Identification and Characterization of Genes Related to Resistance of Autographa californica Nucleopolyhedrovirus Infection in Bombyx mori
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Genome-Wide Association and Population Genetic Analysis
2.3. Virus Gene Expression Profile Analysis based on RNA-Sequencing Data
2.4. Genome and RNA Extraction
2.5. PCR and qRT-PCR
2.6. Synthesis of siRNA
2.7. BmN Cell Culture and Transfection
2.8. Statistical Analysis
3. Results
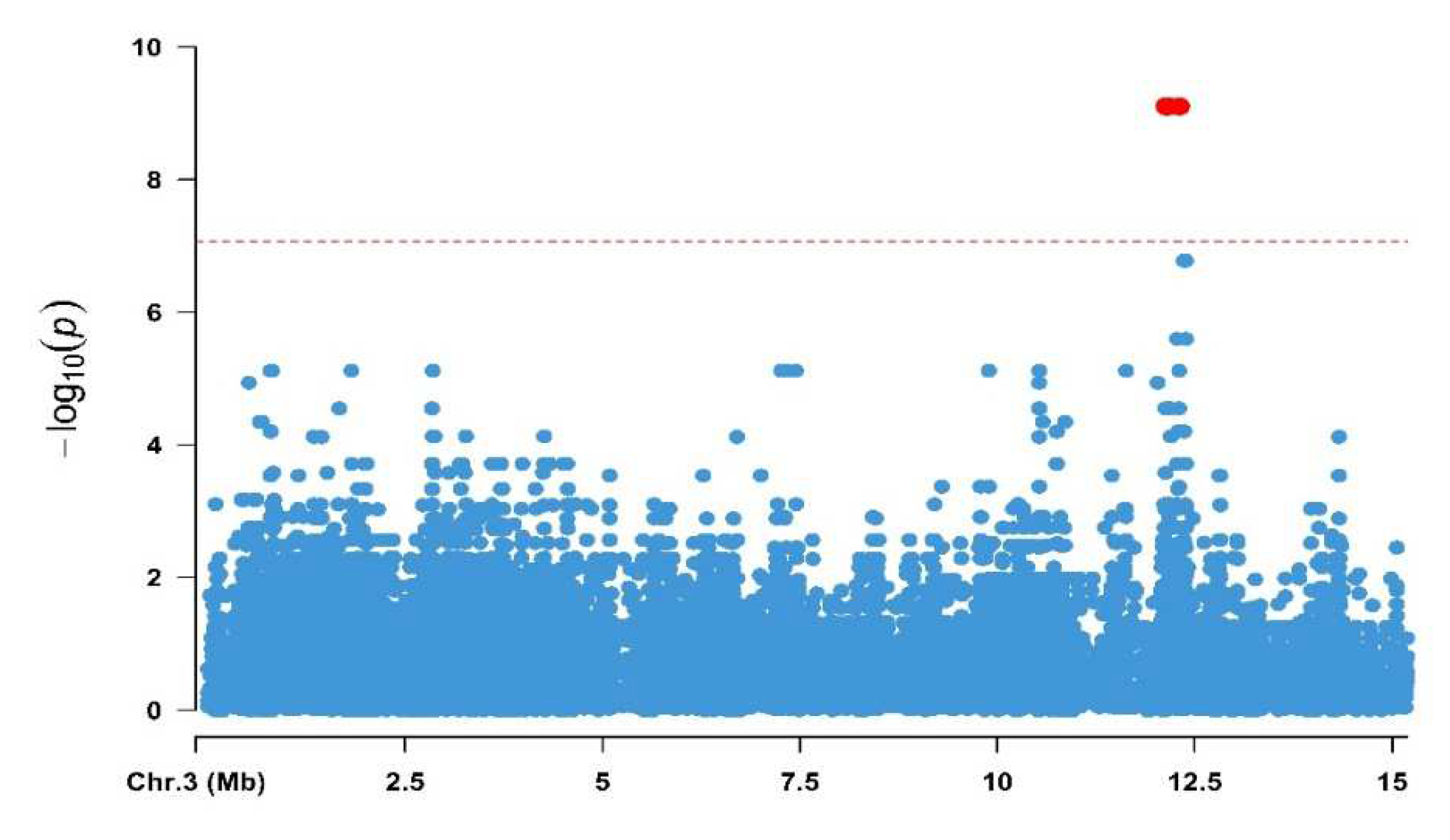
3.1. Genome-Wide Association Analysis
3.2. Several Consistent Amino Acid Mutations among Different Resistance Silkworm Strains
3.3. Inhibition of BmNPC1 Decreases Propagation of the AcMNPV
3.4. Distinct Virus Gene Expression Pattern between C108 and p50 Strain
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Rohrmann, G.F. Baculovirus Molecular Biology, 4th ed.; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2019. [Google Scholar]
- Goulson, D. Can Host Susceptibility to Baculovirus Infection be Predicted from Host Taxonomy or Life History? Environ. Entomol. 2003, 32, 61–70. [Google Scholar] [CrossRef]
- Lu, A.; Miller, L.K. Species-specific effects of the hcf-1 gene on baculovirus virulence. J. Virol. 1996, 70, 5123–5130. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Thiem, S.M. Characterization of Host Range Factor 1 (hrf-1) Expression inLymantria disparM Nucleopolyhedrovirus- and RecombinantAutographa californicaM Nucleopolyhedrovirus-Infected IPLB-Ld652Y Cells. Virology 1997, 227, 420–430. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-J.; Quentin, M.E.; Brennan, L.A.; Kukel, C.; Thiem, S.M. Lymantria dispar Nucleopolyhedrovirus hrf-1 Expands the Larval Host Range of Autographa californica Nucleopolyhedrovirus. J. Virol. 1998, 72, 2526–2531. [Google Scholar] [CrossRef]
- Shikata, M.; Matsumoto, T.; Sano, Y.; Hashimoto, Y.; Shibata, H.; Sakurai, M. The ecdysteroid UDP-glucosyltransferase gene of Autographa californica nucleopolyhedrovirus alters the moulting and metamorphosis of a non-target insect, the silkworm, Bombyx mori (Lepidoptera, Bombycidae). J. Gen. Virol. 1998, 79, 1547–1551. [Google Scholar] [CrossRef][Green Version]
- Yamao, M.; Katayama, N.; Nakazawa, H.; Yamakawa, M.; Hayashi, Y.; Hara, S.; Kamei, K.; Mori, H. Gene targeting in the silkworm by use of a baculovirus. Genes Dev. 1999, 13, 511–516. [Google Scholar] [CrossRef][Green Version]
- Guo, T.; Wang, S.; Guo, X.; Lu, C. Productive infection of Autographa californica nucleopolyhedrovirus in silkworm Bombyx mori strain Haoyue due to the absence of a host antiviral factor. Virology 2005, 341, 231–237. [Google Scholar] [CrossRef]
- Xu, J.; Kusakabe, T.; Yamamoto, K.; Suetsugu, Y.; Mon, H.; Li, Z.; Zhu, L.; Iiyama, K.; Banno, Y.; Yoshimura, K.; et al. A novel third chromosomal locus controls susceptibility to Autographa californica multiple nucleopolyhedrovirus in the silkworm, Bombyx mori. Appl. Microbiol. Biotechnol. 2013, 98, 3049–3058. [Google Scholar] [CrossRef]
- Loftus, S.K.; Morris, J.A.; Carstea, E.D.; Gu, J.Z.; Cummings, C.; Brown, A.; Ellison, J.; Ohno, K.; Rosenfeld, M.A.; Tagle, D.A.; et al. Murine Model of Niemann-Pick C Disease: Mutation in a Cholesterol Homeostasis Gene. Science 1997, 277, 232–235. [Google Scholar] [CrossRef]
- Higgins, M.E.; Davies, J.P.; Chen, F.W.; Ioannou, Y.A. Niemann–Pick C1 Is a Late Endosome-Resident Protein That Transiently Associates with Lysosomes and the Trans-Golgi Network. Mol. Genet. Metab. 1999, 68, 1–13. [Google Scholar] [CrossRef]
- Watari, H.; Blanchette-Mackie, E.J.; Dwyer, N.K.; Glick, J.M.; Patel, S.; Neufeld, E.B.; Brady, R.O.; Pentchev, P.G.; Strauss, J.F. Niemann-Pick C1 protein: Obligatory roles for N-terminal domains and lysosomal targeting in cholesterol mobilization. Proc. Natl. Acad. Sci. USA 1999, 96, 805–810. [Google Scholar] [CrossRef] [PubMed]
- Côté, M.; Misasi, J.; Ren, T.; Bruchez, A.; Lee, K.; Filone, C.M.; Hensley, L.; Li, Q.; Ory, D.; Chandran, K.; et al. Small molecule inhibitors reveal Niemann–Pick C1 is essential for Ebola virus infection. Nature 2011, 477, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Carette, J.E.; Raaben, M.; Wong, A.C.; Herbert, A.S.; Obernosterer, G.; Mulherkar, N.; Kuehne, A.I.; Kranzusch, P.J.; Griffin, A.M.; Ruthel, G.; et al. Ebola virus entry requires the cholesterol transporter Niemann–Pick C1. Nature 2011, 477, 340–343. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, A.; Miller, E.H.; Herbert, A.S.; Ng, M.; Ndungo, E.; Whelan, S.; Dye, J.M.; Chandran, K. Niemann-Pick C1 (NPC1)/NPC1-like1 Chimeras Define Sequences Critical for NPC1’s Function as a Filovirus Entry Receptor. Viruses 2012, 4, 2471–2484. [Google Scholar] [CrossRef] [PubMed]
- Mitroi, D.N.; Pereyra-Gómez, G.; Soto-Huelin, B.; Senovilla, F.; Kobayashi, T.; Esteban, J.A.; Ledesma, M.D. NPC 1 enables cholesterol mobilization during long-term potentiation that can be restored in Niemann–Pick disease type C by CYP 46A1 activation. EMBO Rep. 2019, 20, e48143. [Google Scholar] [CrossRef]
- Li, Z.; Fan, Y.; Wei, J.; Mei, X.; He, Q.; Zhang, Y.; Li, T.; Long, M.; Chen, J.; Bao, J.; et al. Baculovirus Utilizes Cholesterol Transporter NIEMANN–Pick C1 for Host Cell Entry. Front. Microbiol. 2019, 10, 2825. [Google Scholar] [CrossRef]
- Ding, X.-Y.; Wang, X.-Y.; Kong, Y.-H.; Zhao, C.-X.; Qin, S.; Sun, X.; Li, M.-W. Comparative Transcriptome Analysis of Bombyx mori (Lepidoptera) Larval Hemolymph in Response to Autographa californica Nucleopolyhedrovirus in Differentially Resistant Strains. Processes 2021, 9, 1401. [Google Scholar] [CrossRef]
- Xiang, H.; Liu, X.; Li, M.; Zhu, Y.; Wang, L.; Cui, Y.; Liu, L.; Fang, G.; Qian, H.; Xu, A.; et al. The evolutionary road from wild moth to domestic silkworm. Nat. Ecol. Evol. 2018, 2, 1268–1279. [Google Scholar] [CrossRef]
- Li, T.; Wang, X.; Qin, S.; Sun, X.; Wang, S.; Li, M. The hemolymph melanization response is related to defence against the AcMNPV infection in Bombyx mori. Arch. Insect Biochem. Physiol. 2021, 108, e21764. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, H. Exploring single-sample SNP and INDEL calling with whole-genome de novo assembly. Bioinformatics 2012, 28, 1838–1844. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-C.; Yokoi, K.; Yamamoto, K.; Jouraku, A. An update of KAIKObase, the silkworm genome database. Database 2021, 2021, baaa099. [Google Scholar] [CrossRef] [PubMed]
- Poplin, R.; Ruano-Rubio, V.; DePristo, M.A.; Fennell, T.J.; Carneiro, M.O.; Van der Auwera, G.A.; Kling, D.E.; Gauthier, L.D.; Levy-Moonshine, A.; Roazen, D.; et al. Scaling Accurate Genetic Variant Discovery to Tens of Thousands of Samples. BioRxiv 2018, 201178. Available online: https://www.biorxiv.org/content/10.1101/201178v3.abstract (accessed on 17 October 2021).
- Van Der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X.; et al. rMVP: A Memory-efficient, Visualization-enhanced, and Parallel-accelerated Tool for Genome-wide Association Study. Genom. Proteom. Bioinform. 2021, 19, 619–628. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.-S.; Xu, J.-Y.; He, W.-M.; Yang, T.-L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. Feature Counts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; O’Connor, B.D. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra; O’Reilly Media: Sebastopol, CA, USA, 2020. [Google Scholar]
- Ke, X.-X.; Chao, H.; Abbas, M.N.; Kausar, S.; Gul, I.; Ji, H.; Yang, L.; Cui, H. Niemann-Pick type C1 regulates cholesterol transport and metamorphosis in silkworm, Bombyx mori (Dazao). Int. J. Biol. Macromol. 2020, 152, 525–534. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Yin, T.; Cook, D.; Lawrence, M. ggbio: An R package for extending the grammar of graphics for genomic data. Genome Biol. 2012, 13, R77. [Google Scholar] [CrossRef] [PubMed]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Lai, Q.; Zhu, L.; Xu, L.; Yuan, M.; Wu, W.; Yang, K. AcMNPV PKIP is associated with nucleocapsid of budded virions and involved in nucleocapsid assembly. Virus Res. 2019, 268, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Goldsmith, M.R.; Xia, Q. Advances in the Arms Race Between Silkworm and Baculovirus. Front. Immunol. 2021, 12, 628151. [Google Scholar] [CrossRef]
- Feng, M.; Zhang, J.; Xu, W.; Wang, H.; Kong, X.; Wu, X. Bombyx mori nucleopolyhedrovirus utilizes a clathrin and dynamin dependent endocytosis entry pathway into BmN cells. Virus Res. 2018, 253, 12–19. [Google Scholar] [CrossRef]
- Huang, J.; Hao, B.; Cheng, C.; Liang, F.; Shen, X.; Cheng, X. Entry of Bombyx mori nucleopolyhedrovirus into BmN cells by cholesterol-dependent macropinocytic endocytosis. Biochem. Biophys. Res. Commun. 2014, 453, 166–171. [Google Scholar] [CrossRef]
- Rahman, M.; Gopinathan, K.P. Systemic and in vitro infection process of Bombyx mori nucleopolyhedrovirus. Virus Res. 2004, 101, 109–118. [Google Scholar] [CrossRef]
- Katou, Y.; Ikeda, M.; Kobayashi, M. Abortive replication of Bombyx mori nucleopolyhedrovirus in Sf9 and High Five cells: Defective nuclear transport of the virions. Virology 2006, 347, 455–465. [Google Scholar] [CrossRef]
- Luz-Madrigal, A.; Asanov, A.; Camacho-Zarco, A.R.; Sampieri, A.; Vaca, L. A Cholesterol Recognition Amino Acid Consensus Domain in GP64 Fusion Protein Facilitates Anchoring of Baculovirus to Mammalian Cells. J. Virol. 2013, 87, 11894–11907. [Google Scholar] [CrossRef]
- Hao, B.; Nan, W.; Xu, Y.; Liu, L.; Liu, N.; Huang, J. Two Cholesterol Recognition Amino Acid Consensus Motifs of GP64 with Uncleaved Signal Peptide Are Required for Bombyx mori Nucleopolyhedrovirus Infection. Microbiol. Spectr. 2021, 9, 01725-21. [Google Scholar] [CrossRef]
- Takadate, Y.; Kondoh, T.; Igarashi, M.; Maruyama, J.; Manzoor, R.; Ogawa, H.; Kajihara, M.; Furuyama, W.; Sato, M.; Miyamoto, H.; et al. Niemann-Pick C1 Heterogeneity of Bat Cells Controls Filovirus Tropism. Cell Rep. 2020, 30, 308–319.e5. [Google Scholar] [CrossRef] [PubMed]
- Miller, E.H.; Obernosterer, G.; Raaben, M.; Herbert, A.S.; Deffieu, M.S.; Krishnan, A.; Ndungo, E.; Sandesara, R.G.; Carette, J.E.; Kuehne, A.I.; et al. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J. 2012, 31, 1947–1960. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Qian, H.; Zhou, X.; Wu, J.; Wan, T.; Cao, P.; Huang, W.; Zhao, X.; Wang, X.; Wang, P.; et al. Structural Insights into the Niemann-Pick C1 (NPC1)-Mediated Cholesterol Transfer and Ebola Infection. Cell 2016, 165, 1467–1478. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Saha, P.; Li, J.; Blobel, G.; Pfeffer, S.R. Clues to the mechanism of cholesterol transfer from the structure of NPC1 middle lumenal domain bound to NPC2. Proc. Natl. Acad. Sci. USA 2016, 113, 10079–10084. [Google Scholar] [CrossRef] [PubMed]
- Monsma, S.A.; Oomens, A.G.; Blissard, G.W. The GP64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J. Virol. 1996, 70, 4607–4616. [Google Scholar] [CrossRef]
- Oomens, A.; Blissard, G. Requirement for GP64 to Drive Efficient Budding ofAutographa californicaMulticapsid Nucleopolyhedrovirus. Virology 1999, 254, 297–314. [Google Scholar] [CrossRef]




| Primer Names | Sequences (5′–3′) |
|---|---|
| BmNPC1-1 Oligo-1 | GATCACTAATACGACTCACTATAGGGCGTGCTGCAATTACGAACAACTGAATT |
| BmNPC1-1 Oligo-2 | AATTCAGTTGTTCGTAATTGCAGCACGCCCTATAGTGAGTCGTATTAGTGATC |
| BmNPC1-1 Oligo-3 | AACGTGCTGCAATTACGAACAACTGAACCCTATAGTGAGTCGTATTAGTGATC |
| BmNPC1-1 Oligo-4 | GATCACTAATACGACTCACTATAGGGTTCAGTTGTTCGTAATTGCAGCACGTT |
| BmNPC1-2 Oligo-1 | GATCACTAATACGACTCACTATAGGGGAGCAAATACTTGAAGCCAGTTCAATT |
| BmNPC1-2 Oligo-2 | AATTGAACTGGCTTCAAGTATTTGCTCCCCTATAGTGAGTCGTATTAGTGATC |
| BmNPC1-2 Oligo-3 | AAGAGCAAATACTTGAAGCCAGTTCAACCCTATAGTGAGTCGTATTAGTGATC |
| BmNPC1-2 Oligo-4 | GATCACTAATACGACTCACTATAGGGTTGAACTGGCTTCAAGTATTTGCTCTT |
| RFP-Oligo-1 | GATCACTAATACGACTCACTATAGGGGCACCCAGACCATGAGAATTT |
| RFP-Oligo-2 | AAATTCTCATGGTCTGGGTGCCCCTATAGTGAGTCGTATTAGTGATC |
| RFP-Oligo-3 | AAGCACCCAGACCATGAGAATCCCTATAGTGAGTCGTATTAGTGATC |
| RFP-Oligo-4 | GATCACTAATACGACTCACTATAGGGATTCTCATGGTCTGGGTGCTT |
| Chr | Position | Ref | Alt | p-Value | Nearest Gene |
|---|---|---|---|---|---|
| 3 | 12,131,761 | C | T | 7.88 × 10−10 | Chloride intracellular channel |
| 3 | 12,131,766 | A | T | 7.88 × 10−10 | Chloride intracellular channel |
| 3 | 12,156,953 | A | C | 7.88 × 10−10 | Chloride intracellular channel |
| 3 | 12,166,583 | T | C | 7.88 × 10−10 | Chloride intracellular channel |
| 3 | 12,167,055 | T | G | 7.88 × 10−10 | Chloride intracellular channel |
| 3 | 12,306,266 | A | C | 7.88 × 10−10 | Niemann-Pick C1 |
| 3 | 12,306,312 | G | A | 7.88 × 10−10 | Niemann-Pick C1 |
| 3 | 12,306,315 | G | A | 7.88 × 10−10 | Niemann-Pick C1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, Y.; Sun, L.; Tang, Y.; Li, J.; Qin, S.; Li, M. Identification and Characterization of Genes Related to Resistance of Autographa californica Nucleopolyhedrovirus Infection in Bombyx mori. Insects 2022, 13, 435. https://doi.org/10.3390/insects13050435
Kong Y, Sun L, Tang Y, Li J, Qin S, Li M. Identification and Characterization of Genes Related to Resistance of Autographa californica Nucleopolyhedrovirus Infection in Bombyx mori. Insects. 2022; 13(5):435. https://doi.org/10.3390/insects13050435
Chicago/Turabian StyleKong, Yunhui, Lingling Sun, Yaling Tang, Jiashuang Li, Sheng Qin, and Muwang Li. 2022. "Identification and Characterization of Genes Related to Resistance of Autographa californica Nucleopolyhedrovirus Infection in Bombyx mori" Insects 13, no. 5: 435. https://doi.org/10.3390/insects13050435
APA StyleKong, Y., Sun, L., Tang, Y., Li, J., Qin, S., & Li, M. (2022). Identification and Characterization of Genes Related to Resistance of Autographa californica Nucleopolyhedrovirus Infection in Bombyx mori. Insects, 13(5), 435. https://doi.org/10.3390/insects13050435





