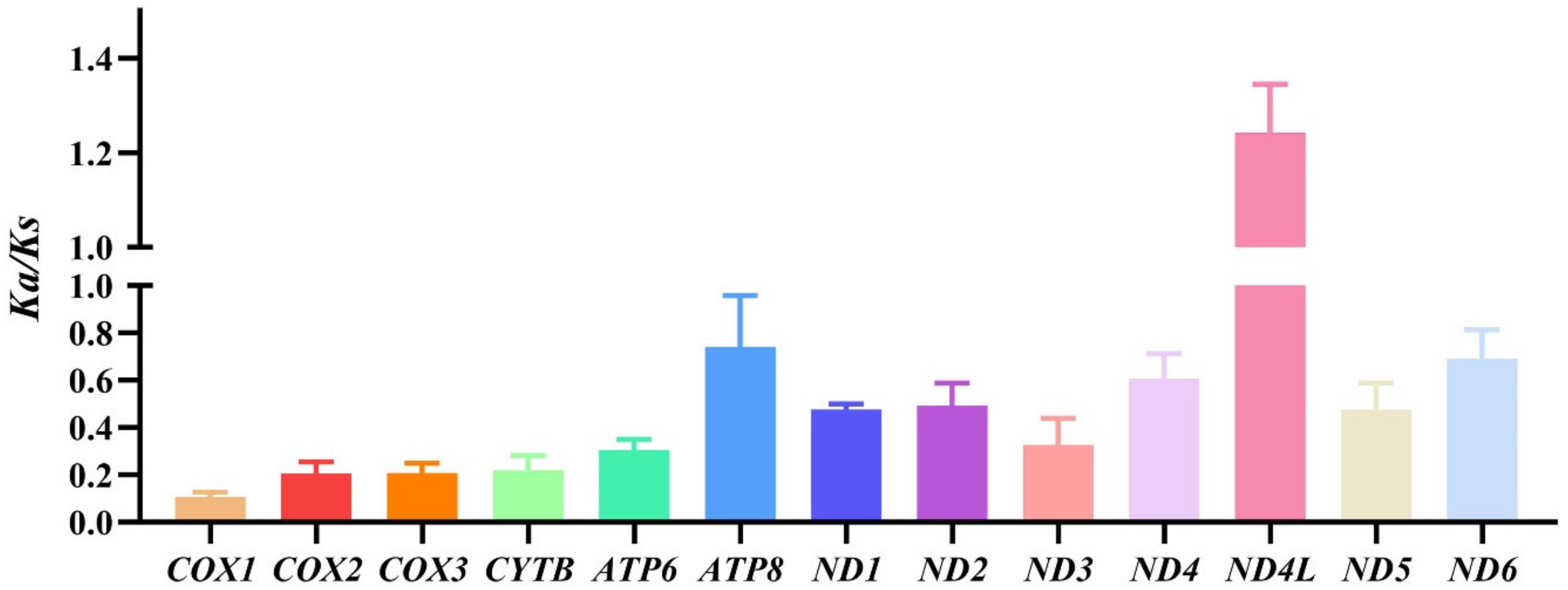
3.2. Protein-Coding Genes and Codon Usage Patterns
3.3. Gene Arrangement
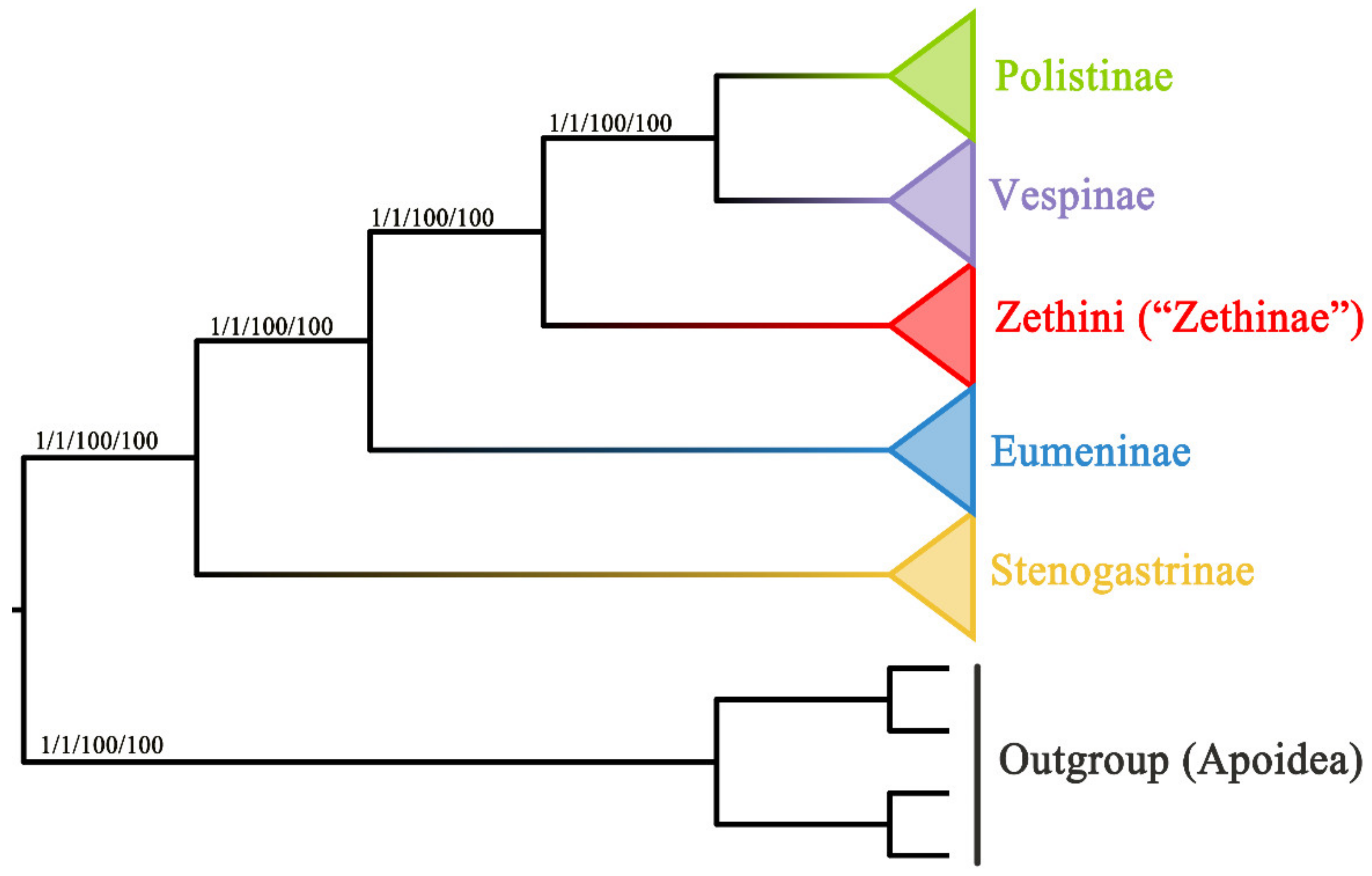
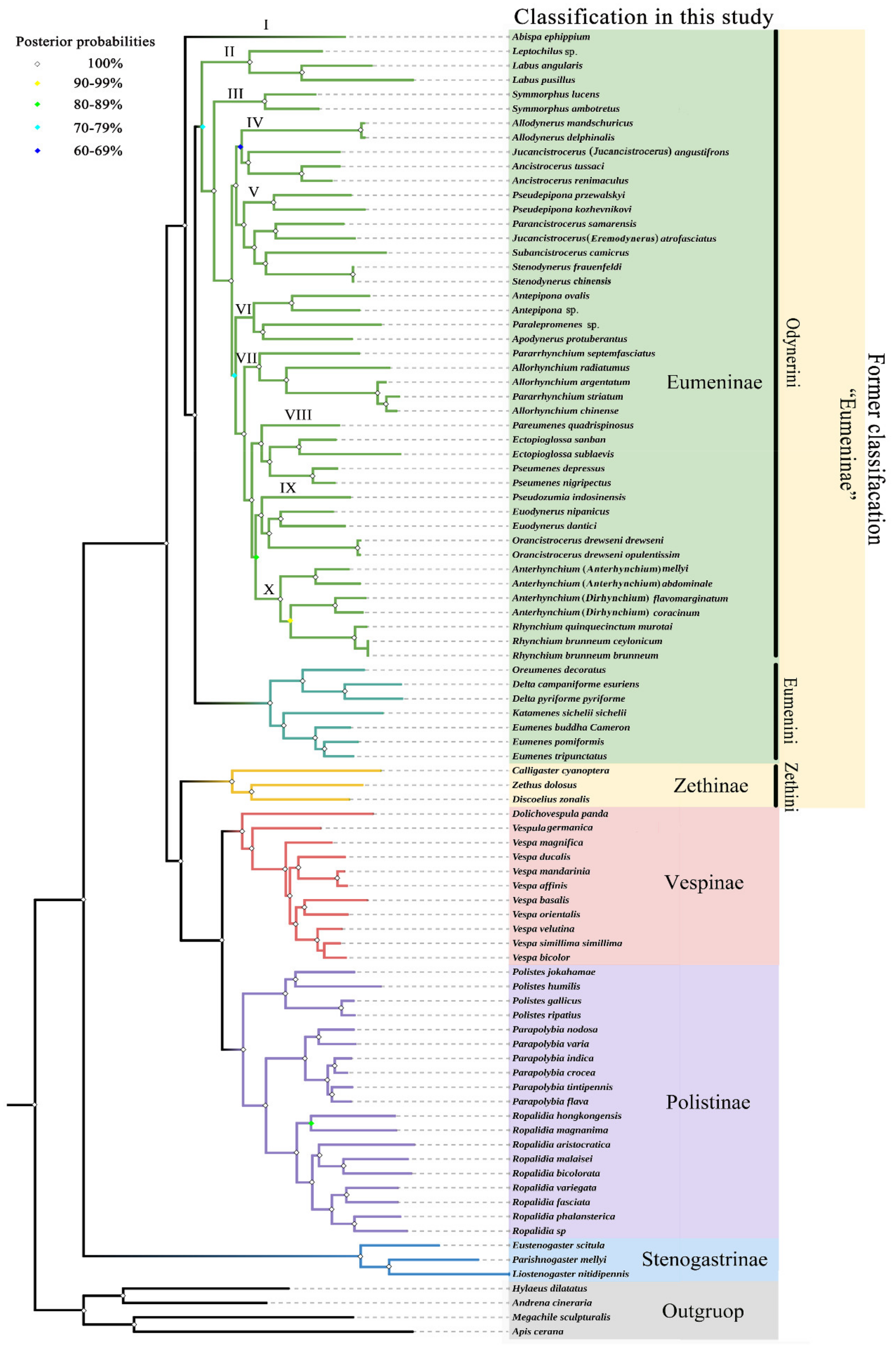
3.4. Phylogenetic Relationship of Vespidae
3.5. Phylogenetic Relationship within Eumeninae
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cowan, D.P. The Social Biology of Wasps; Cornell University Press: Ithaca, NY, USA, 1991; pp. 33–73. [Google Scholar]
- Jennings, D.T.; Houseweart, M.W. Predation by Eumenid Wasps (Hymenoptera: Eumenidae) on Spruce Budworm (Lepidoptera: Tortricidae) and Other Lepidopterous Larvae in Spruce-Fir Forests of Maine. Ann. Entomol. Soc. Am. 1984, 49, 39–45. [Google Scholar] [CrossRef]
- Pannure, A.; Belavadi, V.V.; Carpenter, J.M. Taxonomic studies on potter wasps (Hymenoptera: Vespidae: Eumeninae) of south India. Zootaxa 2016, 4171, 1–50. [Google Scholar] [CrossRef] [PubMed]
- Bohart, R.M.; Stange, L.A. A Revision of the Genus Zethus in the Western Hemisphere (Hymenoptera, Eumenidae); University of California Press: Oakland, CA, USA, 1965; Volume 40, pp. 1–208. [Google Scholar]
- Hines, H.M.; Hunt, J.H.; O’Connor, T.K.; Gillespie, J.J.; Cameron, S.A. Multigene phylogeny reveals eusociality evolved twice in vespid wasps. Proc. Natl. Acad. Sci. USA 2007, 104, 3295–3299. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, J.M.; Cumming, J.M. A character analysis of the North American potter wasps (Hymenoptera: Vespidae; Eumeninae). J. Nat. Hist. 1985, 19, 877–916. [Google Scholar] [CrossRef]
- Latreille, P.A. Histoire Naturelle, Générale et Particulière des Crustacés et des Insects: Ouvrage Faisant Suite aux Oeuvres de Leclerc de Buffon, et Partie du Cours Complet D’historie Naturelle Rédigé par C.S.; Sonnini, F., Ed.; Dufart: Paris, France, 1802. [Google Scholar] [CrossRef]
- de Saussure, H.F. Etudes sur la Famille des Vespides 1. Monographie des Guepes Solitaires ou de la Tribe des Eumeniens; Masson, V., Cherbuliez, J., Eds.; Victor Masson: Paris, France, 1852–1853; pp. 1–286. [Google Scholar]
- de Saussure, H.F. Etudes sur la Famille des Vespides 3. La Monographie des Masariens et un Supplement a la Monographie des Eumeniens; Masson, V., Cherbuliez, J., Eds.; Victor Masson: Paris, France, 1854–1856; pp. 1–352. [Google Scholar]
- Richards, O.W. A Revisional Study of the Masarid Wasps (Hymenoptera, Vespoidea); British Museum (Natural History): London, UK, 1962; pp. 1–294. [Google Scholar]
- Carpenter, J.M. The phylogenetic relationships and natural classification of the Vespoidea (Hymenptera). Syst. Entomol. 1982, 7, 11–38. [Google Scholar] [CrossRef]
- Hermes, M.G.; Melo, G.; Carpenter, J.M. The higher-level phylogenetic relationships of the Eumeninae (Insecta, Hymenoptera, Vespidae), with emphasis onEumenessensu lato. Cladistics 2013, 30, 453–484. [Google Scholar] [CrossRef]
- Schmitz, J.; Moritz, R. Molecular Phylogeny of Vespidae (Hymenoptera) and the Evolution of Sociality in Wasps. Mol. Phylogenetics Evol. 1998, 9, 183–191. [Google Scholar] [CrossRef][Green Version]
- Bank, S.; Sann, M.; Mayer, C.; Meusemann, K.; Donath, A.; Podsiadlowski, L.; Kozlov, A.; Petersen, M.; Krogmann, L.; Meier, R.; et al. Transcriptome and target DNA enrichment sequence data provide new insights into the phylogeny of vespid wasps (Hymenoptera: Aculeata: Vespidae). Mol. Phylogenetics Evol. 2017, 116, 213–226. [Google Scholar] [CrossRef]
- Piekarski, P.K.; Carpenter, J.M.; Lemmon, A.R.; Lemmon, E.M.; Sharanowski, B.J. Phylogenomic Evidence Overturns Current Conceptions of Social Evolution in Wasps (Vespidae). Mol. Biol. Evol. 2018, 35, 2097–2109. [Google Scholar] [CrossRef]
- Heraty, J.; Ronquist, F.; Carpenter, J.M.; Hawks, D.; Schulmeister, S.; Dowling, A.P.; Murray, D.; Munro, J.; Wheeler, W.C.; Schiff, N.; et al. Evolution of the hymenopteran megaradiation. Mol. Phylogenetics Evol. 2011, 60, 73–88. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, J.-J.; Li, W.-H. Complete Mitochondrial Genome of Suwallia teleckojensis (Plecoptera: Chloroperlidae) and Implications for the Higher Phylogeny of Stoneflies. Int. J. Mol. Sci. 2018, 19, 680. [Google Scholar] [CrossRef]
- Mao, M.; Gibson, T.; Dowton, M. Higher-level phylogeny of the Hymenoptera inferred from mitochondrial genomes. Mol. Phylogenetics Evol. 2014, 84, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Li, T.-J.; Chen, B. Descriptions of four new species of Pararrhynchium de Saussure (Hymenoptera: Vespidae: Eumeninae) from China, with one newly recorded species and a key to Chinese species. Orient. Insects 2018, 52, 175–189. [Google Scholar] [CrossRef]
- Li, T.-J.; Barthélémy, C.; Carpenter, J.M. The Eumeninae (Hymenoptera, Vespidae) of Hong Kong (China), with description of two new species, two new synonymies and a key to the known taxa. J. Hymenopt. Res. 2019, 72, 127–176. [Google Scholar] [CrossRef]
- Tan, J.-L.; Carpenter, J.M.; Van Achterberg, C. An illustrated key to the genera of Eumeninae from China, with a checklist of species (Hymenoptera, Vespidae). ZooKeys 2018, 740, 109–149. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.-L.; Carpenter, J.M.; Van Achterberg, C. Most northern Oriental distribution of Zethus Fabricius (Hymenoptera, Vespidae, Eumeninae), with a new species from China. J. Hymenopt. Res. 2018, 62, 1–13. [Google Scholar] [CrossRef]
- Wang, H.-C.; Chen, B.; Li, T.-J. A new species and a new record of the genus Discoelius Latreille, 1809 (Hymenoptera: Vespidae: Eumeninae) from China. Zootaxa 2019, 4686, 297–300. [Google Scholar] [CrossRef]
- Wang, H.-C.; Chen, B.; Li, T.-J. Three new species of the genus Zethus Fabricius, 1804 (Hymenoptera, Vespidae, Eumeninae) from China, with an updated key to the Oriental species. J. Hymenopt. Res. 2019, 71, 209–224. [Google Scholar] [CrossRef]
- Wang, H.-C.; Chen, B.; Li, T.-J. Taxonomy of the genus Ectopioglossa Perkins from China, with two new species and an updated key to the Oriental species (Hymenoptera: Vespidae: Eumeninae). J. Asia-Pacific Entomol. 2020, 23, 253–259. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, B.; Li, T.-J. Taxonomy of the genus Epsilon from China, with a new species and an updated key to the Oriental species (Hymenoptera, Vespidae, Eumeninae). ZooKeys 2020, 910, 131–142. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, B.; Li, T.-J. A taxonomic revision of Allodynerus Blüthgen (Hymenoptera: Vespidae: Eumeninae) from China. Zootaxa 2020, 4750, 545–559. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Chen, B.; Li, T.-J. A new species and two new records of the genus Pseudepipona de Saussure, 1856 (Hymenoptera, Vespidae, Eumeninae) from China, with a key to the Chinese species. J. Hymenopt. Res. 2021, 82, 285–304. [Google Scholar] [CrossRef]
- Bai, Y.; Chen, B.; Li, T.-J. Two newly recorded genera Malayepipona Giordani Soika and Megaodynerus Gusenleitner, with eight new species from China (Hymenoptera, Vespidae, Eumeninae). Zootaxa 2021, 5060, 371–391. [Google Scholar] [CrossRef]
- Li, T.-J.; Bai, Y.; Chen, B. A revision of the genus Jucancistrocerus Blüthgen, 1938 from China, with review of three related genera (Hymenoptera: Vespidae: Eumeninae). Zootaxa 2022, 5105, 401–420. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Leung, H.C.M.; Yiu, S.M.; Chin, F.Y.L. IBDA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.K.; Mukesh, J.; Liu, Z. NGS QC Toolkit: A Toolkit for Quality Control of Next Generation Sequencing Data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef] [PubMed]
- Simon, C.; Buckley, T.R.; Frati, F.; Stewart, J.B.; Beckenbach, A.T. Incorporating Molecular Evolution into Phylogenetic Analysis, and a New Compilation of Conserved Polymerase Chain Reaction Primers for Animal Mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 545–579. [Google Scholar] [CrossRef]
- Altschup, S.; Gish, W.; Miller, W.; Myers, E.; Lipman, D. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Search and contextual analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Laslett, D.; Canbäck, B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2008, 24, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.-S.; Cheng, A.-C.; Wang, M.-S.; Zhao, L.-C. Characterization of Synonymous Codon Usage Bias in the Duck Plague Virus UL35 Gene. Karger 2009, 52, 266–278. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Merkle, D.; Ramsch, D.; Fritzsch, G.; Perseke, M.; Bernhard, D.; Schlegel, M.; Stadler, P.F.; Middendorf, M. CREx: Inferring genomic rearrangements based on common intervals. Bioinformatics 2007, 23, 2957–2958. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlićc, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Abascal, F.; Zardoya, R.; Telford, M.J. TranslatorX: Multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010, 38, W7–W13. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of Phylogenies after Removing Divergent and Ambiguously Aligned Blocks from Protein Sequence Alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Lanfear, R.; Calcott, B.; Ho, S.Y.; Guindon, S. PartitionFinder: Combined Selection of Partitioning Schemes and Substitution Models for Phylogenetic Analyses. Mol. Biol. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Mark, P.; Ayres, D.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.; Huelsenbeck, J. MrBayes 3.2: Efcient Bayesian phyloge netic inference and model choice across a large model space. Syst. Biol. 2015, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Goloboff, P.A.; Catalano, S.A. TNT version 1.5, including a full implementation of phylogenetic morphometrics. Cladistics 2016, 32, 221–238. [Google Scholar] [CrossRef] [PubMed]
- Nixon, K.C. Winclada, version 1.98; Published by the author: Ithaca, NY, USA, 2015.
- Tang, P.; Zhu, J.-C.; Zheng, B.-Y.; Wei, S.-J.; Sharkey, M.; Chen, X.-X. Mitochondrial phylogenomics of the Hymenoptera. Mol. Phylogenetics Evol. 2018, 131, 8–18. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Mao, M.; Valerio, A.; Austin, A.D.; Dowton, M.; Johnson, N.F. The first mitochondrial genome for the wasp superfamily Platygastroidea: The egg parasitoid Trissolcus basalis. Genome 2012, 55, 194–204. [Google Scholar] [CrossRef]
- Korkmaz, E.M.; Dogan, O.; Durel, B.S.; Budak, M.; Basibuyuk, H.H. Mitogenome organization and evolutionary history of the subfamily Cephinae (Hymenoptera: Cephidae). Syst. Entomol. 2018, 43, 606–618. [Google Scholar] [CrossRef]
- Cameron, S.L.; Dowton, M.; Castro, L.R. Mitochondrial genome organization and phylogeny of two vespid wasps. Genome 2008, 51, 800–808. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.M.; Li, W.-H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol. 1986, 24, 28–38. [Google Scholar] [CrossRef]
- Yu, T.; Li, J.; Yang, Y.; Qi, L. Codon usage patterns and adaptive evolution of marine unicellular cyanobacteria Synechococcus and Prochlorococcus. Mol. Phylogenetics Evol. 2012, 61, 206–213. [Google Scholar] [CrossRef]
- Wei, L.; He, J.; Jia, X.; Qi, Q.; Liang, Z.; Zheng, H. Analysis of codon usage bias of mitochondrial genome in Bombyx moriand its relation to evolution. BMC Evol. Biol. 2014, 14, 262. [Google Scholar] [CrossRef] [PubMed]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486. [Google Scholar] [CrossRef]
- Barr, C.M.; Neiman, M.; Taylor, D.R. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 2005, 168, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Dowton, M.; Austin, A.D. Evolutionary dynamics of a mitochondrial rearrangement "hot spot" in the Hymenoptera. Mol. Biol. Evol. 1999, 16, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.B.; Wu, Y.F.; Yang, C.; Gu, X.H.; Wilson, J.J.; Li, H.; Cai, W.Z.; Yang, H.L.; Song, F. Evolution of tRNA gene rearrangement in the mitochondrial genome of ichneumonoid wasps (Hymenoptera: Ichneumonoidea). Int. J. Biol. Macromol. 2020, 164, 540–547. [Google Scholar] [CrossRef]
- Wei, S.-J.; Niu, F.-F.; Du, B.-Z. Rearrangement oftrnQ-trnMin the mitochondrial genome ofAllantus luctifer(Smith) (Hymenoptera: Tenthredinidae). Mitochondrial DNA 2014, 27, 856–858. [Google Scholar] [CrossRef]
- Peters, R.S.; Krogmann, L.; Mayer, C.; Donath, A.; Niehuis, O. Evolutionary History of the Hymenoptera. Curr. Biol. 2017, 27, 1013–1018. [Google Scholar] [CrossRef]
- Huang, P.; Carpenter, J.M.; Chen, B.; Li, T.-J. The first divergence time estimation of the subfamily Stenogastrinae (Hymenoptera: Vespidae) based on mitochondrial phylogenomics. Int. J. Biol. Macromol. 2019, 137, 767–773. [Google Scholar] [CrossRef]
- Ducke, A. Uber Phylogenie und Klassifikation der socialen Vespiden. Zool. Jahr. Abt. Syst. Geogr. Biol. Tiere 1914, 36, 303–330. [Google Scholar]
- Rau, P. The Jungle Bees and Wasps of Barro Colorado Island. Ann. Entomol. Soc. Am. 1933, 2, 376. [Google Scholar] [CrossRef]
- Blüthgen, P. Beiträge zur Kenntnis der paläarktischen und einiger athiopischer Faltenwespen (Hym. Vespidae). Veröffentlichungen Aus Dem Dtsch. Kolon. Und Übersee Mus. Bremen 1939, 2, 233–267. [Google Scholar]



| Species | bp | Gene Number | Accession No. |
|---|---|---|---|
| Allodynerus delphinalis | 16,932 | 38 | ON024142 |
| Allodynerus mandschuricus | 17,449 | 38 | ON012816 |
| Allorhynchium chinense | 16,909 | 38 | MK051021 |
| Allorhynchium argentatum | 17,972 | 38 | MK051022 |
| Allorhynchium radiatumus | 17,349 | 38 | ON055163 |
| Ancistrocerus renimaculus | 15,614 | 36 | ON045342 |
| Ancistrocerus tussaci | 15,679 | 35 | ON012815 |
| Antepipona sp. | 19,040 | 38 | ON012817 |
| Antepipona ovalis | 17,514 | 36 | ON012818 |
| Anterhynchium abdominale | 16,488 | 36 | MK051029 |
| Anterhynchium coracinum | 16512 | 36 | MK051028 |
| Anterhynchium flavomarginatum | 15,196 | 35 | MK051026 |
| Anterhynchium mellyi | 18,692 | 38 | ON012812 |
| Apodynerus protuberantus | 17,943 | 36 | ON045341 |
| Calligaster cyanoptera | 16,316 | 38 | ON012814 |
| Delta pyriforme pyriforme | 14,883 | 35 | ON076029 |
| Delta campaniforme esuriens | 16,126 | 36 | ON055486 |
| Discoelius zonalis | 15,435 | 38 | ON076025 |
| Eumenes buddha | 16,048 | 37 | ON076024 |
| Eumenes tripunctatus | 15,702 | 36 | ON045343 |
| Eumenes pomiformis | 16,520 | 38 | ON076031 |
| Ectopioglossa sublaevis | 16,203 | 36 | ON045340 |
| Ectopioglossa sanban | 16,454 | 37 | ON012813 |
| Euodynerus dantici | 17,493 | 37 | ON076022 |
| Euodynerus nipanicus | 22,088 | 38 | ON076021 |
| Jucancistrocerus atrofasciatus | 18,848 | 38 | ON045348 |
| Jucancistrocerus angustifrons | 19,867 | 38 | ON012819 |
| Katamenes sichelii sichelii | 14,807 | 37 | ON076027 |
| Labus pusillus | 17,409 | 36 | ON076026 |
| Labus angularis | 15,227 | 35 | ON076030 |
| Leptochilus sp. | 14,241 | 36 | ON045339 |
| Orancistrocerus drewseni drewseni | 17,636 | 38 | ON045338 |
| Oreumenes decorates | 15,563 | 37 | ON076028 |
| Paralepromenes sp. | 16,805 | 37 | ON045337 |
| Parancistrocerus samarensis | 17,773 | 38 | ON076023 |
| Pararrhynchium striatum | 20,403 | 38 | ON045347 |
| Pararrhynchium septemfasciatus | 18,003 | 36 | ON055487 |
| Pareumenes quadrispinosus acutus | 17,426 | 38 | ON076020 |
| Pseudepipona kozhevnikovi | 15,650 | 35 | ON076019 |
| Pseudepipona przewalskyi | 20,281 | 37 | ON024141 |
| Pseudozumia indosinensis | 16,376 | 35 | ON045335 |
| Pseumenes nigripectus | 17,773 | 38 | ON045336 |
| Pseumenes depressus | 16,677 | 38 | ON045346 |
| Rhynchium quinquecinctum murotai | 16,317 | 36 | MK051030 |
| Rhynchium brunneum brunneum | 23,251 | 38 | MK051031 |
| Rhynchium brunneum ceylonicum | 23,122 | 38 | MK051032 |
| Stenodynerus frauenfeldi | 17,252 | 36 | ON045334 |
| Stenodynerus chinensis | 17,194 | 38 | ON045345 |
| Subancistrocerus camicrus | 18,035 | 38 | ON045344 |
| Symmorphus ambotretus | 17,280 | 38 | ON076018 |
| Symmorphus lucens | 17,865 | 38 | ON076017 |
| Zethus dolosus | 16,306 | 38 | ON076016 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, L.; Carpenter, J.M.; Chen, B.; Li, T. First Comprehensive Analysis of Both Mitochondrial Characteristics and Mitogenome-Based Phylogenetics in the Subfamily Eumeninae (Hymenoptera: Vespidae). Insects 2022, 13, 529. https://doi.org/10.3390/insects13060529
Luo L, Carpenter JM, Chen B, Li T. First Comprehensive Analysis of Both Mitochondrial Characteristics and Mitogenome-Based Phylogenetics in the Subfamily Eumeninae (Hymenoptera: Vespidae). Insects. 2022; 13(6):529. https://doi.org/10.3390/insects13060529
Chicago/Turabian StyleLuo, Li, James M. Carpenter, Bin Chen, and Tingjing Li. 2022. "First Comprehensive Analysis of Both Mitochondrial Characteristics and Mitogenome-Based Phylogenetics in the Subfamily Eumeninae (Hymenoptera: Vespidae)" Insects 13, no. 6: 529. https://doi.org/10.3390/insects13060529
APA StyleLuo, L., Carpenter, J. M., Chen, B., & Li, T. (2022). First Comprehensive Analysis of Both Mitochondrial Characteristics and Mitogenome-Based Phylogenetics in the Subfamily Eumeninae (Hymenoptera: Vespidae). Insects, 13(6), 529. https://doi.org/10.3390/insects13060529







