Simple Summary
Tudor domain-containing proteins (TDRDs) are a group of evolutionarily conserved and germline-enriched proteins, and most of them function in reproduction and the P-element-induced wimpy testis-interacting RNA (piRNA) pathway. We investigated LmTDRD5, a TDRD5 ortholog in Locusta migratoria. In males, LmTdrd5 knockdown delayed meiosis phase transition and reduced the number of elongated spermatids and sperm count. The expression levels of two haploid germ cell marker genes, LmCREM and LmACT, as well as the sperm tail marker gene LmQrich2 were downregulated. In females, LmTdrd5 knockdown decreased vitellogenin (Vg) and Vg receptor (VgR) transcript levels, thereby affecting ovarian development and oocyte maturation. Therefore, LmTdrd5 may play an important role in locust reproduction, indicative of a conserved primary function of TDRD5.
Abstract
Tudor family proteins exist in all eukaryotic organisms and play a role in many cellular processes by recognizing and binding to proteins with methylated arginine or lysine residues. TDRD5, a member of Tudor domain-containing proteins (TDRDs), has been implicated in the P-element-induced wimpy testis-interacting RNA (piRNA) pathway and germ cell development in some model species, but little is known about its function in other species. Therefore, we identified and characterized LmTDRD5, the TDRD5 ortholog in Locusta migratoria, a hemimetabolous pest. The LmTdrd5 gene has 19 exons that encode a protein possessing a single copy of the Tudor domain and three LOTUS domains at its N-terminus. qRT-PCR analysis revealed a high LmTdrd5 expression level in genital glands. Using RNA interference, LmTdrd5 knockdown in males led to a lag in meiosis phase transition, decreased spermatid elongation and sperm production, and downregulated the expression of the two germ cell-specific transcription factors, LmCREM and LmACT, as well as the sperm tail marker gene LmQrich2. LmTdrd5 knockdown in females reduced the expression levels of vitellogenin (Vg) and Vg receptor (VgR) and impaired ovarian development and oocyte maturation, thus decreasing the hatchability rate. These results demonstrate that LmTdrd5 is essential for germ cell development and fertility in locusts, indicating a conserved function for TDRD5.
1. Introduction
Living organisms continue their species by reproduction, the way an individual produces offspring. The two main types of reproduction are asexual and sexual reproduction. In asexual reproduction, the offspring is produced from only one parent without fertilization, such as budding in yeast, regeneration in hydra and planaria, and fragmentation in blackworms. In sexual reproduction, the dominant way in nature, two parents produce offspring, and it requires a sperm to fertilize an egg, which forms a zygote [1,2]. Insects are a diverse group with a huge population, including prolific agricultural pests. Insect reproduction has broad significance in basic biology and potential application in biological control. Like many animals, most insect species reproduce sexually through several events, from mating to oviposition, and complex processes in each sex, including spermatogenesis in the testis and oogenesis in the ovary. Both spermatogenesis and oogenesis have been extensively studied in the model insect Drosophila melanogaster. Many genes and signaling pathways are revealed in these processes, and they advance our understanding of the underlying molecular mechanisms [3,4,5,6,7,8,9,10,11,12,13,14,15,16,17]. However, in other species, the information is fragmented.
Locusta migratoria, a devastating agricultural pest, causes locust plagues resulting in serious economic losses and shortage of food for both people and livestock in the area [18]. Therefore, keeping their population under control is crucial. L.migratoria, from Orthoptera, has been used as a hemimetabolous model since its genome was sequenced in 2014 [19]. Recent studies focus on ovarian development and oocyte maturation to understand the molecular mechanism of female reproduction in this species [20,21,22,23,24]. Although a few studies have been conducted on the cellular biology of spermatogenesis, such as the fine structure and ultrastructure of sperm [25,26,27], the genes controlling spermatogenesis or male reproduction in this species remain largely elusive.
Tudor proteins are members of a royal superfamily that also contains the Chromo, MBT, PWWP, and plant Agenet domain proteins [28,29]. Since the first Tudor protein discovered in D. melanogaster [30], these proteins are also found in mouse, human, zebrafish, yeast, and plants [31,32,33,34,35,36,37,38,39,40,41]. Tudor proteins bind to proteins with methylated arginine or lysine residues and take part in cellular processes, such as DNA methylation, RNA metabolism, transcription regulation, DNA damage detection and repair, the P-element-induced wimpy testis (PIWI)-interacting RNA (piRNA) pathway, and the maintenance of genomic stability [32,42]. In particular, a special group of Tudor domain-containing proteins (TDRDs) is highly expressed in the gonads and selectively bind methylarginine, implicating a function for these proteins in gametogenesis [43]. Ten TDRD proteins have been identified in mice, and most of them have functions in spermatogenesis and male fertility [32,44,45,46]. For example, TDRD6 interacts directly with MILI and MIWI, the components of the chromatoid body (CB) and the piRNA pathway, and is required for spermatogenesis. Tdrd6-knockout mice did not produce elongated spermatids but round spermatids with defective CBs [47]. TDRD7 is also essential for CB assembly and spermatid development, but the underlying mechanism might differ from TDRD6, in which TDRD7 suppressed the retrotransposon LINE1 but TDRD6 did not [45]. Several studies focusing on TDRD5 have revealed that it is a component of intermitochondrial cement (IMC) and CB and is an important factor for piRNA biogenesis [48,49,50,51,52]. Tdrd5-deficient males were sterile because of defective spermiogenesis caused by disorganized IMCs and CBs, in which the key components TDRD1/6/7/9 and MILI/MIWI/MIWI2 were mislocalized. TDRD proteins are evolutionarily conserved and also found in other species. In D. melanogaster, the TDRD5 ortholog Tejas is localized to the nuage, the counterpart of CB in mammals, and required for germline piRNA production and retroelement repression [9]. Loss of function of Tejas led to sterile females but fertile males, indicating a role of Tejas in oogenesis [9].
In this study, we identified a TDRD5 ortholog in L. migratoria, named LmTDRD5, and addressed the question of whether LmTDRD5 plays a role in spermatogenesis and oogenesis. By using RNA interference (RNAi) technology, we showed that LmTdrd5 knockdown in males decreased the number of elongated spermatids and eventually reduced the sperm count, whereas LmTdrd5 knockdown in females caused defective ovarian development and oocyte maturation. These data demonstrate that LmTDRD5 is essential for both spermatogenesis and oogenesis in locusts.
2. Materials and Methods
2.1. Insects
L. migratoria eggs were obtained from the Insect Protein Co., Ltd., Cangzhou, Hebei, China, and were incubated in sandy soil covered with a clingfilm at 30 ± 1 °C. After hatching, the nymphs were fed with fresh wheat sprouts and oat meal in a climate-controlled cabinet (Yiheng, Shanghai, China) with a light–dark cycle of 14 h:10 h and 40 ± 10% relative humidity at 30 ± 1 °C. Instar nymphs at different stages and adult locusts were used in this study.
2.2. Isolation of LmTdrd5 Gene and Bioinformatics Analysis
Tejas, the TDRD5 ortholog in D. melanogaster, was chosen as a query for local BLAST searches of transcriptome data of L. migratoria [53]. The obtained cDNA was sequence confirmed and then blasted in the Locusta migratoria genome at NCBI to obtain the gene structure [19]. To predict the open reading frame (ORF) and physicochemical properties of LmTdrd5, the EXPASy online tools (http://www.expasy.org/tools/dna.html (accessed on 20 May 2020)) were used. For analysis of the domain structure, SMART (http://smart.embl-heidelberg.de/ (accessed on 10 August 2021)) was applied.
2.3. RNA Preparation and Real-Time Quantitative Reverse Transcription PCR (qRT-PCR)
Whole bodies at different developmental stages, from egg to adult, and tissues from the foregut (FG), midgut (MG), hindgut (HG), integument (IN), Malpighian tubule (MT), gastric caecum (GC), fat body (FB), testis (TE), and ovary (OV) of the fifth-instar nymphs at day 3 (N5D3) were sampled and frozen immediately in liquid nitrogen and stored at −80 °C for total RNA preparation. Samples from three replicates were collected.
Total RNA was extracted with TRIZOL reagent (Invitrogen, Carlsbad, CA, USA). First-strand cDNA was synthesized using Reverse Transcriptase M-MLV (TaKaRa Biotechnology, Co., Ltd., Shiga, Japan). qRT-PCR was performed in a Light Cycler480 (Roche Applied Science, Mannheim, Germany) with a protocol of 94 °C for 2 min, followed by two-step PCR, 40 cycles at 94 °C for 15 s, and at 60 °C for 31 s. Each PCR assay contained 10 μL of 2× SYBR® qPCR MasterMix, 0.8 μL of primers (10 μM), and 4 μL cDNA template in a final volume of 20 µL. Two technical repeats were performed. β-actin was used as an internal control. The primers used are listed in Table S1. The 2−△△Ct method was used to analyze the qRT-PCR data. Statistical analyses were performed by one-way analysis of variance, followed by Tukey’s honestly significant difference (HSD) test with SPSS v16.0 (IBM Corporation, Armonk, NY, USA).
2.4. Double-Stranded RNA (dsRNA) Synthesis and Injection
dsLmTdrd5 and dsGFP were synthesized using T7 RiboMAX™ Express RNAi System (Promega, Madison, USA) as described previously [18]. The dsRNA was injected into the locust at the internode membrane of the second and third abdominal segments by using an injection needle. The injection was administered on the first day of the fifth-instar nymphs (N5D1), then at N5D4, on the first day of adult (AD1), and at AD4, with 12 μg dsRNA per nymph and 15 μg per adult, respectively.
2.5. Histology and Microscopy
Testes of locusts from N5D3, N5D5, AD2, AD5, and AD6 were dissected in phosphate buffered saline (PBS) and fixed in 4% paraformaldehyde fixative. To prepare the slide, the seminiferous tubules were randomly selected from the fixed testis, rinsed three times with PBS, stained with improved phenol fuchsin stain (Yuanye Bio-technology Co., Ltd., Shanghai, China) for 10–15 min at room temperature, and squashed under a coverslip. All images were taken under a microscope (Leica Microsystems, Wetzlar, Germany).
Ovaries of adult female locusts at 4, 6, and 8 days post-adult eclosion (PAE) were excised in PBS. After the outer membrane was removed, the size of ovarioles and primary oocytes was measured and photographed under a Leica M205C microscope. The data for primary oocyte size were analyzed by the two-sample and two-tailed t-test and presented as a scatter dot plot (prepared by GraphPad Prism 8). All differences were considered significant at p < 0.05.
2.6. Analysis of Hatchability
To investigate the hatchability, three types of combinations were prepared. In type I, there were 30 dsLmTdrd5-treated or 30 dsGFP-treated males with 30 wild type (WT) females. In type II, there were 30 dsLmTdrd5-treated or 30 dsGFP-treated females paired with 30 WT males. In type III, there were 30 males and 30 females injected with dsLmTdrd5. The control is the equal number of males and females injected with dsGFP. Each combination group of adult locusts was incubated in a cage with moist sandy soil under the same conditions mentioned in Section 2.1. After mating behavior was observed, the egg pods were collected every other day from each group until all adults stopped mating.
Egg pods were kept for 10 days in bottles filled with moist soil at room temperature. Eggs were mixed from the egg pods of the same group. One hundred eggs were randomly selected from the same group and were incubated in a container with sterile, moist sandy soil. The first-instar nymphs in the containers were counted daily until no nymphs appeared. The hatchability rate was calculated as a percentage, i.e., the number of nymphs from 100 eggs. Five counting processes were repeated for each group.
3. Results
3.1. LmTDRD5 Is a LOTUS-Containing Protein Highly Expressed in the Testes and Ovaries
To identify the LmTdrd5 gene, Tejas, the TDRD5 ortholog in D. melanogaster, was used as a query to perform local BLAST searches of transcriptome data of L. migratoria [53]. A 4749-bp sequence was obtained and further cloned by RT-PCR. This sequence was then used to blast the L. migratoria genome at NCBI (taxid:7004). The LmTdrd5 gene consists of 20 exons and 19 introns (Figure 1a). The cDNA included an 18-bp 5′ untranslated region (UTR) and a 4731-bp ORF encoding 1577 amino acid residues (GenBank accession no. OM 324402). The theoretical molecular weight was 179.20 kDa, and the pI was 5.10. Domain architecture analysis by SMART showed that LmTDRD5 had four conserved domains, namely one single copy of the Tudor domain and three LOTUS domains at its N-terminus (Figure 1b).
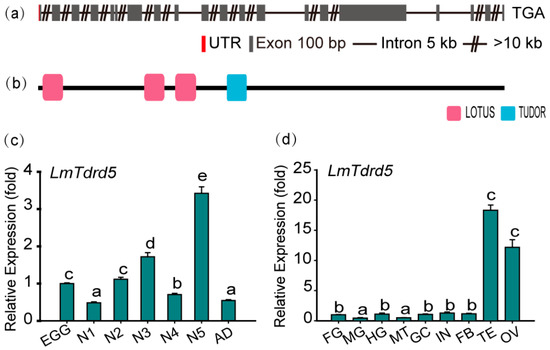
Figure 1.
Gene structure and expression profile of LmTdrd5. (a) Schematic representation of the exon–intron structure of LmTdrd5. Exons contributed to the coding sequence (CDS) marked in goldenrod, red represents the untranslated region (UTR), the black line denotes an intron, and the double slashes indicate a break of introns. Gene structures were deduced from the analysis on the L. migratoria genome database (http://www.locustmine.org/index.html (accessed on 12 September 2021)). (b) LmTDRD5 was analyzed by SMART (Smart.embl-heidelberg.de), showing the four conserved domains marked in two colors. The first three in pink are the LOTUS domains, and the TUDOR domain is in blue. (c) LmTdrd5 expression at different locust developmental stages. Egg (EGG), first-instar (N1), second-instar (N2), third-instar (N3), fourth-instar (N4), fifth-instar (N5) nymphs, and adult (AD). (d) LmTdrd5 mRNA expression in different tissues of fifth-instar nymphs. FG, foregut; MG, midgut; HG, hindgut; MT, Malpighian tubules; GC, gastric caecum; IN, integument; FB, fat body; TE, testis; OV, ovary. Letters on the bars represent significant differences among the developmental stages or different tissues based on Tukey’s HSD multiple comparison test (p < 0.05).
To obtain the LmTdrd5 expression profile, a series of qRT-PCR assays was performed in various tissues at different developmental stages. The results showed that LmTdrd5 was expressed in all examined stages, from egg to adult, with a peak at the fifth-instar nymphs (Figure 1c). Interestingly, LmTdrd5 was most highly expressed in the testes and ovaries (Figure 1d), suggesting a role of this gene in the reproductive phase.
3.2. LmTdrd5 Knockdown Reduced the Hatchability Rate in Female Locusts
To test the role of LmTdrd5 in the reproductive phase, we investigated the hatchability rate for these dsLmTdrd5- or dsGFP-treated locusts in three types of combination groups (100 eggs were counted in each group). In type I, only males were dsRNA-treated and females were wild type (WT). The hatchability rate in the dsLmTdrd5-treated group was 60.1%, a significant reduction compared with 93% in the control dsGFP-treated group (Figure 2a), indicative of male fertility defect. In type II, only females were injected with dsRNA, and males were WT. In this combination, the rate from dsLmTdrd5-treated group reached 73.1%, still lower than 91% in the control (Figure 2b), suggesting female fertility defect. In type III, both males and females were treated with dsRNA. Interestingly, the rate of dsLmTdrd5-treated males and females was 45%, much lower than the control 91% (Figure 2c). These data suggest reproductive defects in both males and females.
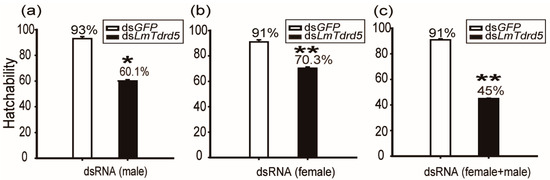
Figure 2.
Analysis of hatchability. Hatchability was calculated as a percentage. (a) DsLmTdrd5 or dsGFP injected only in males. (b) DsLmTdrd5 or dsGFP injected only in females. (c) Both males and females injected with either dsLmTdrd5 or dsGFP. Statistically significant differences are indicated as follows: * p < 0.05; ** p < 0.01.
3.3. DsLmTdrd5-Treated Males Produced a Lower Number of Sperms
To find the reproductive defect in dsLmTdrd5-treated males, the testes from these fifth-instar nymphs and adult locusts were dissected. The testis size was not affected by the dsLmTdrd5 injection (Figure S1). We focused on the transition zone of seminiferous tubules in the stained testes. An obvious difference was observed between dsLmTdrd5-treated and dsGFP-treated males: the meiotic phase lagged in the dsLmTdrd5-treated group compared with the dsGFP-treated group. For example, dsGFP-treated males were already in metaphase II but dsLmTdrd5-treated males were still in prophase II (Figure 3a,b). Later, many spermatids were formed and elongated in dsGFP-treated males, whereas only a portion of spermatids was elongated (others were still in the round spermatid stage) and the spermatid count was reduced by almost half in dsLmTdrd5-treated males (Figure 3c,d). In the advanced developmental stage, bunches of the needle-formed sperms were observed in the dsGFP-treated males but the round spermatids were still found in dsLmTdrd5-treated males (Figure 3e,f). At least one-third of round spermatids did not elongate. Therefore, LmTdrd5 may be involved in meiosis, spermatid elongation, and sperm maturation in male locusts.
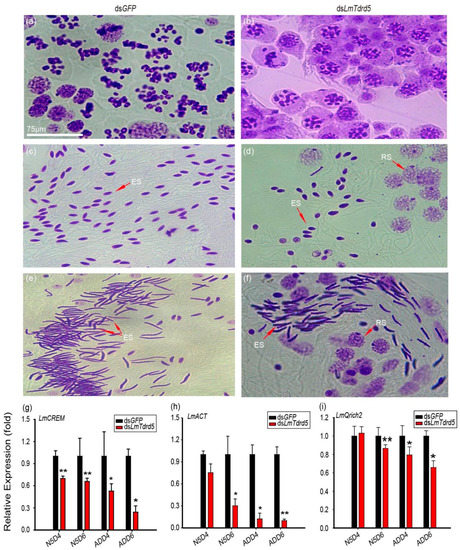
Figure 3.
Defects in spermatogenesis of dsLmTdrd5-treated locusts. (a–f) Cytological phenotype of three representative stages during spermatogenesis observed at the transition zone of seminiferous tubules from testes stained with the improved phenol fuchsin stain. Purple represents chromatin. (a,c,e) Haploid germ cells in the meiotic II stage and elongated spermatids from dsGFP-treated testes. (b,d,f) Haploid germ cells in the meiotic II stage and elongated spermatids from dsLmTdrd5-treated testes. Meiosis phase transition was delayed, and the sperm count was significantly reduced. RS, round spermatids; ES, elongated spermatids. (g–i) Effect of dsLmTdrd5 injection on LmCREM expression levels (g), LmACT (h), and LmQrich2 (i) in the testes on days N5D4, N5D6, ADD4, and ADD6 compared with the control dsGFP-treated locusts. Data are presented as mean ± standard error of the mean (n = 9–15). Statistically significant differences are indicated as follows: * p < 0.05; ** p < 0.01.
In the mouse Tdrd5−/− mutant, the expression level of ACT, an activator of CREM (a germ cell-specific transcription factor involved in the cAMP-dependent signaling pathway), was significantly reduced in the testis [51]. Because of phenotypic similarities in spermatogenesis between the mouse Tdrd5−/− mutant and LmTdrd5 knockdown, we examined LmCREM and LmACT expression levels in LmTdrd5 knockdown. Our qRT-PCR results indicated a significant decrease in LmCREM and LmACT expression in dsLmTdrd5-treated locusts at N5D6, ADD4, and ADD6 (Figure 3g,h; Supplemental File S1). In addition, we examined a sperm tail marker gene, glutamine rich 2 (Qrich2) [54], and found that the expression level of its ortholog in L. migratoria LmQrich2 was suppressed by 13%, 21%, and 34% at N5D6, ADD4, and ADD6, respectively (Figure 3i; Supplemental File S1). Therefore, LmTdrd5 plays a role in spermatogenesis.
3.4. LmTdrd5 Knockdown Impaired Ovarian Development and Oocyte Maturation
To investigate the role of LmTdrd5 in female reproduction, the same amount of dsRNA as used in males was used in females. We dissected ovaries from these dsRNA-treated locusts and focused on the phenotype of ovarian development and oocyte maturation. At four days PAE (ADD4), almost all ovaries in dsLmTdrd5-treated locusts were smaller than those in the dsGFP-treated locusts, and the primary oocytes were also remarkably smaller (Figure 4a). At six days PAE (ADD6), the dsLmTdrd5-treated females produced two types of ovaries, i.e., two levels of retarded ovarian development, and primary oocyte maturation with smaller primary oocyte length. Type I: 70% of dsLmTdrd5-treated females had yellow ovaries and primary oocytes, which were thinner and shorter than those of the controls. Type II: the rest (30%) showed a serious phenotype with small ovaries and white primary oocytes, which appeared at a standstill (Figure 4b). On average, the primary oocyte length in types I and II was 3.64 mm and 1.34 mm, respectively (Figure 4c). Both were significantly reduced compared with the control dsGFP-treated locusts (5.77 mm) (Figure 4d). At eight days PAE (ADD8), the primary oocytes in the control ovaries matured normally and those in the type I ovaries were at the stage of reaching maturity, although a bit slower than those in the control. However, the primary oocytes were still underdeveloped in type II (Figure 4c).
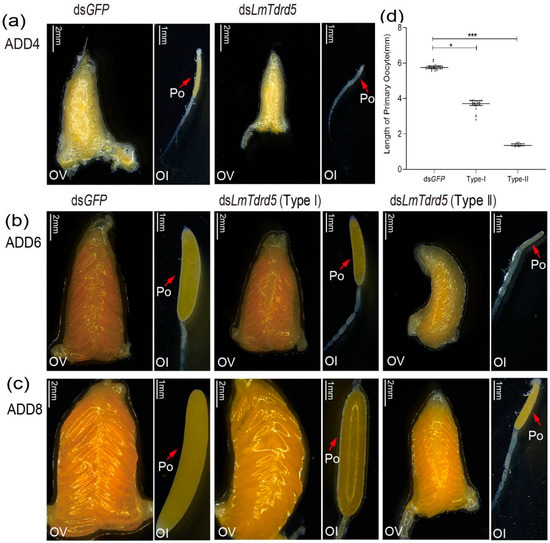
Figure 4.
Effects of dsLmTdrd5 injection on ovarian development and oocyte maturation. (a–c) Morphology of ovaries and ovarioles in female locusts after dsGFP or dsLmTdrd5 treatment. Ol, ovariole; Ov, ovary; Po, primary oocyte. ADD4, adult of 4 days; ADD6, adult of 6 days; ADD8, adult of 8 days. Scale bars: Ov, 2 mm; Ol, 1 mm. (d) Primary oocyte length of locusts injected with dsLmTdrd5 or dsLmGFP at ADD4, ADD6, ADD8. Data are presented as scatter dot plot. The horizontal lines depict the median with range. Statistically significant differences are indicated as follows: * p < 0.05; *** p < 0.001. n = 11–33.
3.5. Vg and VgR Expression Levels Were Affected in dsLmTdrd5-Treated Females
Vitellogenin (Vg) and vitellogenin receptor (VgR) are two key factors for oocyte development and maturation in insects. To explain the reason why LmTdrd5 knockdown led to defective oocyte development and maturation, we examined the expression levels of LmVgA and LmVgB as well as LmVgR in dsLmTdrd5-treated female locusts. qRT-PCR data showed 88.2% and 86.3% reductions in LmVgA and LmVgB expression levels, respectively, at ADD4, whereas the LmVgR expression level decreased only 10.3% (Figure 5a–c). At ADD6 and ADD8, the dsLmTdrd5-treated females had two types of ovaries. For type I females, the relative expression levels of LmVgA and LmVgB in fat bodies more or less reached the level of the control (Figure 5d,e). However, the LmVgR expression level was only 21.1% of that in the control at ADD6 and reached 83.7% at ADD8 (Figure 5f). For type II, dsLmTdrd5 treatment reduced the LmVgA expression level in the control to 17.5% and 33.9% at ADD6 and ADD8, respectively (Figure 5g). Similarly, the LmVgB expression level reduced to 20.2% and 35.1% (Figure 5h). However, the LmVgR expression level differed from that in type I, with a significant reduction (54%) only found at ADD8 (Figure 5i). Therefore, LmTdrd5 might regulate ovarian development and oocyte maturation by mediating in LmVg and LmVgR expression.
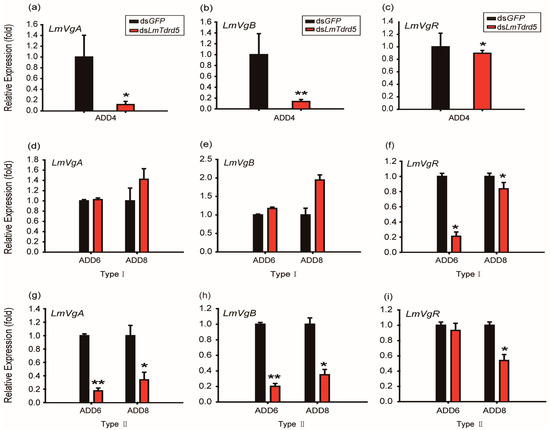
Figure 5.
Effect of dsLmTdrd5 injection on the transcript levels of vitellogenin (Vg) and its receptor. (a–c) Relative expression levels (fold) of LmVgA (a), LmVgB (b), and LmVgR (c) in the fat bodies and ovaries in locusts at ADD4. (d–f) Type I: Relative expression levels (fold) of LmVgA (d), LmVgB (e), and LmVgR (f) at ADD6 and ADD8. (g–i) Type II: Relative expression levels (fold) of LmVgA (g), LmVgB (h), and LmVgR (i) at ADD6 and ADD8. Data in all panels are presented as mean ± standard error of the mean (n = 9–15). Statistically significant differences are indicated as follows: * p < 0.05; ** p < 0.01.
4. Discussion
We identified and characterized LmTdrd5, the Tdrd5 ortholog, in L. migratoria. Our data showed that LmTDRD5 had evolutionarily conserved domains and functions, implying that it may share the same or similar molecular mechanism as other TDRD5s to exert its function.
Like mouse TDRD5, LmTDRD5 possesses only one repeat of the Tudor domain with three LOTUS domains at its N-terminus. This Tudor domain in mouse TDRD5 is an extended Tudor domain (eTudor/eTud) containing 180 amino acids that consists of a canonical Tudor domain (60 amino acids), α-helix linker, and a staphylococcal nuclease-like domain. These three structural elements are required for TDRD5 binding to the PIWI proteins [55,56,57]. eTudor domains are also found in other TDRD proteins, such as the 36 eTudor domains present in the 12 human TDRD proteins. These eTudor domains specifically recognize the RA/RG repeats in the N-termini of PIWI proteins, and symmetric arginine demethylation of RA/RG is essential for binding to them [55]. Although the Smart program recently recognized the Tudor domain as a canonical domain in LmTDRD5, considering the sequence and topology similarity between LmTDRD5 and mouse TDRD5, this single Tudor domain in LmTDRD5 might also be an eTudor domain. When we used the CDD Search program on the NCBI website (https://www.ncbi.nlm.nih.gov/structure/cdd/ (accessed on 15 December 2021)), it revealed a large Tudor domain of 129 amino acids in LmTDRD5, supporting our assumption. LmTDRD5 could bind PIWI proteins by the eTudor domain. We further examined the N-termini of PIWI proteins in L. migratoria and they all contained RA/RG repeats (Supplemental File S2), indicating that this feature of PIWI proteins is also conserved.
The LOTUS domain (also known as OST-HTH) is an RNA-binding domain found in germline proteins, such as Oskar, TDRD5, TDRD7, and MARF1 (meiosis arrest female 1), and is mainly associated with the small RNA pathway [58,59,60]. This domain comprises 80–100 amino acids that form a winged helix-turn-helix fold. The LOTUS domain is conserved in bacteria, fungi, and plants but experimentally studied only in animals [60]. The LOTUS domain selectively binds to G-rich RNAs and the RNA G-quadruplex (G4) secondary structure in vitro [61]. However, whether the LOTUS domain in LmTDRD5 has the same specificity needs further investigation.
Mouse TDRD5 can associate with pachytene piRNA precursors by its LOTUS domain and is thus essential for pachytene piRNA biogenesis [50,62], which contains a low level of transposon sequences, suggesting that they regulate mRNA and long non-coding RNA in mouse testis [63,64,65] and are required for spermatogenesis.
In addition, in the mouse tdrd5-deficient mutant, the expression of ACT, which encodes a coactivator of the germline transcription factor CREM, was greatly reduced in the round spermatids, and the expression of CREM target genes, such as Prm1, Prm2, Tnp1, Calspermin, and RT7, was also reduced [51,66,67,68], but not the expression of CREM itself. Mouse TDRD5 may thus function independently of piRNA in posttranscriptional gene regulation, and defective spermatogenesis in the mouse tdrd5−/− mutant may be caused by the downregulation of these CREM target genes. Intriguingly, LmTdrd5 knockdown led to downregulated orthologs of both ACT and CREM in the testis of L. migratoria (Figure 3g,h), implying that LmTDRD5 may also have a function in posttranscriptional gene regulation. However, we cannot conclude that the LmTdrd5 knockdown phenotype in males was indeed caused by the downregulation of ACT and CREM.
Tejas and Tapas, the orthologs of TDRD5 and TDRD7 in D. melanogaster, have also been implicated in the piRNA pathway, and both interact with PIWI proteins. However, the precise molecular mechanisms underlying the function of these two genes during oogenesis are unknown. Tejas and Tapas bind to germline protein Vasa by their LOTUS domains [68]. Vasa is located in the nuage, the counterpart of CB in mammals, and involved in the piRNA pathway. Thus, Tejas and Tapas might form a complex with Vasa and thereby function in piRNA-mediated transcript repression [68]. In the spermatogenesis of L. migratoria, CBs appeared in the spermatids [26]. Thus, LmTDRD5 accumulated in CBs may also bind to Vasa and function in the piRNA pathway. Like Tejas, the detailed molecular mechanism of LmTDRD5 in locust reproduction requires further investigation.
5. Conclusions
We characterized LmTDRD5, the TDRD5 ortholog in L. migratoria. LmTdrd5 was highly expressed in the testes and ovaries. LmTdrd5 knockdown in males delayed meiosis phase transition, impaired spermatid elongation, and eventually reduced the sperm count. In line with these phenotypes, the expression levels of the two haploid germline marker genes, LmCREM and its activator LmACT, as well as the sperm tail marker gene LmQrich2 were reduced. LmTdrd5 knockdown in females decreased LmVg and LmVgR expression and blocked ovarian development and oocyte maturation. In short, LmTdrd5 plays an important role in locust reproduction, both spermatogenesis and oogenesis, indicative of a conserved function in TDRD5 family members. However, this study provides only some information on LmTdrd5 and the molecular mechanism by which it exerts its function remains to be answered.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/insects13030227/s1. Table S1: Primers used in this study. Figure S1: Phenotype of the testis from dsGFP- and dsLmTdrd5-treated males at ADD6. Figure S2: LmTdrd5 RNA interference efficiency in the testes, ovaries, and fat bodies of dsLmTdrd5-treated males and females. Supplemental File S1: The coding sequences of LmACT, LmCREM and LmQrich2. Supplemental File S2: Amino acid sequences of PIWI proteins from L. migratoria.
Author Contributions
Conceptualization, S.X. and S.D.; methodology, J.W. and S.D.; investigation: S.D.; data curation, S.D., J.W. and S.X.; writing—original draft preparation, S.D. and S.X.; writing—review and editing, S.X., E.M. and J.Z.; supervision, S.X., J.Z. and E.M.; funding acquisition, J.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China (No. 31730074).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data produced from this study are included in this published paper.
Acknowledgments
We thank our lab members for their valuable comments and suggestions during preparation of this manuscript. We are particularly grateful for the start-up financial support from Shanxi University (2017–2019).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Crow, J.F. Advantages of sexual reproduction. Dev. Genet. 1994, 15, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Schurko, A.M.; Neiman, M.; Logsdon, J.M., Jr. Signs of sex: What we know and how we know it. Trends Ecol. Evol. 2009, 24, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.W.; Luo, Y.C.; Aravin, A.A. RDC complex executes a dynamic piRNA program during Drosophila spermatogenesis to safeguard male fertility. PLoS Genet. 2021, 17, e1009591. [Google Scholar] [CrossRef] [PubMed]
- Blatt, P.; Martin, E.T.; Breznak, S.M.; Rangan, P. Post-transcriptional gene regulation regulates germline stem cell to oocyte transition during Drosophila oogenesis. Curr. Top. Dev. Biol. 2020, 140, 3–34. [Google Scholar] [CrossRef]
- Ibaraki, K.; Nakatsuka, M.; Ohsako, T.; Watanabe, M.; Miyazaki, Y.; Shirakami, M.; Karr, T.L.; Sanuki, R.; Tomaru, M.; Takano-Shimizu-Kouno, T. A cross-species approach for the identification of Drosophila male sterility genes. G3 2021, 11, jkab183. [Google Scholar] [CrossRef]
- Rivard, E.L.; Ludwig, A.G.; Patel, P.H.; Grandchamp, A.; Arnold, S.E.; Berger, A.; Scott, E.M.; Kelly, B.J.; Mascha, G.C.; Bornberg-Bauer, E.; et al. A putative de novo evolved gene required for spermatid chromatin condensation in Drosophila melanogaster. PLoS Genet. 2021, 17, e1009787. [Google Scholar] [CrossRef]
- Tanaka, T.; Tani, N.; Nakamura, A. Receptor-mediated yolk uptake is required for oskar mRNA localization and cortical anchorage of germ plasm components in the Drosophila oocyte. PLoS Biol. 2021, 19, e3001183. [Google Scholar] [CrossRef]
- Vedelek, V.; Bodai, L.; Grézal, G.; Kovács, B.; Boros, I.M.; Laurinyecz, B.; Sinka, R. Analysis of Drosophila melanogaster testis transcriptome. BMC Genom. 2018, 19, 697. [Google Scholar] [CrossRef]
- Patil, V.S.; Kai, T. Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr. Biol. 2010, 20, 724–730. [Google Scholar] [CrossRef]
- Handler, D.; Olivieri, D.; Novatchkova, M.; Gruber, F.S.; Meixner, K.; Mechtler, K.; Stark, A.; Sachidanandam, R.; Brennecke, J. A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J. 2011, 30, 3977–3993. [Google Scholar] [CrossRef]
- Liu, L.; Qi, H.; Wang, J.; Lin, H. PAPI, a novel TUDOR-domain protein, complexes with AGO3, ME31B and TRAL in the nuage to silence transposition. Development 2011, 138, 1863–1873. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, J.; Koppetsch, B.S.; Wang, J.; Tipping, C.; Ma, S.; Weng, Z.; Theurkauf, W.E.; Zamore, P.D. Heterotypic piRNA Ping-Pong requires qin, a protein with both E3 ligase and Tudor domains. Mol. Cell 2011, 44, 572–584. [Google Scholar] [CrossRef]
- Anand, A.; Kai, T. The Tudor domain protein Kumo is required to assemble the nuage and to generate germline piRNAs in Drosophila. EMBO J. 2012, 31, 870–882. [Google Scholar] [CrossRef]
- Patil, V.S.; Anand, A.; Chakrabarti, A.; Kai, T. The Tudor domain protein Tapas, a homolog of the vertebrate Tdrd7, functions in the piRNA pathway to regulate retrotransposons in germline of Drosophila melanogaster. BMC Biol. 2014, 12, 61. [Google Scholar] [CrossRef]
- Sato, K.; Iwasaki, Y.W.; Shibuya, A.; Carninci, P.; Tsuchizawa, Y.; Ishizu, H.; Siomi, M.C.; Siomi, H. Krimper enforces an antisense bias on piRNA pools by binding AGO3 in the Drosophila germline. Mol. Cell 2015, 59, 553–563. [Google Scholar] [CrossRef]
- Andress, A.; Bei, Y.; Fonslow, B.R.; Giri, R.; Wu, Y.; Yates, J.R., III; Carthew, R.W. Spindle-E cycling between nuage and cytoplasm is controlled by Qin and PIWI proteins. J. Cell Biol. 2016, 213, 201–211. [Google Scholar] [CrossRef]
- Ryazansky, S.S.; Kotov, A.A.; Kibanov, M.V.; Akulenko, N.V.; Korbut, A.P.; Lavrov, S.A.; Gvozdev, V.A.; Olenina, L.V. RNA helicase Spn-E is required to maintain Aub and AGO3 protein levels for piRNA silencing in the germline of Drosophila. Eur. J. Cell Biol. 2016, 95, 311–322. [Google Scholar] [CrossRef]
- Wang, X.; Li, J.; Wang, T.; Yang, Y.; Zhang, H.; Zhou, M.; Kang, L.; Wei, L. A novel non-invasive identification of genome editing mutants from insect exuviae. Insect Sci. 2022, 29, 21–32. [Google Scholar] [CrossRef]
- Wang, X.; Fang, X.; Yang, P.; Jiang, X.; Jiang, F.; Zhao, D.; Li, B.; Cui, F.; Wei, J.; Ma, C.; et al. The locust genome provides insight into swarm formation and long-distance flight. Nat. Commun. 2014, 5, 2957. [Google Scholar] [CrossRef]
- Santos, C.G.; Humann, F.C.; Hartfelder, K. Juvenile hormone signaling in insect oogenesis. Curr. Opin. Insect Sci. 2019, 31, 43–48. [Google Scholar] [CrossRef]
- Zheng, H.; Zeng, B.; Shang, T.; Zhou, S. Identification of G protein-coupled receptors required for vitellogenesis and egg development in an insect with panoistic ovary. Insect Sci. 2021, 28, 1005–1017. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Li, D.; Wang, Z.; Guo, W.; Kang, L.; Zhou, S. Juvenile hormone differentially regulates two Grp78 genes encoding protein chaperones required for insect fat body cell homeostasis and vitellogenesis. J. Biol. Chem. 2017, 292, 8823–8834. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Guo, W.; Jiang, F.; Kang, L.; Zhou, S. Argonaute 1 is indispensable for juvenile hormone mediated oogenesis in the migratory locust, Locusta migratoria. Insect Biochem. Mol. Biol. 2013, 43, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Yang, L.; He, Q.; Zhou, S. Regulatory mechanisms of vitellogenesis in insects. Front. Cell Dev. Biol. 2021, 8, 593613. [Google Scholar] [CrossRef]
- Gibbons, I.R.; Brandfield, J.R. The fine structure of nuclei during sperm maturation in the locust. J. Biophys. Biochem. Cytol. 1957, 3, 133–140. [Google Scholar] [CrossRef]
- Szöllösi, A. Electron microscope study of spermiogenesis in Locusta migratoria (Insect Orthoptera). J. Ultrastruct. Res. 1975, 50, 322–346. [Google Scholar] [CrossRef]
- Szöllösi, A.; Marcaillou, C. The apical cell of the locust testis: An ultrastructural study. J. Ultrastruct. Res. 1979, 69, 331–342. [Google Scholar] [CrossRef]
- Chen, C.; Nott, T.J.; Jin, J.; Pawson, T. Deciphering arginine methylation: Tudor tells the tale. Nat. Rev. Mol. Cell Biol. 2011, 12, 629–642. [Google Scholar] [CrossRef]
- Lasko, P. Tudor domain. Curr. Biol. 2010, 20, R666–R667. [Google Scholar] [CrossRef][Green Version]
- Boswell, R.E.; Mahowald, A.P. tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell 1985, 43, 97–104. [Google Scholar] [CrossRef]
- Jin, J.; Xie, X.; Chen, C.; Park, J.G.; Stark, C.; James, D.A.; Olhovsky, M.; Linding, R.; Mao, Y.; Pawson, T. Eukaryotic protein domains as functional units of cellular evolution. Sci. Signal. 2009, 2, ra76. [Google Scholar] [CrossRef]
- Siomi, M.C.; Mannen, T.; Siomi, H. How does the Royal Family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev. 2010, 24, 636–646. [Google Scholar] [CrossRef]
- Huang, H.; Houwing, S.; Kaaij, L.J.T.; Meppelink, A.; Redl, S.; Gauci, S.; Vos, H.; Draper, B.W.; Moens, C.B.; Burgering, B.M.; et al. Tdrd1 acts as a molecular scaffold for Piwi proteins and piRNA targets in zebrafish. EMBO J. 2011, 30, 3298–3308. [Google Scholar] [CrossRef]
- Dai, X.; Shu, Y.; Lou, Q.; Tian, Q.; Zhai, G.; Song, J.; Lu, S.; Yu, H.; He, J.; Yin, Z. Tdrd12 Is essential for germ cell development and maintenance in Zebrafish. Int. J. Mol. Sci. 2017, 18, 1127. [Google Scholar] [CrossRef]
- Li, J.; Xue, X.; Ruan, J.; Wu, M.; Zhu, Z.; Zang, J. Cloning, purification, crystallization and preliminary crystallographic analysis of the tandem tudor domain of Sgf29 from Saccharomyces cerevisiae. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 902–904. [Google Scholar] [CrossRef]
- Brasil, J.N.; Cabral, L.M.; Eloy, N.B.; Primo, L.M.F.; Barroso-Neto, I.L.; Grangeiro, L.P.P.; Gonzalez, N.; Inzé, D.; Ferreira, P.C.G.; Hemerly, A.S. AIP1 is a novel Agenet/Tudor domain protein from Arabidopsis that interacts with regulators of DNA replication, transcription and chromatin remodeling. BMC Plant Biol. 2015, 15, 270. [Google Scholar] [CrossRef]
- Gutierrez-Beltran, E. Genome-wide analysis of uncapped mRNAs under heat stress in Arabidopsis. Genom. Data 2015, 5, 7–8. [Google Scholar] [CrossRef][Green Version]
- Chou, H.; Tian, L.; Kumamaru, T.; Hamada, S.; Okita, T.W. Multifunctional RNA binding protein OsTudor-SN in storage protein mRNA transport and localization. Plant Physiol. 2017, 175, 1608–1623. [Google Scholar] [CrossRef]
- Gutierrez-Beltran, E.; Elander, P.H.; Dalman, K.; Dayhoff, G.W., II; Moschou, P.N.; Uversky, V.N.; Crespo, J.L.; Bozhkov, P.V. Tudor staphylococcal nuclease is a docking platform for stress granule components and is essential for SnRK1 activation in Arabidopsis. EMBO J. 2021, 40, e105043. [Google Scholar] [CrossRef]
- Huang, J.; Tan, S. Piccolo NuA4-catalyzed acetylation of nucleosomal histones: Critical roles of an Esa1 Tudor/chromo barrel loop and an Epl1 enhancer of polycomb A (EPcA) basic region. Mol. Cell Biol. 2013, 33, 159–169. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, S.; Shi, H.; Ma, J.; Jing, M.; Han, Y. The TSN1 binding protein RH31 is a component of stress granules and participates in regulation of salt-stress tolerance in Arabidopsis. Front. Plant Sci. 2021, 12, 804356. [Google Scholar] [CrossRef] [PubMed]
- Pek, J.W.; Anand, A.; Kai, T. Tudor domain proteins in development. Development 2012, 139, 2255–2266. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Ki, B.S.; Hong, K.; Park, S.-P.; Ko, J.-J.; Choi, Y. Tudor domain containing protein TDRD12 expresses at the acrosome of spermatids in mouse testis. Asian-Australas. J. Anim. Sci. 2016, 29, 944–951. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Saxe, J.P.; Tanaka, T.; Chuma, S.; Lin, H. Mili interacts with Tudor domain-containing protein 1 in regulating spermatogenesis. Curr. Biol. 2009, 19, 640–644. [Google Scholar] [CrossRef]
- Tanaka, T.; Hosokawa, M.; Vagin, V.V.; Reuter, M.; Hayashi, E.; Mochizuki, A.L.; Kitamura, K.; Yamanaka, H.; Kondoh, G.; Okawa, K.; et al. Tudor domain containing 7 (Tdrd7) is essential for dynamic ribonucleoprotein (RNP) remodeling of chromatoid bodies during spermatogenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 10579–10584. [Google Scholar] [CrossRef]
- Hosokawa, M.; Shoji, M.; Kitamura, K.; Tanaka, T.; Noce, T.; Chuma, S.; Nakatsuji, N. Tudor-related proteins TDRD1/MTR-1, TDRD6 and TDRD7/TRAP: Domain composition, intracellular localization, and function in male germ cells in mice. Dev. Biol. 2007, 301, 38–52. [Google Scholar] [CrossRef]
- Vasileva, A.; Tiedau, D.; Firooznia, A.; Müller-Reichert, T.; Jessberger, R. Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Curr. Biol. 2009, 19, 630–639. [Google Scholar] [CrossRef]
- Sun, Y.H.; Jiang, F.; Li, X.Z. Disruption of Tdrd5 decouples the stepwise processing of long precursor transcripts during pachytene PIWI-interacting RNA biogenesis. Biol. Reprod. 2018, 99, 684–685. [Google Scholar] [CrossRef]
- Sun, Y.H.; Zhu, J.; Xie, L.H.; Li, Z.; Meduri, R.; Zhu, X.; Song, C.; Chen, C.; Ricci, E.P.; Weng, Z.; et al. Ribosomes guide pachytene piRNA formation on long intergenic piRNA precursors. Nat. Cell Biol. 2020, 22, 200–212. [Google Scholar] [CrossRef]
- Ding, D.; Liu, J.; Midic, U.; Wu, Y.; Dong, K.; Melnick, A.; Latham, K.E.; Chen, C. Tdrd5 binds piRNA precursors and selectively enhances pachytene piRNA processing in mice. Nat. Commun. 2018, 9, 127. [Google Scholar] [CrossRef]
- Yabuta, Y.; Ohta, H.; Abe, T.; Kurimoto, K.; Chuma, S.; Saitou, M. Tdrd5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice. J. Cell Biol. 2011, 192, 781–795. [Google Scholar] [CrossRef]
- Smith, J.M.; Bowles, J.; Wilson, M.; Teasdale, R.D.; Koopman, P. Expression of the tudor-related gene Tdrd5 during development of the male germline in mice. Gene Expr. Patterns 2004, 4, 701–705. [Google Scholar] [CrossRef]
- Zhang, X.; Kang, X.; Wu, H.; Silver, K.; Zhang, J.; Ma, E.; Zhu, K.Y. Transcriptome-wide survey, gene expression profiling and exogenous chemical-induced transcriptional responses of cytochrome P450 superfamily genes in migratory locust (Locusta migratoria). Insect Biochem. Mol. Biol. 2018, 100, 66–77. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, F.; Li, F.; Jiang, X.; Yang, Y.; Li, X.; Li, W.; Wang, X.; Cheng, J.; Liu, M.; et al. Loss-of-function mutations in QRICH2 cause male infertility with multiple morphological abnormalities of the sperm flagella. Nat. Commun. 2019, 10, 433. [Google Scholar] [CrossRef]
- Gan, B.; Chen, S.; Liu, H.; Min, J.; Liu, K. Structure and function of eTudor domain containing TDRD proteins. Crit. Rev. Biochem. Mol. Biol. 2019, 54, 119–132. [Google Scholar] [CrossRef]
- Liu, K.; Chen, C.; Guo, Y.; Lam, R.; Bian, C.; Xu, C.; Zhao, D.Y.; Jin, J.; MacKenzie, F.; Pawson, T.; et al. Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain. Proc. Natl. Acad. Sci. USA 2010, 107, 18398–18403. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, K.; Izumi, N.; Huang, H.; Ding, D.; Ni, Z.; Sidhu, S.S.; Chen, C.; Tomari, Y.; Min, J. Structural basis for arginine methylation-independent recognition of PIWIL1 by TDRD2. Proc. Natl. Acad. Sci. USA 2017, 114, 12483–12488. [Google Scholar] [CrossRef]
- Anantharaman, V.; Zhang, D.; Aravind, L. OST-HTH: A novel predicted RNA binding domain. Biol. Direct 2010, 5, 13. [Google Scholar] [CrossRef]
- Callebaut, I.; Mornon, J.-P. LOTUS, a new domain associated with small RNA pathways in the germline. Bioinformatics 2010, 26, 1140–1144. [Google Scholar] [CrossRef] [PubMed]
- Kubíková, J.; Reinig, R.; Salgania, H.K.; Jeske, M. LOTUS-domain proteins—Developmental effectors from a molecular perspective. Biol. Chem. 2020, 402, 7–23. [Google Scholar] [CrossRef]
- Ding, D.; Wei, C.; Dong, K.; Liu, J.; Stanton, A.; Xu, C.; Min, J.; Hu, J.; Chen, C. LOTUS domain is a novel class of G-rich and G-quadruplex RNA binding domain. Nucleic Acids Res. 2020, 48, 9262–9272. [Google Scholar] [CrossRef] [PubMed]
- Goh, W.S.S.; Falciatori, I.; Tam, O.H.; Burgess, R.; Meikar, O.; Kotaja, N.; Hammell, M.; Hannon, G.J. piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes Dev. 2015, 29, 1032–1044. [Google Scholar] [CrossRef] [PubMed]
- Gou, L.; Dai, P.; Yang, J.; Xue, Y.; Hu, Y.; Zhou, Y.; Kang, J.; Wang, X.; Li, H.; Hua, M.; et al. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 2014, 24, 680–700. [Google Scholar] [CrossRef]
- Watanabe, T.; Cheng, E.; Zhong, M.; Lin, H. Retrotransposons and pseudogenes regulate mRNAs and lncRNAs via the piRNA pathway in the germline. Genome Res. 2015, 25, 368–380. [Google Scholar] [CrossRef]
- Fimia, G.M.; De, C.D.; Sassone-Corsi, P. CBP-independent activation of CREM and CREB by the LIM-only protein ACT. Nature 1999, 398, 165–169. [Google Scholar] [CrossRef]
- Blendy, J.A.; Kaestner, K.H.; Weinbauer, G.F.; Nieschlag, E.; Schütz, G. Severe impairment of spermatogenesis in mice lacking the CREM gene. Nature 1996, 380, 162–165. [Google Scholar] [CrossRef]
- Nantel, F.; Sassone-Corsi, P. CREM: A transcriptional master switch during the spermatogenesis differentiation program. Front. Biosci. 1996, 1, d266–d269. [Google Scholar] [CrossRef][Green Version]
- Jeske, M.; Müller, C.W.; Ephrussi, A. The LOTUS domain is a conserved DEAD-box RNA helicase regulator essential for the recruitment of Vasa to the germ plasm and nuage. Genes Dev. 2017, 31, 939–952. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).